Abstract
PNO1 has been reported to be involved in tumorigenesis, however, its role in glioma remains unexplored. In the present study, PNO1 expression in glioma from on-line databases, cDNA, and tissue microarrays was upregulated and associated with poor prognosis. PNO1 knockdown inhibits tumor cell growth and invasion both in vitro and in vivo; whereas PNO1 overexpression promoted cell proliferation and invasion in vitro. Notably, PNO1 interacted with THBS1 and the promotion of glioma by PNO1 overexpression could be attenuated or even reversed by simultaneously silencing THBS1. Functionally, PNO1 was involved in activation of FAK/Akt pathway. Moreover, overexpressing MYC increased PNO1 promoter activity. MYC knockdown decreased PNO1 and THBS1 expression, while inhibited cell proliferation and invasion. In conclusion, MYC-mediated upregulation of PNO1 contributes to glioma progression by activating THBS1/FAK/Akt signaling. PNO1 was reported to be a tumor promotor in the development and progression of glioma and may act as a candidate of therapeutic target in glioma treatment.
Similar content being viewed by others
Introduction
Glioma is one of the most common primary malignant tumors in the central nervous system and is divided by The World Health Organization into four grades: low-grade (Grades I and II) that are well-differentiated and have low malignancy and high-grade (Grades III and IV) that are poorly differentiated and highly malignant.1 And high-grade gliomas, which account for more than 50% of all gliomas, have been considered as a major threat to human health.2 Nowadays, despite great advances in therapeutic interventions, such as surgical resection, oral alkylating agents, and radiation, the prognosis in glioma patients remains poor.3,4 Therefore, exploring the underlying molecular mechanisms of glioma development is urgently needed. Significantly, there is growing evidence that identification of novel prognostic biomarkers is important for glioma treatment.
Thrombospondin1 (THBS1), one of the important components of extracellular matrix, was first discovered in platelets but has now been involved in regulating tumor development, including glioma.5,6,7 It has been reported that THBS1 acts as a stimulator of focal adhesion kinase (FAK, also known as PTK2).8,9 Importantly, a role for FAK activation in GBM cell invasion and migration has been reported.10 Thus, THBS1 and FAK may serve as the potential therapeutic target for GBM.
In this study, we found that ribosome assembly factor PNO1 was upregulated in glioma tissues and promoted cell growth of glioma cells. As is known, the RNA-binding protein partner of NOB1” (PNO1) is essential in ribosome biogenesis.11,12,13 In addition, increasing evidence suggests that ribosome biogenesis plays important roles in cell growth and proliferation of many cancers, including glioma.14,15,16,17 Thus, inhibiting ribosome biogenesis might provide new opportunities for cancer treatment. However, there are few studies of the ribosome-related function of PNO1 in cancers so far. Therefore, cDNA microarray analysis, high-throughput sequencing technologies, on-line databases analysis, biochemistry, and molecular biology were applied to evaluate PNO1 as a prognostic biomarker and its underlying molecular mechanisms of glioma development.
Results
Identification of PNO1 as a novel target in glioma
In our study, we first utilized three GBM tissues and three normal brain tissues for microarray analysis and found that a total of 3461 genes were differentially expressed, of which 1652 were upregulated and 1809 were downregulated (Fig. 1A, B). To identify potential oncogenes, we focused on six upregulated genes that have not been studied in glioma-related research, including MS4A7, DPYSL3, CYBB, PNO1, STOM, and HNMT (Fig. 1C). We knocked down the selected genes in U251 cells with lentivirus-transduced target-shRNA and the cell growth were detected by high-content screening (Fig. 1D, E). The results showed that the cell growth was significantly inhibited in shPNO1, shSTOM, and shHNMT groups (Supplementary Fig. S1A). PNO1 was chosen for subsequent studies because of its most obvious effect on cell growth. We found that PNO1 was upregulated in glioma cells both in protein and mRNA levels (Supplementary Fig. S1B, C); lentivirus encoding shRNAs specific against PNO1 were effective, of which, shPNO1-2 was the most effective shRNA (Supplementary Fig. S1D, E).
mRNA microarray analysis was used to identify differentially expression genes (DEGs) between three GBM tissues (T) and three normal tissues (N). High-content screening and target-shRNAs delivered by lentivirus, were used to assess the effects of six selected genes on glioma cell growth. Hierarchical clustering plots (A) and volcano plots (B) were used to show the DEGs from mRNA microarray analysis (|logFC | >1 and FDR < 0.05). C Heatmap of six selected DEGs. D Representative images of U251 cell growth are shown. U251 cells were infected with lentivirus encoding shRNAs against these 6 DEGs. E Heatmap(left) and growth curves (right) showing the growth of U251 cells. Data were normalized to cell number on day 1 and are represented as fold change. ***P < 0.001 versus shCtrl.
