Abstract
N6-methyladenosine (m6A) is the most common epigenetic RNA modification with essential roles in cancer progression. However, roles of m6A and its regulator METTL3 on non-coding RNA in gastric cancer are unknown. In this study, we found elevated levels of m6A and METTL3 in gastric cancer. Increased METTL3 expression indicated poor outcomes of patients and high malignancy in vitro and in vivo. Mechanically, m6A facilitated processing of pri-miR-17-92 into the miR-17-92 cluster through an m6A/DGCR8-dependent mechanism. The m6A modification that mediated this process occurred on the A879 locus of pri-miR-17-92. The miR-17-92 cluster activated the AKT/mTOR pathway by targeting PTEN or TMEM127. Compared with those with low levels of METTL3, METTL3-high tumors showed preferred sensitivity to an mTOR inhibitor, everolimus. These results reveal a perspective on epigenetic regulations of non-coding RNA in gastric cancer progression and provide a theoretical rationale for use of everolimus in the treatment of m6A/METTL3-high gastric cancer.
Similar content being viewed by others
Introduction
Gastric cancer is one of the most common malignancies and the third leading cause of cancer-related death worldwide1. With few specific symptoms in early stages and with low rates of gastroscopy, most patients have already reached an advanced stage at the time of initial diagnosis. Even among patients who underwent curative resection, 60% suffered recurrences and distant metastasis, with a median overall survival (mOS) of <12 months2. Moreover, gastric cancers with peritoneal metastasis respond rarely to any treatments, leading to an extremely inferior prognosis with life expectancy <6 months3. The effect of targeted therapy is quite limited by the lack of dominant driver genes in gastric cancer. Trastuzumab is the only target drug approved for the first-line treatment of advanced gastric cancer based on the TOGA trial, but its usage was confined in a small part of the patients with ERBB2 amplification4. The anti-angiogenic drug bevacizumab only improves overall survival in non-Asian patients as the first-line treatment5. Up to now, the immune checkpoint inhibitors, pembrolizumab (KEYNOTE-059 cohort 1)6 and nivolumab (ATTRACTION-02)7, are only approved for third-line and later treatment in gastric cancer, with response rates <15%. Therefore, an in-depth investigation of the molecular mechanisms in gastric cancer oncogenesis and progression is critical to allow early diagnosis, innovative therapeutic methods, and ultimately improved prognosis and quality of life for patients.
N6-Methyladenosine (m6A) is the most common epigenetic modification in eukaryotic messenger RNA (mRNA)8 and non-coding RNA (ncRNA)9. It plays a crucial role in gene expression by participating in almost every stage of mRNA metabolism and exerts vital and specific roles in the pathogenesis of various cancers10. Recently, several studies reported that METTL3, the core methyltransferase for m6A modification, promotes gastric cancer progression11,12. However, these studies only focused on m6A of mRNA, and few studies investigated m6A of ncRNA in gastric cancer or implied the possible clinical translational value of m6A/METTL3 related signaling pathways.
Here, we investigated the biological function of METTL3/m6A in regulating ncRNA and defined a novel pathway for m6A-dependent primary microRNA (miRNA) maturation and AKT/mTOR activation in gastric cancer, which could be counteracted by everolimus.
Materials and methods
Cell culture and transfection
AGS, HGC-27, and MKN-45 cell lines were obtained from the Cell Bank, Chinese Academy of Science (Shanghai, China). All cells were authenticated and tested for mycoplasma contamination. Overexpressing plasmids were transfected by Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA). Lentivirus containing METTL3 and shRNA against METTL3 (shM3: CCGGCGTCAGTATCTTGGGCAAGTTCTCGAGAACTTGCCCAAGATACTGACGTTTTTG and shM3-2: CCGGGCTGCACTTCAGACGAATTATCTCGAGATAATTCGTCTGAAGTGCAGCTTTTTG) were used for stable transfection, followed by puromycin selection. The miniMIR17HG plasmid was constructed by inserting the 961–1917th nucleotides of the miR-17-92a-1 RNA (GenBank #: NR_027350.1) into the GV146 vector.
Clinical samples
Fresh tissues were obtained from gastric cancer patients who underwent radical resections at Qilu Hospital of Shandong University between January 2017 and August 2018. Pathologically confirmed gastric cancer paraffin-embedded tissues between 2009 and 2014 were obtained from the Department of Pathology, Qilu Hospital of Shandong University. Only patients with evidence of survival and recurrence were included for OS and RFS analysis, respectively.
RNA m6A quantification
Total RNA was isolated from 10 mm3 of fresh tissue or 106 cells using TRIzol (Invitrogen, CA, USA). The m6A RNA Methylation Quantification Kit (Abcam, Cambridge, UK) was used to quantify the m6A content according to the manufacturer’s instructions. The optical absorbance was measured by a SpectraMax Plus384 Microplate Spectrophotometer (Molecular Device, Sunnyvale, CA, USA).
