Abstract
Ferroptosis is a type of cell death induced by the iron-dependent accumulation of lipid hydroperoxides and reactive oxygen species (ROS) in cells. Inhibiting ferroptosis is important for improving the survival of transplanted bone marrow-derived mesenchymal stem cells (BMSCs). Although it is known that NOP2/Sun RNA methyltransferase 5 (NSUN5) post-transcriptionally regulates ferroptosis in BMSCs through RNA methylation, the precise mechanisms underlying these effects have not been reported. In this study, we demonstrate that NSUN5 is downregulated in erastin-induced ferroptosis in BMSCs. Ferroptosis was inhibited by the overexpression of NSUN5 or ferritin heavy chain/light-chain (FTH1/FTL) and was enhanced by NSUN5 knockdown. RNA immunoprecipitation experiments revealed that NSUN5 binds to FTH1/FTL, while NSUN5 depletion reduced the levels of 5-methylcytosine in FTH1/FTL RNA and increased intracellular iron concentrations, resulting in the downregulation of glutathione peroxidase 4 (GPX4) and the accumulation of ROS and lipid peroxidation products. Co-immunoprecipitation experiments demonstrated that the recognition of FTH1 and FTL by NSUN5 is dependent on the recruitment of tumor necrosis factor receptor-associated protein 1 (TRAP1). These results suggested that the NSUN5-FTH1/FTL pathway mediates ferroptosis in BMSCs and that the therapeutic targeting of components of this pathway may promote resistance to ferroptosis and improve the survival of transplanted BMSCs.
Similar content being viewed by others
Introduction
Mesenchymal stem cell (MSC) transplantation is an important treatment option for acute liver failure as well as for cardiac and neurodegenerative diseases [1,2,3]. Bone marrow-derived mesenchymal stem cells (BMSCs) can home to injured tissues and contribute to tissue repair [4]. Perturbation of the intracellular environment and metabolism are major causes of treatment failure in patients following transplantation. The latter can lead to iron overload and the toxic accumulation of reactive oxygen species (ROS) and lipid hydroperoxides, resulting in a form of controlled cell death known as ferroptosis [5]. Inhibiting ferroptosis in BMSCs can potentially enhance the survival of transplanted cells.
Ferritins are the only proteins capable of storing large quantities of iron, thereby playing a critical role in the regulation of cellular iron metabolism. The heavy (H)- and light (L)-chain subunits of ferritin (FTH and FTL, respectively) are responsible for intracellular iron storage [6] and recent evidence suggests that they are involved in ferroptosis and apoptosis [7, 8].
RNA-binding proteins participate in cellular metabolism and have also been implicated in ferroptosis [9]. Mitochondrial RNA-binding protein tumor necrosis factor receptor-associated protein 1 (TRAP1), also known as heat-shock protein 75-kDa, is highly expressed in a variety of cancer types and is known to induce cell death [10,11,12]. In particular, TRAP1-associated, chaperone-mediated autophagy can promote ferroptosis [13], while TRAP1 was shown to define a common regulatory nodal between necroptosis and ferroptosis [14]. Additionally, studies have reported that TRAP1 is an iron-binding protein associated with the plasma membrane [15]. Combined, these observations suggested that TRAP1 may also be involved in ferroptosis in BMSCs.
RNA modifications play an important role in the pathogenesis of many diseases. Methylation is a common chemical modification found in most RNAs. 5-Methylcytosine (5mC) is associated with the regulation of protein translation, RNA processing, metabolism (including iron metabolism), the stress response, and cell differentiation [16,17,18]. RNA-modifying enzymes that are involved in transcriptional regulation have been shown to modulate ferroptosis in stem cells. Several members of the NOP2/Sun RNA methyltransferase (Nsun) protein family have been shown to possess putative 5mC methyltransferase activity [19] and some Nsun family proteins are presumed to regulate ferroptosis in BMSCs. Nsun2 catalyzes the methylation of tRNAs and may also target mRNAs and noncoding RNAs [20]. NSUN5 is a conserved RNA (C5-cytosine) methyltransferase with a role in lifespan Modulation [21]. However, it is not known whether NSUN5 is also involved in iron metabolism.
Here, we tested the possibility that NSUN5 may mediate 5mC RNA modification in the regulation of iron metabolism and ferroptosis using BMSCs treated with erastin, a small-molecule known to induce ferroptosis. Our results provide evidence for NSUN5-mediated 5mC modification of FTH1 and FTL mRNA as a novel mechanism underlying ferroptosis in BMSCs.
Results
Erastin-induced ferroptosis in BMSCs
We evaluated erastin-induced BMSC ferroptosis by measuring the intracellular levels of Fe, lipid peroxidation products, and ROS. The flow cytometric analysis showed that the fraction of dead BMSCs was larger in the erastin group than in the untreated control group (Fig. 1A, B; Supplementary Fig. 1). Iron is a mediator of ferroptosis, and total iron levels were significantly higher in BMSCs treated with erastin than in control cells (Fig. 1C). ROS levels were also higher in the erastin group than in the control group (Fig. 1D). To confirm the induction of oxidative stress by erastin, we assessed lipid peroxidation in BMSCs using the C11-BODIPY 581/591 assay, and found that lipid peroxide levels were significantly higher in erastin-treated BMSCs than in control cells (Fig. 1E–G). These results suggested that erastin induces the ferroptosis of BMSCs.