PNO1 is highly expressed in glioma tissues and predicts poor prognosis
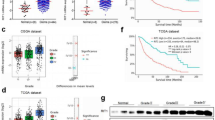
To further confirm the expression pattern of PNO1 in glioma, we first investigated the PNO1 protein expression levels in 141 glioma tissues (55 LGG and 86 GBM) and 24 normal tissues by IHC. The results showed that PNO1 was obviously upregulated in tumor tissues and the higher PNO1 expression was significantly correlated with recurrence patients and advanced pathological grading (Fig. 2A–D). Based on detailed survival information from the glioma patients, the cutoff value for PNO1 was set by the median method for further survival analysis. The results indicated that the high expression of PNO1 was positively associated with the poor prognosis of glioma patients, such as overall survival (OS, P = 0.000) and disease-free survival (DFS, P = 0.000) (Fig. 2E). Bioinformatic analyses of public datasets using GEPIA2 and CGGA revealed that PNO1 was upregulated at mRNA levels in GBM and predicted poor prognosis, which was consistent with the previous IHC results (Fig. 2F–H). The PNO1 protein and mRNA expression levels were also elevated in glioma tissues by RT-PCR and western blot (Fig. 2I, J).
A Representative images of normal tissues and glioma tissues showing PNO1 expression (50 µm). B PNO1 protein levels in 141 glioma tissues (55 LGG and 86 GBM) and 24 normal tissues were determined by IHC. C Correlation analysis between the PNO1 IHC score and tumor recurrence. D Correlation analysis between the PNO1 IHC score and tumor Grade. E Kaplan–Meier survival curve analysis showing the correlation between the PNO1 IHC score and OS (left) and DFS (right) in glioma patients. F PNO1 mRNA expression in 163 GBM tissues and 207 normal tissues were analyzed using online glioma database from GEPIA2. *P < 0.05. G Correlation analysis between the PNO1 mRNA expression and tumor Grade using online glioma database from CGGA. H Kaplan–Meier survival curve analysis showing the correlation between the PNO1 mRNA expression and OS in primary or recurrent glioma patients using online glioma database from CGGA. Western Blotting (I) and RT-PCR (J) showing PNO1 protein and mRNA levels between three GBM tissues (T) and three normal tissues (N). n = 3, *P < 0.05, **P < 0.01, ***P < 0.001.
PNO1 knockdown inhibits GBM cell proliferation and metastasis in vivo
Mice xenograft models were used through subcutaneous injection of U87 cell models to verify the effect of U87 knockdown on tumor growth in vivo. The measurement of tumor volume started at day 5 post-tumor inoculation and end up with day 18. The results of which illustrated a slower increase in tumor volume and less weight compared with controls (Fig. 3A, B). The expression of PNO1 at the protein level in tumors was shown in Fig. 3C. In vivo imaging of this model confirmed the effect of PNO1 on suppressing tumor growth (Fig. 3D). Moreover, Ki-67 staining was used to detecting proliferative activity of tumors, which was obviously weaker in shPNO1 group than shCtrl group (Fig. 3E). In addition, we established metastasis mouse models by intravenous injection. After six weeks, the mice were sacrificed and images were taken by a Living Image System (PerkinElmer). We found that U87 cells infected with shCtrl metastasized to the brain and lung, whereas there was no visual metastasis in shPNO1 group (Fig. 3F).
Mice tumor volume growth curves (A) and weight change curves (B) for subcutaneous xenografts. n = 10, ***p < 0.001. C Western blotting analyses of PNO1 expression in the indicated tumors. GAPDH was used as a loading control. Each bar represents the mean values ± SD of three independent experiments. ***p < 0.001. D Images of the subcutaneous xenografts from the shPNO1 and shCtrl groups. Images were taken by PerkinElmer IVIS Spectrum (D, respectively). n = 10, *p < 0.05. E Representative images of sections sliced from the indicated tumors and stained with anti-Ki67, respectively. F Images of the intravenous xenografts from the shPNO1 and shCtrl groups. Images were taken by PerkinElmer IVIS Spectrum (respectively).
PNO1 promotes GBM cell proliferation and invasion in vitro
In cultured U251, U87, and SHG-44 cells, PNO1 knockdown decreased cell growth and increased cell apoptosis (Fig. 4A, B). PNO1 overexpression increased colony formation, cell viability, and cell invasion; while PNO1 knockdown shown the opposite (Fig. 4C–E). The PNO1 expression at protein and mRNA levels in U251, U87, and SHG-44 cell lines were verified by western blotting and RT-PCR, which indicated the target cells were successfully infected with lentivirus (Fig. 4F, G).
A Effects of PNO1 on growth of three glioma cells infected with shCtrl or shPNO1. n = 3, **P < 0.01. B Effects of PNO1 on cell apoptosis of three glioma cells infected with shCtrl or shPNO1. n = 3, **P < 0.01. C Plate clone formation assay was used to detect PNO1 effect on three glioma cell lines infected with shCtrl, shPNO1, or PNO1. n = 3, **P < 0.01, ***P < 0.001. D CCK8 assays showing cell growth in three glioma cell lines infected with shCtrl, shPNO1, or PNO1. n = 3, **P < 0.01, ***P < 0.001. E Transwell assays were used to detect invasive ability of three glioma cell lines infected with shCtrl, shPNO1, or PNO1. n = 3, ***P < 0.001. (F and G) Western Blotting (C) and RT-PCR (D) showing PNO1 protein and mRNA levels in three glioma cell lines infected with shCtrl, shPNO1, or PNO1(PNO1 overexpression). n = 3, *P < 0.05, **P < 0.01, ***P < 0.001.
THBS1 acts as the potential target of PNO1 in the regulation of glioma
To further explored the mechanism of PNO1 on glioma, microarray analysis between shPNO1 and shCtrl groups of U251 cells was performed and revealed 1122 DEGs, including 650 downregulated DEGs and 472 upregulated DEGs. KEGG Pathway Enrichment Analysis of these DEGs using Cytoscape indicated that PNO1 may participate in several functional pathways involved in cell proliferation and metastasis, including focal adhesion and PI3K-Akt signaling pathway (Fig. 5A). Many intersection DEGs, including CCND1, RAC1, PRKCA, THBS1, and AKT3, have been reported to involve in glioma tumorigenesis18,19,20,21,22,23 and were further confirmed by RT-PCR in U251-shPNO1 cells (Fig. 5B). Through the STRING program, THBS1 appeared to be the potential target of PNO1 (Fig. 5C).
A KEGG Pathway Enrichment Analysis of these DEGs from microarray analysis. B RT-PCR showing the intersection DEGs from KEGG Pathway Enrichment Analysis. n = 3, *P < 0.05, **P < 0.01, ***P < 0.001. C Using the STRING program to analysis potential interactions between PNO1 and the five intersection DEGs. FAK, also known as PTK2, was the central of the DEGs. Co-immunoprecipitation (IP) analysis in U251 cells after PNO1 overexpression was performed to determine the binding of PNO1 to THBS1 using anti-FLAG antibody (D, FLAG-tag overexpressing PNO1) or anti-THBS1 antibody (E). F Western blotting showing the protein expression levels in U251 cell lines infected with PNO1 overexpression or the control (vector), and transduced with or without shTHBS1 or shCtrl. G Cell viability was determined using the CCK-8 assay. U251 cells were infected with PNO1 overexpression or the control (vector), and transduced with or without shTHBS1 or shCtrl. n = 3, **, ##P < 0.01, ***, ###P < 0.001, * vs. shCtrl + vector, # vs. shTHBS1+ vector. H Cell invasion was determined using the transwell assay. U251 cells were infected with PNO1 overexpression or the control (vector), and transduced with or without shTHBS1 or shCtrl. n = 3, ***, ###P < 0.001, * vs. shCtrl + vector, # vs. shTHBS1+ vector.
Co-immunoprecipitation (CO-IP) assay and western blot analysis suggested that PNO1 overexpression led to an increase of THBS1 in U251 cells (Fig. 5D–F). Furthermore, we confirmed that the promotion of PNO1 overexpression on glioma cell growth and invasion could be rescued by downregulated THBS1 (Fig. 5G, H). Moreover, the results from on-line datasets analysis indicated that THBS1 was upregulated in GBM patients and predicts poor prognosis, which was also positively correlated with PNO1 expression in glioma patients (Supplementary Fig. S2A–D). In summary, THBS1 showed similar regulatory effects on the development of glioma with PNO1.
PNO1 activates FAK-Akt signaling through THBS1
Consistent with our array data, we confirmed the protein expression of THBS1, phosphorylation levels of FAK, Akt, and the focal adhesion maker Paxillin by western blotting (Fig. 6A and Supplementary Fig. S3A). Next, we performed PF562271 (FAK inhibitor, 10 µM) and MK2206 (Akt inhibitor, 10 µM) to inhibit the FAK/Akt signaling. As shown in Fig. 6B and Supplementary Fig. S3B, PF562271 distinctly downregulated the phosphorylation levels of both FAK and Akt in U251 cell lines, respectively. In addition, MK2206 visibly decreased the phosphorylation levels of Akt, but it failed to attenuate the phosphorylation levels of FAK, suggesting that Akt might serve as downstream of FAK in this study (Fig. 6C and Supplementary Fig. S3C). Moreover, PF562271 has no observable effects on cell viability (Fig. 6D) while MK2206 overtly decreased cell growth (Fig. 6F). Notably, overexpression of PNO1-induced increased cell invasion was profoundly reversed by treatment with PF562271 or MK2206 in U251 cell lines (Fig. 6E, G).
A Western blotting showing the protein expression levels in U251 cell lines infected with shPNO1 or shCtrl (vector). B Western blotting showing the protein expression levels in U251 cells infected with PNO1 overexpression or the control (vector), and treatment with or without PF562271 (FAK inhibitor). C Western blotting showing the protein expression levels in U251 cells infected with PNO1 overexpression or the control (vector), and treatment with or without MK2206 (Akt inhibitor). D CCK8 assay showing cell growth in U251 cells infected with PNO1 overexpression or the control (vector), and treatment with or without PF562271 (FAK inhibitor). E Transwell assay showing cell invasion in U251 cells infected with PNO1 overexpression or the control (vector), and treatment with or without PF562271 (FAK inhibitor). F CCK8 assay showing cell growth in U251 cells infected with PNO1 overexpression or the control (vector), and treatment with or without MK2206 (Akt inhibitor). G Transwell assay showing cell invasion in U251 cells infected with PNO1 overexpression or the control (vector), and treatment with or without MK2206 (Akt inhibitor).