NSUN5 is downregulated in erastin-induced BMSC ferroptosis
We screened for the mRNAs and proteins of some RNA methyltransferases that were differentially expressed during BMSC ferroptosis using RT-qPCR and western blotting, respectively. Of the seven RNA methyltransferases evaluated, only NSUN5 and NSUN7 mRNA expression levels differed significantly between control and erastin-treated BMSCs (Fig. 2A–H). RT-qPCR analysis showed that NSUN5 transcript levels were downregulated by erastin treatment (Fig. 2A), and the same was true for protein levels as determined by western blotting (Fig. 2H). Immunofluorescence labeling and confocal microscopy analysis revealed that the NSUN5 fluorescence signal was weaker in erastin-treated BMSCs than in control cells (Fig. 2I, J). These results suggested that NSUN5 is downregulated in erastin-induced BMSC ferroptosis and may be a key mediator of this process.
A–H Compared with control cells, NSUN5 and Nsun7 mRNA (A–G) and NSUN5 protein (H) expression levels were downregulated in erastin-induced BMSC ferroptosis as determined by RT-qPCR and western blotting, respectively. Data are presented as means ± SD. *p < 0.05. I, J Fluorescence signals associated with NSUN5 protein were lower in erastin-treated BMSCs than in control cells, *p < 0.05 (A–J).
NSUN5 inhibits BMSC ferroptosis
To clarify the function of NSUN5 in BMSC ferroptosis, NSUN5 was overexpressed or silenced using lentiviral vectors. The stable knockdown or overexpression of NSUN5 in BMSCs was confirmed by western blotting (Fig. 3A; Supplementary Fig. 2). Compared with untreated controls, the expression levels of glutathione peroxidase 4 (GPX4) were lower and higher with NSUN5 knockdown and overexpression, respectively, in BMSCs treated with erastin (Fig. 3B). Total iron and Fe2+ levels were enhanced by NSUN5 depletion (Fig. 3C, D), whereas NSUN5 overexpression abrogated the erastin-induced increases in total iron and Fe2+ levels (Fig. 3C, D). ROS levels were also increased by NSUN5 knockdown and decreased by NSUN5 overexpression (Fig. 3E, F), and the same effect was observed for lipid peroxidation products (Fig. 3G, H). These data suggested that NSUN5 inhibits ferroptosis in BMSCs.
A NSUN5 knockdown and overexpression in BMSCs confirmed by western blotting. *p < 0.05. B Compared with control cells, GPX4 expression was downregulated by NSUN5 depletion in BMSCs treated with erastin. *p < 0.05. C, D Total iron concentrations (C) and Fe2+ levels (D) were enhanced by NSUN5 knockdown. *p < 0.05. E–H Erastin-induced ROS production (E, F) and lipid peroxidation (G, H) were increased by NSUN5 depletion and reduced by NSUN5 overexpression, *p < 0.05 (C–H).
FTH1 and FTL are substrates of NSUN5
To clarify the mechanism by which NSUN5 inhibits ferroptosis, RIP and RT-qPCR were employed to identify its target transcripts. The results showed that NSUN5 bound FTH1 and FTL mRNA in BMSCs (Fig. 4A, B). Meanwhile, western blotting analysis indicated that NSUN5 overexpression increased the expression levels of FTH1 and FTL (Fig. 4C; Supplementary Fig. 3), whereas those of both ferritin subunits were downregulated following si-NSUN5 transfection (Fig. 4D; Supplementary Fig. 3). Immunofluorescence analysis confirmed the colocalization of NSUN5, FTH1, and FTL in the cytoplasm of BMSCs (Fig. 4E; Supplementary Figs. 4, 5). These observations suggested that FTH1 and FTL are potential targets or substrates of NSUN5.
A, B NSUN5 binds to FTH1 and FTL mRNA in bone marrow-derived stem cells (BMSCs), as revealed by RNA immunoprecipitation (RIP), **p < 0.05. C, D NSUN5 overexpression increased the expression levels of FTH1 and FTL (C), whereas both ferritin subunits were downregulated by si-NSUN5 (D), as determined by western blotting. *p < 0.05. E NSUN5, FTH1, and FTL co-localized in the cytoplasm of BMSCs, as revealed by immunofluorescence analysis, *p < 0.05. F FTH1 and FTL overexpression attenuated the erastin-induced increase in total iron concentrations, *p < 0.05. G, H Erastin-induced ROS production (G) and lipid peroxidation (H) were attenuated by FTH1 and FTL overexpression in BMSCs. *p < 0.05 (A–H).
To determine whether FTH1 and FTL mediate the inhibitory effect of NSUN5 on BMSC ferroptosis, we overexpressed FTH1 and FTL in erastin-treated BMSCs and found that the increase in total iron concentration induced by erastin was abolished (Fig. 4F). This was accompanied by a decrease in ROS levels (Fig. 4G) and the partial abrogation of lipid peroxidation (Fig. 4H), both of which were induced by erastin. The results of the dot blot assay showed that NSUN5 knockdown reduced 5mC levels, whereas NSUN5 overexpression had the opposite effect (Fig. 5A, B; Supplementary Fig. 6). Immunofluorescence analysis of 5mC levels using confocal microscopy revealed that 5mC-associated fluorescence was weaker in NSUN5-depleted BMSCs than in control cells (Fig. 5C).