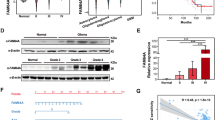
Transcription factor MYC positively regulates PNO1 expression in glioma
To further explored the upstream mechanism of PNO1 on glioma, we identified MYC as a potential transcription factor of PNO1 using TFBIND screening of transcription factor binding sites combined with DEGs from KEGG Pathway Enrichment Analysis. As shown in Fig. 7A, knockdown of PNO1 not only suppressed PNO1 expression but also reduced MYC expression in U251 cells. Moreover, the dual luciferase assay indicated that PNO1 promoter activity is increased when MYC was overexpressed in U251 cells (Fig. 7B). These findings demonstrated that MYC was regarded as an upstream transcription factor of PNO1 in glioma. Furthermore, we found that MYC knockdown overtly decreased expression of PNO1 and THBS1 at mRNA levels in glioma cells (Fig. 7C). MYC knockdown in U251 cells visibly induced cell apoptosis and decreased cell viability (Fig. 7D, E). It also inhibited cell invasion in U251 cells (Fig. 7F). Besides, MYC knockdown reduced the protein levels of PNO1 and THBS1, as well as the genes involved in the FAK/Akt signaling pathway (Fig. 7G). In addition, according to the data analysis of online tools (GEPIA2 and CGGA databases), we found that MYC expression was positively correlated with PNO1 (Supplementary Fig. S2E–G). All of above results suggest that MYC overexpression may help drive glioma by triggering PNO1 overexpression.
A RT-PCR showing PNO1 and MYC mRNA levels in U251 cells infected with shCtrl or shPNO1. n = 3, *P < 0.05, **P < 0.01, ***P < 0.001. B A dual luciferase assay was performed to determine the effect of MYC overexpression on transcriptional activity of PNO1 in U251 cells. C RT-PCR showing MYC, PNO1 and THBS1 mRNA levels in U251 cells infected with shCtrl or shMYC. n = 3, **P < 0.01, ***P < 0.001. D Effects of MYC on cell apoptosis of U251 cells infected with shCtrl or shMYC. n = 3, ***P < 0.001. E Effects of MYC on growth of U251 cells infected with shCtrl or shMYC. n = 3, ***P < 0.001. F Effects of MYC on cell invasion of U251 cells infected with shCtrl or shMYC. n = 3, ***P < 0.001. G Western Blotting showing MYC, PNO1, THBS1 and FAK/AKT signaling protein levels in U251 cells infected with shCtrl or shMYC. H Schematic depicting the regulation of PNO1 in GBM cells.
Discussion
Several studies have proven that PNO1 acts as an essential role in tumorigenesis and development;24,25,26,27 however, its role in glioma remains poorly defined. In this study, we provided evidence from clinical specimens and glioma cell lines that PNO1, driven by the transcription factor MYC, significantly contributes to glioma cell proliferation and metastasis via regulating the expression of THBS1, leading to the phosphorylation of FAK and Akt (Fig. 7H). These findings revealed a new oncogenic signal axis, MYC/PNO1/THBS1/FAK/Akt, in the modulation of cell proliferation and metastasis and provided new insights into how PNO1 works in the progression of glioma. Consequently, interfering in this signal axis could be an efficient way to inhibit glioma development and this is the first report showing the roles and mechanism of PNO1 as an oncogene in glioma.
To further explore the underlying mechanism of oncogenic effects of PNO1 in glioma, we applied cDNA microarray analysis combined with KEGG pathway enrichment analysis to identify these DEGs in U251 cells after treated with shPNO1 or shCtrl. Our results suggested that PNO1 may participate in several functional pathways involved in cell proliferation and migration, such as focal adhesion and PI3K-Akt signaling pathway. Coincidentally, the focal adhesion kinase (FAK)/Akt signaling was proved to participate in mediating the inhibition of cell migration and invasion of GBM cells.28 However, the regulatory molecular mechanism of PNO1 on these pathways have not yet been fully determined. Intriguingly, the CO-IP experiments in this study indicated that THBS1 interacted with PNO1 in U251 cells, which has been reported to be associated with tumor development and progression in several types of cancer.9 Notably, THBS1 was highly expressed in high-grade glioma patients, and THBS1 silencing inhibited GBM cell growth and invasion.23,29 In our study, we observed that the inhibition of THBS1 knockdown on glioma cell growth and invasion could be rescued by PNO1 overexpression. Therefore, we provide evidence that THBS1 acts as a novel target of PNO1 in the progression of glioma. Whereas, the mechanisms for THBS1 regulation in glioma was still limited. Previous studies have proven that THBS1 could modulate FAK phosphorylation to regulate focal adhesion dynamics.9,30 Importantly, inhibition of PI3K activity led to both a decrease in FAK phosphorylation and a decrease in the ability of cells to migrate in response to a directed gradient of THBS1.31 Other study reported that FAK had since been shown to transduce many of the matrix-dependent effects on cell proliferation, migration, and survival.32 Similarly, high FAK expression and activity in GBM is corrected with poor overall survival.28,33 Mechanistically, PNO1 interacted with THBS1. Here we provide evidence that PNO1 positively regulated the THBS1/FAK/Akt signaling in GBM cells, which was further confirmed by using individual inhibitors targeting FAK or Akt. Therefore, our study indicates PNO1 as a novel regulator in THBS1/FAK/Akt signaling pathway-mediated effects on GBM cell proliferation and metastasis.
To further explored the upstream mechanism of PNO1 on glioma, we identified MYC as a potential transcription factor of PNO1 using web-based screening (TFBIND) of transcription factor binding sites combined with DEGs from KEGG Pathway Enrichment Analysis. It has been demonstrated that MYC regulates transcriptional amplification by super enhancers, which was a main hallmark of cancer.34We uncovered that MYC knockdown overtly decreased expression levels of PNO1 and THBS1 mRNA and protein in glioma cells, leading to the phosphorylation of FAK and Akt. Recent studies had shown that MYC overexpression was positively correlated with glioma grade and that increased MYC level was observed in approximately 60-80% gliomas.35 Inhibition of MYC repressed the proliferation of tumor cells, damages cell activity, and promoted apoptosis.36 Indeed, our data showed that MYC knockdown in U251 cells visibly decreased anti-apoptosis, cell viability, and cell invasion. MYC is a key integrator of growth-regulatory and oncogenic signaling pathways. We also identified that MYC expression correlated obviously with PNO1 based on data from on-line glioma databases analysis. Our study is the first to reveal the relationship between PNO1 and MYC. Relevantly, others have proposed that deficient of THBS1 is critical for MYC-induced metastatic phenotypes in medulloblastoma.37 Similarly, MYC amplification may play a crucial role in the angiogenic phenotype through downregulating THBS1 in angiosarcomas.38 All of above results suggest that MYC overexpression may help drive glioma by triggering PNO1 or THBS1 overexpression. It was consistent with previous studies that activation of MYC was involved in tumor formation by targeting ribosome biogenesis.23,35,39,40,41,42,43 However, how MYC regulates PNO1 expression needs further in-depth study.
In conclusion, our findings show that MYC-mediated upregulation of PNO1 contributes to glioma progression by activating THBS1/FAK/Akt signaling. In addition, several small compounds targeting FAK, such as PF562271 and VS-4718, have been launched for conducting clinical trials. Therefore, understanding the roles and mechanism of PNO1 will provide novel therapeutic strategies in glioma.
Materials and methods
Ethics statement
This study was compiled with the ethical guidelines of the Helsinki Declaration and approved by the Ethics Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology (IACUC-2020-02-A).
Patients and tissue specimens
Three glioma tissues and three normal brain tissues were obtained from (Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology). Tissue chips were purchased from OUTDO BIOTECH (shanghai, China). Clinicopathologic characteristics of patients could be seen in Supplementary Table S1. Samples stained with hematoxylin and eosin were verified by experienced pathologists.
Microarray analysis
The detection of gene expression profile in glioma tissues (n = 3) and normal brain tissues (n = 3), as well as in U251 cells infected with shCtrl or shPNO1 by microarray analysis. Total RNA was extracted by the RNeasy kit (Sigma). The concentration of total RNA was determined by Nanodrop 2000 (Thermo Fisher Scientific). RIN value was evaluated with Agilent 2100 and Agilent RNA 6000 Nano Kit (Agilent). RNA sequencing was performed with Affymetrix human GeneChip PrimeView according to the manufacturer’s instruction and the outcomes were scanned by Affymetrix Scanner 3000 (Affymetrix). Raw data statistical significance assessment was accomplished using a Welch t-test with Benjamin–Hochberg FDR. Significant difference analysis and functional analysis based on KEGG pathway enrichment analysis were executed.
Cell culture and transfection
U87, U251, SHG-44, and HEB cell lines were purchased from BeNa Technology (Beijing, China). Cells were identified by STR and the test results for mycoplasma were negative. All cell lines were grown in 90% DMEM with 10% FBS. Cells were grown at 37 ˚C in a humidified atmosphere of 5% CO2. All the plasmids involved in this paper were obtained from Genechem (Shanghai, China). The information of sequences could be found in Supplementary Table S2. Glioma cell lines in logarithmic phase were infected with lentivirus vectors with polybrene at a multiplicity of infection of 10 for 12 h, and then maintained in the new fresh complete medium. After 72 h infection, when the infection efficiency of lentivirus on cells was observed over 85% by fluorescence microscope (Olympus), the cells were harvested for the follow-up experiments.