A, B The results of the dot blot assay showed that NSUN5 knockdown reduced 5mC levels, whereas NSUN5 overexpression elicited the opposite effect. *p < 0.05. C Compared with control cells, 5mC levels were reduced in NSUN5-depleted cells, as determined by immunofluorescence analysis. *p < 0.05. D 5mC antibody enriched FTH1 and FTL mRNA, whereas NSUN5 knockdown reduced 5mC levels in FTH1 and FTL mRNA, as determined by RNA immunoprecipitation (RIP) and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis. *p < 0.05. E Methylation sites of FTH1 and FTL mRNA in control of NSUN5-depleted BMSCs were mainly in the 5′ or 3′ UTR. F, G Bisulfite-sequencing analysis revealed that NSUN5 knockdown abolished the C-to-T conversion at FTH1 and FTL mRNA cytosine methylation sites, *p < 0.05 (D).
We next evaluated the functional significance of the NSUN5-mediated 5mC modification of FTH1 and FTL mRNA. The results of the 5mC RIP assay and RT-qPCR showed that the anti-5mC antibody enriched the mRNA of FTH1 and FTL. NSUN5 silencing reduced the 5mC levels in both transcripts, suggesting that NSUN5 regulates FTH1 and FTL expression via 5mC RNA modification (Fig. 5D).
We also assessed the methylation status of FTH1 and FTL mRNAs in BMSCs by bisulfite sequencing using FTH1- and FTL-specific primers and found that they were methylated in the 5′ (FTH1) and 3′ (FTL) untranslated regions (UTRs) (Fig. 5E–G). The nucleotide was the 5mC methylation site for FTH1, and the nucleotide 628 was the 5mC methylation site for FTL (Fig. 5F, G). Sequence analysis indicated that NSUN5 depletion induced the C-to-T conversion at FTH1 and FTL mRNA cytosine methylation sites, lead to the reduce of the methylate modification. These results suggested that 5mC levels in FTH1 and FTL RNA are decreased with NSUN5 knockdown, leading to a reduction in FTH1 and FTL protein synthesis.
NSUN5 recruits TRAP1 to modify FTH1 and FTL
We investigated whether other proteins are involved in the NSUN5-mediated modification of FTH1 and FTL. Using silver staining and western blotting, we found a protein band among the pulldown products of BMSCs expressing NSUN5 that was absent in those of NSUN5-depleted cells (Fig. 6A), which was identified as TRAP1. The same result was obtained in the LC–MS analysis (Supplementary Materials LC–MS report). The specific enrichment of TRAP1 in lysates of NSUN5-expressing BMSCs was confirmed by western blotting, and vice versa (Fig. 6A, Supplementary Fig. 7). TRAP1 is a mitochondrial paralog of heat-shock protein 90 (Hsp90) that has been reported to induce cell death and may also regulate ferroptosis in BMSCs. We speculated that NSUN5 might regulate FTH1 and FTL through the recruitment of TRAP1. RIP analysis identified an abundance of NSUN5–TRAP1 complexes (Fig. 6B), whileL through the recruitment oimmunofluorescence analysis showed that NSUN5 and TRAP1 proteins co-localized in the cytoplasmL through the recruitment oof BMSCs (Fig. 6C; Supplementary Fig. 8). The interaction between TRAP1 and FTH1 and FTLL through the recruitment owas confirmed by a co-IP assay (Fig. 6D). To examine the role of NSUN5 in the TRAP1-FTH1/FTLL through the recruitment ointeraction, we silenced NSUN5 expression in BMSCs, and found that the amount of FTH1/FTLL through the recruitment oprecipitated in TRAP1 complexes was reduced. This suggested that, in the absence of NSUN5, theL through the recruitment ointeraction between TRAP1 and FTH1/FTL was abolished (Fig. 6E, F). In contrast, NSUN5 knockdown had no effect on the protein (Fig. 6G) or mRNA (Fig. 6H, I) levels of TRAP1 and vice versa. These findings suggested that NSUN5 recruits TRAP1 to modify FTH1 and FTL (Fig. 7; Supplementary Fig. 9).
A Silver-stained gel showing a protein band among the pulldown products of bone marrow-derived stem cells (BMSCs) expressing NSUN5 that was absent in those of NSUN5-depleted cells; western blot analysis confirmed TRAP1 enrichment in the former group. B Detection of NSUN5/TRAP1 binding complexes by RNA immunoprecipitation (RIP). *p < 0.05. C Immunofluorescence analysis confirmed that NSUN5 and TRAP1 protein co-localized in the cytoplasm of BMSCs. *p < 0.05. D Interaction between TRAP1 and FTH1/FTL revealed by co-immunoprecipitation (co-IP). E, F NSUN5 knockdown reduced the amount of precipitated FTH1/FTL, indicating that the interaction between TRAP1 and FTH1/FTL was abolished. G–I NSUN5 silencing did not affect the mRNA and protein levels of TRAP1 and vice versa, *p < 0.05 (B).