Real-time quantitative PCR
Total RNA from the cell lines or tissue samples was isolated, respectively using TRIZOL (Sigma) following the manufacturer’s instructions. Reverse transcription of RNA (2.0 µg) to cDNA was performed using the PrimeScript RT reagent kit (Takara) following the manufacturer’s instructions. RT-PCR was carried out using the SYBR Premix Ex Tag (Takara) on VII7 real time PCR instrument. 2-ΔΔCt Method was used to analyze the relative quantitative of target genes with GAPDH as the internal reference. Primers for genes were synthesized by Sangon and shown in Supplementary Table S3.
Immunohistochemical staining
Tissue sections were incubated with PNO1 antibody (1:100, Sigma) at 4 °C overnight, then washed with phosphate-buffered saline (PBS) for 3 times, and incubated with horseradish peroxidase (HRP) conjugated goat anti-mouse IgG polyclonal antibody for 30 min at room temperature. DAB and hematoxylin were used to stain tissue slides. IHC scoring of patients was determined based on the total of the staining intensity and staining extent scores (0, undetectable; 1, weak; 2, moderate; 3, strong). Antibodies used in IHC staining were listed in Supplementary Table S4.
Western blot
Cells and tissues were lysed in ice-cold RIPA buffer (Millipore) and protein concentration was detected by BCA Protein Assay Kit (HyClone-Pierce). The same amount of total protein from each group was separated by 12% SDS-PAGE and transferred onto a PVDF membrane (Bio-Rad Laboratory). The membranes were blocked with primary antibodies in TBST plus 5% non-fat milk. After washed with TBST, membranes were incubated with the appropriate secondary antibody. The signals were visualized using ECL-PLUS Kit (Amersham). Band intensities were quantified using Image J software (NIH). Antibodies were detailed in Supplementary Table S4.
Celigo cell counting assay
72 h after the infection, cells were seeded onto a 96-well plate with the cell density of 2000 cells/well/100 µL in DMEM medium containing 10% FBS and further cultured for 5 days. Cell counting was accomplished every day by Celigo image cytometer (Nexcelom Bioscience) and the cell proliferation curve was drawn.
CCK8 assay
Lentivirus infected glioma cells were measured by CCK8 assay. Cells in the logarithmic phase were seeded onto 96-well plates (2000 cells/well). CCK8 solution (10 µL, MCE) was incubated with cells for 3 h. The OD450 was measured by microplate reader (Tecan) on day 1, 2, 3, 4, and 5 which reflects the number of viable cells. The cell viability ratio was calculated.
Cell apoptosis
LV-shPNO1 and LV-Ctrl infected cells were inoculated in a six-well plate and cells were harvested when the cell density reached 70%. After washed with 4 °C ice-cold PBS, cells were centrifuged at 1300 rpm and resuspended with 200 µL binding buffer. Evaluation of apoptosis was performed by Annexin V-APC staining flow cytometry method (eBioscience) according to the manufacturer’s protocol.
Colony formation assay
Lentivirus infected cells were inoculated in a 6-well plate with 800 cells/well/ 2 mL DMEM with 20% FBS. Medium was exchanged every 2–3 days. Cell clones were photographed by an Olympus digital camera, then fixed and stained by 4% paraformaldehyde and Giemsa (Solarbio), respectively. The numbers of colonies were counted.
Transwell invasion assay
Invasion assay was performed using Corning matrigel invasion chamber. Lentivirus infected U87, U251 and SHG-44 cells were seeded in a 24-well plate with 1×105 cells/well the upper chambers filled with 100 µL serum-free medium. 600 µL DMEM medium plus 30% FBS were filled in the lower chamber and incubated at 37°C for 36 h. At the end of incubation, floating cells were removed and cells in lower chamber were fixed and stained with Giemsa. Images of cells were taken and analyzed using NIH image J software.
Mouse models
Xenograft model in female BALB/c nude mice (4–6 weeks old) were formulated by subcutaneously injection of 0.2 mL exponentially growing lentivirus infected U87 cell suspensions at a destiny of 2 × 107 cell/ml. Metastasis mouse models were established by intravenous injection of 0.1 ml lentivirus infected U87 cell suspensions at a destiny of 1 × 107 cell/ml. After 6 weeks, the mice were sacrificed and taken in vivo images. Tumor images were obtained by a Living Image System (PerkinElmer). Mice were purchased from Shanghai Ling chang Experimental Animals Co., Ltd and randomly divided into two groups (10 mouse/group) using a computer-based random order generator. Tumor growth was assessed twice a week using a caliper and tumor volumes (V) were estimated as V = π/6 × L × W × W (L and W was tumor length and width, respectively). Due to overt tumor volume, the experimenter could not be blinded to whether the animal was injected with shPNO1 or shCtrl. The tumor tissues were removed for Ki-67 immunostaining. If the animal dies prematurely, preventing the collection of tumor volume and histological data, the data will be excluded from the analysis.