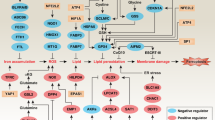
After added erastin, NSUN5 methylated FTH1/FTL mRNA by targeting on its 5′UTR/3′UTR region. NSUN5 recruits trap1 and binds to FTH1/FTL mRNA in BMSCs, and then induced to the degrade of FTH1/FTL. After FTH1/FTL is downregulated, it further promotes the low level of iron storage. Eventually, FTH1/FTL is maintained at an extremely low level, resulting in the accumulation of free ferrous ions, ROS levels, lipid peroxidation products and a reduction in the GPX4 and eventually leading to cell death due to ferroptosis.
Discussion
MSC transplantation is used for the treatment of several diseases; however, the ferroptosis of BMSCs can lead to transplant failure, thereby limiting their use. Consequently, it is important to elucidate the mechanisms underlying ferroptosis so as to identify ways of preventing its occurrence and improving the success rate of transplantations. Here, we used erastin treatment to investigate the roles of NSUN5 in BMSC ferroptosis. The major findings of the present study were as follows: (1) NSUN5 was downregulated in erastin-induced BMSC ferroptosis, while NSUN5 overexpression elicited the opposite effects; (2) FTH1 and FTL are substrates of NSUN5; and (3) NSUN5 recruits TRAP1 to modify FTH1 and FTL. These findings suggest that the impairment of the NSUN5-FTH1/FTL pathway could be a causative event leading to the ferroptosis of BMSCs, and that the NSUN5-FTH1/FTL pathway might be a key target for the development of drugs that can protect against ferroptosis in transplanted BMSCs.
In this study, we used erastin to induce ferroptosis in BMSCs. Erastin functions as a small-molecule inducer of ferroptosis by inhibiting GPX4, which leads to the depletion of glutathione and, eventually, cell death [22,23,24]. Erastin has been shown to induce ferroptosis in both SH-SY5Y and tumor cells [25, 26]. NSUN5 was found to be downregulated in erastin-treated BMSCs. Ferroptosis was alleviated by NSUN5 overexpression and was exacerbated by NSUN5 knockdown, suggesting that RNA methyltransferase activity regulates this process. Some Nsun proteins are presumed to regulate BMSC ferroptosis. NSUN5 is a conserved RNA (5C-cytosine) methyltransferase with a role in the modulation of longevity and cell death [21]. Considering the function of NSUN5 and its potential downstream mechanisms, investigations into the role of NSUN5 in BMSC ferroptosis may help to reveal the molecular basis underlying NSUN5 functions in BMSCs and potentially improve the survival of transplanted BMSCs.
Ferroptosis is believed to be related to the regulation of lysosomal function, glutamate metabolism, the transsulfuration pathway, and iron overload [24, 27,28,29]. Dysregulation of iron and redox homeostasis, autophagic degradation, and iron metabolism barriers also play important roles in the mechanism of ferroptosis [30, 31]. Iron overload affects ferritin deposition and systemic iron homeostasis [32, 33]. High concentrations of iron can lead to the overproduction and aggregation of iron-containing ferritin, which has been observed in the liver and spleen [34]. Ferritin comprises heavy- and light-chain subunits, FTH and FTL, which are responsible for intracellular iron storage [6], play important roles in cell death, and may also be involved in ferroptosis [35, 36]. In our study, FTH1 and FTL levels—which were positively correlated with those of NSUN5—were reduced in erastin-induced ferroptosis, and this was associated with increased intracellular iron concentrations in BMSCs. Conversely, ferroptosis was attenuated by FTH1/FTL overexpression. We further determined that NSUN5 binds to FTH1/FTL mRNA and that the latter are targets for the RNA methyltransferase activity of NSUN5. Interestingly, FTH1 underwent 5mC modification in its 5′ UTR, whereas that of FTL occurred in the 3′ UTR. Nevertheless, the functional significance of this observation remains to be determined.
We identified TRAP1 as an interaction partner of NSUN5 in BMSC cells. The co-IP assay showed that NSUN5 recruits TRAP1 to bind to FTH1 and FTL mRNA. Taken together, our findings suggest a mechanism in which the downregulation of NSUN5 leads to reduced 5mC levels in FTH1/FTL mRNA and a consequent increase in intracellular iron concentrations, decreased GPX4 levels and activity, and accumulation of ROS and lipid peroxidation products, all of which promote ferroptosis.
Our findings have several important clinical implications. First, the treatment of acute liver failure, neurodegenerative diseases, and numerous other conditions are limited by the low survival rate of transplanted BMSCs [37], highlighting that methods to improve survival rates are urgently needed. We found that NSUN5 was downregulated in erastin-induced BMSC ferroptosis and that its overexpression inhibited BMSC ferroptosis. These observations indicate that NSUN5 may be a potential biomarker for the status of BMSCs, and could be used to improve the survival rate of transplanted BMSCs. Second, our results support that the regulation of the NSUN5-FTH1/FTL pathway represents a novel putative therapeutic target for the prevention of ferroptosis in transplanted BMSCs.
In conclusion, our study is the first to demonstrate that NSUN5 suppresses ferroptosis in BMSCs through interaction with TRAP1 and 5mC modification of FTH1 and FTL mRNA. The therapeutic targeting of NSUN5-FTH1/FTL pathway components has the potential to improve the survival of transplanted BMSCs.
Materials and methods
Extraction and culture of BMSCs
Sprague Dawley rats (4 weeks old) were obtained from the Animal Center of Guangzhou University of Chinese Medicine and were housed in standard laboratory rooms under controlled conditions. All animal experiments were performed according to international guidelines for the care and use of laboratory animals and were approved by the Experimental Animal Committee of Guangzhou University of Chinese Medicine. BMSCs were extracted and cultured according to our previously described protocol24. Briefly, bone marrow plugs were extracted from the bones of the hind legs of the rats, and the cells were centrifuged and resuspended twice in complete medium (Cyagen Biosciences, Santa Clara, CA, USA). Subsequently, 5 × l08 cells in 9 mL of complete medium were transferred to 100-mm culture dishes. Three days later, the medium was refreshed and only adherent cells were maintained in complete medium supplemented with glucose and 10% (v/v) fetal bovine serum (Invitrogen, Carlsbad, CA, USA) until treatment with erastin (MedChemExpress, Monmouth Junction, NJ, USA).
Lentivirus overexpression and knockdown experiments
A lentiviral vector (lenti dCAS-VP64) harboring the green fluorescence protein (GFP) coding sequence was used to stably express NSUN5 in BMSCs. We obtained Nsun5 from a different vector and then inserted the sequence into the lenti dCAS-VP64 vector along with the GFP from pLVX-AcGFP, along with one containing a scrambled sequence as a negative control for the overexpression experiments (lenti-dCas9-Vp64-AcGFP). A total of 5 × 105 cells/well were seeded in 6-well plates and incubated for 12 h. At 50% confluence, the cells were infected with either lenti-dCas9-Vp64-NSUN5-AcGFP (NSUN5 overexpression group) or lenti-dCas9-Vp64-AcGFP (negative control group), or not infected. The cells were incubated for 24 h before the medium was replaced with virus-free medium. Three days later, the infection efficiency was evaluated based on GFP signal intensity, and the cells were harvested for western blot and real-time quantitative PCR (RT-qPCR) analyses. For the knockdown experiments, BMSCs were transfected with siRNAs targeting NSUN5 and TRAP1. The siRNAs were designed and synthesized by RiboBio (Guangzhou, China).
C11-BODIPY 581/591 assay
The C11-BODIPY assay was used to characterize the degree of lipid peroxidation, as previously described [38, 39]. BMSCs were first incubated in basal medium without (control) or with erastin (20 μM) for 12 h and then with C11-BODIPY (2.5 μM, Molecular Probes, Carlsbad, CA, USA) for 30 min. The cells were observed and imaged under a fluorescence microscope (Olympus) or analyzed using flow cytometry (Beckman Coulter, CA, USA).
Measurement of intracellular iron concentrations
To measure intracellular iron concentrations, BMSCs were seeded at 1 × 104 cells/well in 96-well plates. After allowing them to adhere for 24 h, the cells were divided into groups as in the C11-BODIPY assay. The iron concentration was quantified using the QuantiChrom Iron Assay Kit (Bioassay Systems, Hayward, CA, USA), and the reduction of Fe3+ to Fe2+ in the presence of the chromogen was quantified by measuring the absorbance at 590 nm using a spectrophotometer (BioTek, VT, USA).
Measurement of ROS levels
To measure ROS production, BMSCs were seeded at 1 × 106 cells/well in 12-well plates. After allowing them to adhere for 24 h, the cells were divided into groups as in the C11-BODIPY assay. The cells were incubated with 10 μM DCF-DA (Molecular Probes) and ROS levels were determined using flow cytometry (Beckman Coulter).
RT-qPCR
NSUN5 transcript levels were evaluated using RT-qPCR. Total RNA was extracted from BMSCs using TRIzol (Invitrogen). RNA purity and concentration were determined by measuring the absorbance at 280 and 260 nm using a NanoDrop-2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The RNA was converted into cDNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Primers for amplifying NSUN5, Trap1, Fth1, Ftl, and beta-actin were obtained from Sangon Biotech (Shanghai, China). The sequences of the primers used are shown in Supplementary Table 1. qPCR was performed in a LightCycler 480 II instrument (Roche Applied Science, Basel, Switzerland) using a SYBR Green Master Mix.
Western blot
Proteins were extracted from BMSCs using RIPA lysis buffer containing a protease inhibitor cocktail (Sigma, MO, USA). Proteins (30 µg) were separated on precast 10% SDS–polyacrylamide gels (Bio-Rad, USA) and then transferred to nitrocellulose membranes (Millipore Sigma, USA) by semi-dry blotting. After blocking with 5% skim milk, the cells were incubated for 12 h at 4 °C with primary antibodies targeting NSUN5 (Proteintech, IL, USA), FTH1, FTL, β-actin, 5mC (Abcam, Cambridge, MA, USA), or DDX25 (Santa Cruz, CA, USA). The next day, the membranes were incubated with an horseradish peroxidase (HRP)-conjugated secondary antibody. Protein bands were visualized by enhanced chemiluminescence (Millipore, MA, USA).