Ki-67 staining assay
Tissue slides were blocked with 3% PBS-H2O2 and were incubated with primary antibody Ki-67 at 4 °C overnight. Then slides were incubated with HRP goat anti-rabbit IgG at room temperature for 2 h. Finally, all slides were stained by Hematoxylin (# BA4041, Baso) and Eosin (# BA4022, Baso).
Promoter activity assay
Briefly, U251 cells were co-transfected with MYC overexpression plasmid (500 ng), Renilla luciferase reporter (20 ng), and luciferase reporter plasmid (500 ng) using Lipofectamine™ 3000 Transfection reagents (Thermo). After transduced for 48 h, cells were washed twice with PBS and then lysed in 1 × PLB from the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) for 30 min. Aliquots of supernatant (20 µL) were added to 96-well plates, followed by 40 µL luciferase assay reagent (Promega) at room temperature. Luciferase activity was measured immediately using a luminometer (Orion II Microplate Luminometer, Berthold Detection Systems, Pforzheim, Germany). Data were normalized to the results obtained for the internal control Renilla luciferase.
Statistical analysis
Cell experiments were performed in triplicate and data were expressed as the mean ± SD. Student’s t test was used to analyze the statistical significance. Multiple groups were compared by one-way ANOVA. All statistical analysis was performed using SPSS 22.0 (IBM, SPSS) and GraphPad Prism 8.01 (GraphPad Software). P < 0.05 was considered statistically significant. Kaplan–Meier survival analysis was performed using the log-rank test.
Change history
28 June 2024
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1038/s41419-024-06847-8
References
Ostrom, Q. T. et al. Cbtrus statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro Oncol. 20, iv1–iv86 (2018).
Cheng, M. et al. Super-enhancers: a new frontier for glioma treatment. Biochim. Biophys. Acta Rev. Cancer 188353, 2020 (1873).
Davis, M. E. Glioblastoma: overview of disease and treatment. Clin. J. Oncol. Nurs. 20, S2–S8 (2016).
Cheng, F. & Guo, D. Met in glioma: signaling pathways and targeted therapies. J. Exp. Clin. Cancer Res. 38, 270 (2019).
Gahtan, V. et al. Thrombospondin-1 induces activation of focal adhesion kinase in vascular smooth muscle cells. J. Vasc. Surg. 29, 1031–1036 (1999).
Adams, J. C. & Lawler, J. The thrombospondins. Int. J. Biochem. Cell Biol. 36, 961–968 (2004).
Firlej, V. et al. Thrombospondin-1 triggers cell migration and development of advanced prostate tumors. Cancer Res. 71, 7649–7658 (2011).
Wang, X. J. et al. Thrombospondin-1-induced migration is functionally dependent upon focal adhesion kinase. Vasc. Endovasc. Surg. 42, 256–262 (2008).
Shen, J. et al. Hippo component yap promotes focal adhesion and tumour aggressiveness via transcriptionally activating thbs1/fak signalling in breast cancer. J. Exp. Clin. Cancer Res. 37, 175 (2018).
Fan, Z. et al. A tropomyosin-like meretrix meretrix linnaeus polypeptide inhibits the proliferation and metastasis of glioma cells via microtubule polymerization and fak/akt/mmps signaling. Int. J. Biol. Macromol. 145, 154–164 (2020).
Wang, X. et al. Pno1 tissue-specific expression and its functions related to the immune responses and proteasome activities. PLoS One 7, e46093 (2012).
Zhou, G. J. et al. Cloning and characterization of a novel human rna binding protein gene pno1. DNA Seq. 15, 219–224 (2004).
Miura, M., Hirose, M., Miwa, T., Kuwae, S. & Ohi, H. Cloning and characterization in pichia pastoris of pno1 gene required for phosphomannosylation of n-linked oligosaccharides. Gene 324, 129–137 (2004).
Tang, X. et al. Hdac8 cooperates with smad3/4 complex to suppress sirt7 and promote cell survival and migration. Nucleic Acids Res. 48, 2912–2923 (2020).
Pelletier, J., Thomas, G. & Volarevic, S. Ribosome biogenesis in cancer: New players and therapeutic avenues. Nat. Rev. Cancer 18, 51–63 (2018).
Collins, J. C. et al. Ribosome biogenesis factor ltv1 chaperones the assembly of the small subunit head. J. Cell Biol. 217, 4141–4154 (2018).
Saito, K. et al. Functional analysis of a novel glioma antigen, eftud1. Neuro Oncol. 16, 1618–1629 (2014).
Turner, K. M. et al. Genomically amplified akt3 activates DNA repair pathway and promotes glioma progression. Proc. Natl Acad. Sci. U.S.A. 112, 3421–3426 (2015).
Feng X., et al. Hypoxia-induced acetylation of pak1 enhances autophagy and promotes brain tumorigenesis via phosphorylating atg5. Autophagy, 1–20 (2020)
Liu, P. et al. Par6 regulates cell cycle progression through enhancement of akt/pi3k/gsk-3beta signaling pathway activation in glioma. FASEB J. 34, 1481–1496 (2020).