Fluorescence immunocytochemistry
BMSCs were fixed in 4% formalin, washed in phosphate-buffered saline, and then permeabilized with 0.25% Triton X-100. Nonspecific binding was blocked using serum-supplemented antibody dilution buffer for 30 min. Immunofluorescence labeling was performed using monoclonal antibodies against NSUN5, TRAP1, FTH1, and FTL diluted 1:100 in 5% goat serum. After incubation for 12 h, the cells were rinsed and incubated for 1 h at room temperature with fluorochrome-conjugated secondary antibodies (Alexa Fluor 488, Alexa Fluor 555, Alexa Fluor 594, or Alexa Fluor 647; Abcam). After counterstaining with DAPI and washing, the cells were imaged using a confocal microscope.
RNA immunoprecipitation assay
RNA immunoprecipitation (RIP) was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Merck Millipore, Billerica, MA, USA) according to the manufacturer’s protocol. Briefly, cultured BMSCs were collected and lysed in RIP lysis buffer. Thewhole-cell extracts were incubated with RIP buffer containing magnetic beads cross-linked with antibodies targeting NSUN5, 5mC, or control IgG at 4 °C for 8 h. The beads were washed and the complexes were incubated with proteinase K and SDS at 55 °C for 30 min to remove proteins. Co-precipitated RNAs were amplified using RT-qPCR.
Dot blot assay
RNA was diluted to 200, 100, 50, and 4 ng/µL. Nitrocellulose membranes were predipped in methyl alcohol and a dot blot apparatus was used to spot the RNA onto the membranes. Approximately 200, 100, 50, and 4 ng of RNA was probed to measure 5mC modification levels. The RNA was cross-linked to the membranes using ultraviolet irradiation and the membranes were incubated with blocking solution, followed by an anti-5mC antibody diluted in blocking buffer, and then an HRP-conjugated secondary antibody. The blots were visualized using enhanced chemiluminescence.
Methylation analysis by bisulfite sequencing
Total RNA was extracted from BMSCs and purified using an RNA extraction kit (Zymo Research, Irvine, CA, USA) according to the manufacturer’s instructions. The RNA was treated with bisulfite and PCR was performed under the following conditions: predenaturation at 95 °C for 10 min, followed by 40 cycles of 95 °C for 30 s, 60 °C for 40 s, and 72 °C for 40 s, with a final extension at 72 °C for 10 min. The methylation status of the Fth1 and Ftl genes was qualitatively analyzed using gel electrophoresis (1× TAE) and methylation-specific PCR, gene sequencing, and direct bisulfite sequencing. The primer sequences are listed in Supplementary Table 2.
Co-immunoprecipitation assay
Co-immunoprecipitation (co-IP) was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. Briefly, BMSCs were collected from culture plates and lysed in co-IP lysis buffer. The lysates were centrifuged at 13,000 × g at 4 °C for 15 min. The extracted proteins were incubated at 4 °C for 12 h with magnetic beads cross-linked with nonspecific mouse IgG (5 mg) or anti-NSUN5/Trap1 antibodies and treated with coupling buffer. After magnetic separation, the precipitates were washed, the proteins were eluted and neutralized, and western blotting and silver staining were performed according to standard protocols.
Silver staining assay and liquid chromatography–mass spectrometric analysis
Co-IP products were analyzed using vertical gel electrophoresis on 12.5% polyacrylamide nondenaturing gels at 300 V for 45 min. The gels were subjected to silver staining and sent to BGI Technology Co., Ltd for analysis by liquid chromatography–tandem mass spectrometry (LC–MS).
Statistical analysis
Each experiment was performed in triplicate. Data were expressed as means ± standard deviation and were analyzed using GraphPad Prism v7.0 (GraphPad Software, La Jolla, CA, USA) or SPSS v19.0 (SPSS Inc, Chicago, IL, USA). The Student’s t test was used to compare means between two groups and a p value of <0.05 was considered statistically significant.
Data availability
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
References
Kong D, Xu H, Chen M, Yu Y, Qian Y, Qin T, et al. Co-encapsulation of HNF4α overexpressing UMSCs and human primary hepatocytes ameliorates mouse acute liver failure. Stem Cell Res Ther. 2020;11:449.
Lee NH, Myeong SH, Son HJ, Hwang JW, Na DL. Ethionamide preconditioning enhances the proliferation and migration of human Wharton’s Jelly-derived mesenchymal stem cells. Int. J. Mol. Sci. 2020;21:7013.
Fields L, Ito T, Kobayashi K, Ichihara Y, Podaru M, Hussain M, et al. Epicardial placement of human MSC-loaded fibrin sealant films for heart failure: preclinical efficacy and mechanistic data. Mol Ther J Am Soc Gene Ther. 2021;29:2554–70.
Feng Y, Xu Q, Yang Y, Shi W, Meng W, Zhang H, et al. The therapeutic effects of bone marrow-derived mesenchymal stromal cells in the acute lung injury induced by sulfur mustard. Stem Cell Res Ther. 2019;10:90.
Yamada N, Karasawa T, Wakiya T, Sadatomo A, Ito H, Kamata R, et al. Iron overload as a risk factor for hepatic ischemia-reperfusion injury in liver transplantation: potential role of ferroptosis. Am J Transplant. 2020;20:1606–18.
Honarmand Ebrahimi K, Hagedoorn P, Hagen W. Unity in the biochemistry of the iron-storage proteins ferritin and bacterioferritin. Chem Rev. 2015;115:295–326.
Wang Z, Ma J, Li X, Wu Y, Shi H, Chen Y, et al. Quercetin induces p53-independent cancer cell death through lysosome activation by the transcription factor EB and reactive oxygen species-dependent ferroptosis. Br J Pharmacol. 2021;178:1133–48.
Asperti M, Bellini S, Grillo E, Gryzik M, Cantamessa L, Ronca R, et al. H-ferritin suppression and pronounced mitochondrial respiration make hepatocellular carcinoma cells sensitive to RSL3-induced ferroptosis. Free Radic Biol Med. 2021;169:294–303.
Munteanu A, Mukherjee N, Ohler U. SSMART: sequence-structure motif identification for RNA-binding proteins. Bioinformatics. 2018;34:3990–8.
Aust S, Bachmayr-Heyda A, Pateisky P, Tong D, Darb-Esfahani S, Denkert C, et al. Role of TRAP1 and estrogen receptor alpha in patients with ovarian cancer -a study of the OVCAD consortium. Mol Cancer. 2012;11:69.
Agliarulo I, Matassa D, Amoroso M, Maddalena F, Sisinni L, Sepe L, et al. TRAP1 controls cell migration of cancer cells in metabolic stress conditions: Correlations with AKT/p70S6K pathways. Biochim Biophys Acta. 2015;1853:2570–9.
Basit F, van Oppen L, Schöckel L, Bossenbroek H, van Emst-de Vries S, Hermeling J, et al. Mitochondrial complex I inhibition triggers a mitophagy-dependent ROS increase leading to necroptosis and ferroptosis in melanoma cells. Cell Death Dis. 2017;8:e2716.
Zhou B, Liu J, Kang R, Klionsky DJ, Kroemer G, Tang D. Ferroptosis is a type of autophagy-dependent cell death. Semin Cancer Biol. 2020;66:89–100.
Wu Z, Geng Y, Lu X, Shi Y, Wu G, Zhang M, et al. Chaperone-mediated autophagy is involved in the execution of ferroptosis. Proc Natl Acad Sci USA. 2019;116:2996–3005.
Kovár J, Stýbrová H, Novák P, Ehrlichová M, Truksa J, Koc M, et al. Heat shock protein 90 recognized as an iron-binding protein associated with the plasma membrane of HeLa cells. Cell Physiol Biochem. 2004;14:41–6.
Leenen F, Muller C, Turner J. DNA methylation: conducting the orchestra from exposure to phenotype? Clin Epigenet. 2016;8:92.
Liu R, Long T, Li J, Li H, Wang E. Structural basis for substrate binding and catalytic mechanism of a human RNA:m5C methyltransferase NSun6. Nucleic Acids Res. 2017;45:6684–97.
Ispada J, da Fonseca Junior A, de Lima C, Dos Santos E, Fontes P, Nogueira M, et al. Tricarboxylic acid cycle metabolites as mediators of DNA methylation reprogramming in bovine preimplantation embryos. Int J Mol Sci. 2020;21:6868.
Li Q, Li X, Tang H, Jiang B, Dou Y, Gorospe M, et al. NSUN2-Mediated m5C methylation and METTL3/METTL14-mediated m6A methylation cooperatively enhance p21 translation. J Cell Biochem. 2017;118:2587–98.
Squires J, Patel H, Nousch M, Sibbritt T, Humphreys D, Parker B, et al. Widespread occurrence. of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–33.
Schosserer M, Minois N, Angerer T, Amring M, Dellago H, Harreither E, et al. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat Commun. 2015;6:6158.
Li Y, Zeng X, Lu D, Yin M, Shan M, Gao Y. Erastin induces ferroptosis via ferroportin-mediated iron accumulation in endometriosis. Hum Reprod. 2021;36:951–64.
Liu M, Fan Y, Li D, Han B, Meng Y, Chen F, et al. Ferroptosis inducer erastin sensitizes NSCLC cells to celastrol through activation of the ROS-mitochondrial fission-mitophagy axis. Mol Oncol. 2021;15:2084–2105.
Liu J, Li X, Cai R, Ren Z, Zhang A, Deng F, et al. SSimultaneous study of anti-ferroptosis and antioxidant mechanisms of butein and ()-Butin. Molecules. 2020;25:674.
Cong L, Dong X, Wang Y, Deng Y, Li B, Dai R. On the role of synthesized hydroxylated chalcones as dual functional amyloid-β aggregation and ferroptosis inhibitors for potential treatment of Alzheimer’s disease. Eur J Med Chem. 2019;166:11–21.
Zhao Y, Li Y, Zhang R, Wang F, Wang T, Jiao Y. The role of erastin in ferroptosis and its prospects in cancer therapy. Onco Targets Ther. 2020;13:5429–41.