Liu, Y. et al. Mtorc2/rac1 pathway predisposes cancer aggressiveness in idh1-mutated glioma. Cancers 12, 787 (2020).
Slegers, R. J. & Blumcke, I. Low-grade developmental and epilepsy associated brain tumors: a critical update 2020. Acta Neuropathol. Commun. 8, 27 (2020).
Tsutsui T., et al. Glioma-derived extracellular vesicles promote tumor progression by conveying wt1. Carcinogenesis 1238–1245 (2020)
Dai, H., Zhang, S., Ma, R. & Pan, L. Celecoxib inhibits hepatocellular carcinoma cell growth and migration by targeting pno1. Med. Sci. Monit. 25, 7351–7360 (2019).
Shen, A. et al. Ebf1-mediated upregulation of ribosome assembly factor pno1 contributes to cancer progression by negatively regulating the p53 signaling pathway. Cancer Res. 79, 2257–2270 (2019).
Lin, C. et al. Importance of pno1 for growth and survival of urinary bladder carcinoma: role in core-regulatory circuitry. J. Cell Mol. Med. 24, 1504–1515 (2020).
Liu, D. et al. Pno1, which is negatively regulated by mir-340-5p, promotes lung adenocarcinoma progression through notch signaling pathway. Oncogenesis 9, 58 (2020).
Jiang, K. et al. Mob2 suppresses gbm cell migration and invasion via regulation of fak/akt and camp/pka signaling. Cell Death Dis. 11, 230 (2020).
Daubon, T. et al. Deciphering the complex role of thrombospondin-1 in glioblastoma development. Nat. Commun. 10, 1146 (2019).
Orr, A. W., Pallero, M. A., Xiong, W.-C. & Murphy-Ullrich, J. E. Thrombospondin induces rhoa inactivation through fak-dependent signaling to stimulate focal adhesion disassembly. J. Biol. Chem. 279, 48983–48992 (2004).
Lymn Joanne, S. et al. Phosphatidylinositol 3-kinase and focal adhesion kinase are early signals in the growth factor–like responses to thrombospondin-1 seen in human vascular smooth muscle. Arterioscler. Thromb. Vasc. Biol. 19, 2133–2140 (1999).
Parsons, J. T. Focal adhesion kinase: the first ten years. J. Cell Sci. 116, 1409 (2003).
Shao, Y. et al. Alternative splicing-derived intersectin1-l and intersectin1-s exert opposite function in glioma progression. Cell Death Dis. 10, 431 (2019).
Kress, T. R., Sabò, A. & Amati, B. Myc: Connecting selective transcriptional control to global rna production. Nat. Rev. Cancer 15, 593–607 (2015).
Chen, X. et al. Mir-9 promotes tumorigenesis and angiogenesis and is activated by myc and oct4 in human glioma. J. Exp. Clin. Cancer Res. 38, 99 (2019).
Annibali, D. et al. Myc inhibition is effective against glioma and reveals a role for myc in proficient mitosis. Nat. Commun. 5, 4632 (2014).
Zhou, L. et al. Silencing of thrombospondin-1 is critical for myc-induced metastatic phenotypes in medulloblastoma. Cancer Res. 70, 8199 (2010).
Italiano, A. et al. The mir-17-92 cluster and its target thbs1 are differentially expressed in angiosarcomas dependent on myc amplification. Genes, Chromosomes Cancer 51, 569–578 (2012).
Bywater, M. J. et al. Inhibition of rna polymerase i as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell 22, 51–65 (2012).
Devlin, J. R. et al. Combination therapy targeting ribosome biogenesis and mrna translation synergistically extends survival in myc-driven lymphoma. Cancer Discov. 6, 59–70 (2016).
Hald, O. H. et al. Inhibitors of ribosome biogenesis repress the growth of mycn-amplified neuroblastoma. Oncogene 38, 2800–2813 (2019).
Destefanis, F., Manara, V. & Bellosta, P. Myc as a regulator of ribosome biogenesis and cell competition: a link to cancer. Int. J. Mol. Sci. 21, 4037 (2020).
Kamijo, H. et al. Thrombospondin-1 promotes tumor progression in cutaneous t-cell lymphoma via cd47. Leukemia 34, 845–856 (2020).
Acknowledgements
Thanks are due to Jenny for assistance with the English editing. This work was supported by State Scholarship Fund of China Scholarship Council (20180851007, 201708515096).
Author information
Authors and Affiliations
Contributions
X.C. designed this program. X.C. operated the experiments. Z-Q.G. and D.C. conducted the data collection and analysis. Y.C. and X.C. produced the manuscript which was checked by J.C. All the authors have confirmed the submission of this manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1038/s41419-024-06847-8"
Edited by B. Joseph
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, X., Guo, ZQ., Cao, D. et al. RETRACTED ARTICLE: MYC-mediated upregulation of PNO1 promotes glioma tumorigenesis by activating THBS1/FAK/Akt signaling. Cell Death Dis 12, 244 (2021). https://doi.org/10.1038/s41419-021-03532-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-021-03532-y
- Springer Nature Limited