Turcu A, Versini A, Khene N, Gaillet C, Cañeque T, Müller S, et al. DMT1 inhibitors kill cancer stem cells by blocking lysosomal iron translocation. Chemistry. 2020;26:7369–413.
Chen S, Bu D, Zhu J, Yue T, Guo S, Wang X, et al. Endogenous hydrogen sulfide regulates xCT stability through persulfidation of OTUB1 at cysteine 91 in colon cancer cells. Neoplasia. 2021;23:461–72.
Chen X, Comish P, Tang D, Kang R. Characteristics and biomarkers of ferroptosis. Front Cell Dev Biol. 2021;9:637162.
Jiang L, Kon N, Li T, Wang S, Su T, Hibshoosh H, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62.
Yoshida G. Therapeutic strategies of drug repositioning targeting autophagy to induce cancer cell death: from pathophysiology to treatment. J Hematol Oncol. 2017;10:67.
Garringer H, Irimia J, Li W, Goodwin C, Richine B, Acton A, et al. Effect of systemic iron overload and a chelation therapy in a mouse model of the neurodegenerative disease hereditary ferritinopathy. PLoS ONE. 2016;11:e0161341.
Morris G, Berk M, Carvalho A, Maes M, Walker A, Puri B. Why should neuroscientists worry about iron? The emerging role of ferroptosis in the pathophysiology of neuroprogressive diseases. Behav Brain Res. 2018;341:154–75.
Basu T, Panja S, Shendge A, Das A, Mandal N. A natural antioxidant, tannic acid mitigates iron-overload induced hepatotoxicity in Swiss albino mice through ROS regulation. Environ Toxicol. 2018;33:603–18.
Gao M, Monian P, Pan Q, Zhang W, Xiang J, Jiang X. Ferroptosis is an autophagic cell death process. Cell Res. 2016;26:1021–32.
Mai T, Hamaï A, Hienzsch A, Cañeque T, Müller S, Wicinski J, et al. Salinomycin kills cancer stem cells by sequestering iron in lysosomes. Nat Chem. 2017;9:1025–33.
Hensley M, Tang J, Woodruff K, Defrancesco T, Tou S, Williams C, et al. Intracoronary allogeneic cardiosphere-derived stem cells are safe for use in dogs with dilated cardiomyopathy. J Cell Mol Med. 2017;21:1503–12.
Forkink M, Basit F, Teixeira J, Swarts H, Koopman W, Willems P. Complex I and complex III inhibition specifically increase cytosolic hydrogen peroxide levels without inducing oxidative stress in HEK293 cells. Redox Biol. 2015;6:607–16.
Aguirre P, García-Beltrán O, Tapia V, Muñoz Y, Cassels B, Núñez M. Neuroprotective effect of a new 7,8-dihydroxycoumarin-based Fe/Cu chelator in cell and animal models of Parkinson’s disease. ACS Chem Neurosci 2017;8:178–85.
Acknowledgements
We thank Yuhong Dou at the Shenzhen Hospital of Integrated Traditional Chinese and Western Medicine for providing equipment.
Funding
This work was supported by the Natural Science Foundation of Guangdong (grant no. 2017A030312009) and Discipline construction project of Guangdong Medical University (No. 4SG21009G); the Joint Research Project of Guangdong Provincial Department of Science and Technology and Guangdong Academy of Traditional Chinese Medicine (grant no. 2017A020213036); the Bao’an District Project (grant no. 2020JD426); the Bao’an TCM Development Foundation (grant no. 2020KJCX-KTYJ-130); and the Shenzhen Science and Technology Innovation Committee (grant no. JCYJ20190807112405520).
Author information
Authors and Affiliations
Contributions
WK, MZ designed the study. JL, LY, and LZ performed the experiments. ZR, YL, and YJ organized and analyzed the data. JL, ZR wrote the paper. HL, CB, and DC revised and polished the paper. All authors read and approved the final paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All experiments in this paper have been performed in accordance with the regulations of the ethics committee of Guangdong Medical University.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, J., Ren, Z., Yang, L. et al. The NSUN5-FTH1/FTL pathway mediates ferroptosis in bone marrow-derived mesenchymal stem cells. Cell Death Discov. 8, 99 (2022). https://doi.org/10.1038/s41420-022-00902-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41420-022-00902-z
- Springer Nature Limited
This article is cited by
-
Study on the differential hepatotoxicity of raw polygonum multiflorum and polygonum multiflorum praeparata and its mechanism
BMC Complementary Medicine and Therapies (2024)
-
Type 2 diabetic mellitus related osteoporosis: focusing on ferroptosis
Journal of Translational Medicine (2024)
-
Ferroptosis: a promising candidate for exosome-mediated regulation in different diseases
Cell Communication and Signaling (2024)
-
APE1 inhibition enhances ferroptotic cell death and contributes to hepatocellular carcinoma therapy
Cell Death & Differentiation (2024)
-
Iron promotes ovarian cancer malignancy and advances platinum resistance by enhancing DNA repair via FTH1/FTL/POLQ/RAD51 axis
Cell Death & Disease (2024)