Immunohistochemistry (IHC)
In clinical studies, paraffin-embedded sections were blocked by goat serum and stained with anti-METTL3 antibody (Abcam, Cambridge, UK) using an IHC staining kit (Zsbio, Beijing, China). Cell nuclei were stained with hematoxylin. In animal studies, tumor xenografts or peritoneal tumors were fixed and processed with a similar procedure with anti-METTL3, anti-Ki67 (Abcam, Cambridge, UK), and anti-PTEN antibody (CST, Danvers, MA).
Proliferation and colony-formation assays
For proliferation assays, cells were seeded in 6-well plates (10,000 or 20,000 cells per well) and counted every 24 h. For colony-formation assays, suspended single cells were seeded in 6-well plates (1000 cells per well), and colonies were counted within 14 days.
Wound-healing, migration, and invasion assays
For wound-healing assays, wounds were made by scratching a line using a 200 µL tip, and the intervals were measured within 72 h. For migration and invasion assays, a Transwell system (Corning, NY, USA) was used as previously described13. Migrated and invaded cells were stained with crystal violet (Beyotime, Shanghai, China) and photographed.
Subcutaneous xenograft and peritoneal implant models
Six-week female BALB/c Nude Mice were purchased from Vital River Laboratory (Beijing, China). For subcutaneous xenograft models, 0.1 mL of cell suspension containing 106 cells were injected subcutaneously into the right flank of mice (n = 6 for each group). Mice were sacrificed at 21- or 32-day after injection. For peritoneal implant models, a cell suspension (5 × 106 cells) was injected intraperitoneally. All mice were sacrificed 4 weeks after injection (n = 4 for each group). Mice were randomly allocated into each group with no blinding. The animal studies were performed following the ARRIVE guidelines and were approved by the Animal Ethical Committee of Qilu Hospital of Shandong University.
Western blot
Approximately 25 µg of protein was separated by 10% SDS-PAGE, transferred to 0.22 μm polyvinylidene difluoride membranes (Thermo Fisher Scientific), and probed by primary antibodies. The anti-METTL3, anti-TMEM127, and anti-GAPDH antibodies were purchased from Abcam (Cambridge, UK). The anti-PTEN, anti-phospho-S6K, anti-phospho-S6, and anti-phospho-4E-BP1 antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA).
RNA immunoprecipitation (RIP)
A Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Darmstadt, Germany) was used for RIP. Briefly, cells were lysed and mixed with anti-m6A (Abcam, Cambridge, UK), anti-DGCR8 (Abcam, Cambridge, UK) antibodies, or isotype controls (Abcam, Cambridge, UK). The antibody-binding RNA was pulled down by protein A/G magnetic beads and quantified by real-time PCR.
Quantitative RT-PCR
Quantification of miRNA was performed using an All-in-One™ miRNA qRT-PCR Detection Kit (GeneCopoeia, San Diego, CA, USA). Quantification of primary miRNA and mRNA was performed using an All-in-One™ First-Strand cDNA Synthesis Kit and an All-in-One qPCR Kit (GeneCopoeia, San Diego, CA, USA). Commercial primers for quantitation of miR-17, miR-18, miR-19a, miR-19b-1, miR-20a, miR-92a-1, snRNA U6, METTL3, TMEM127, PTEN, and GAPDH were purchased from GeneCopoeia (San Diego, CA, USA). Primers for pri-miR-17-92 included 5′-CATCTACTGCCCTAAGTGCTCCTT and 5′-GCTTGGCTTGAATTATTGGATGA. Primers for 5S RNA were 5′-TCTCGTCTGATCTCGGAAGC and 5′-AGCCTACAGCACCCGGTATT.
Everolimus-sensitivity assays
For the in vitro study, everolimus (APExBIO, Houston, TX, USA) was added to cells with final concentrations of 5 or 50 µg/mL. Cell viability was measured using a CCK-8 kit (BestBio, Shanghai, China). For the in vivo study, subcutaneous xenograft models with METTL3-high (n = 3) and control cells (n = 3) were established as above. Control cells were inoculated two days before METTL3-high cells. When the tumor sizes were similar, volume-matched mice received everolimus (50 µg/day intragastrically) or solvent for 17 days and were sacrificed at day 18. Mice were randomly allocated to each group with no blinding.
In silico analyses
All datasets used in this study were derived from public databases. Enrichment analyses were performed by DAVID (https://david.ncifcrf.gov) and TAM (http://www.cuilab.cn/tam). Prediction of miRNA targets was performed by the online tool miRDB (http://mirdb.org/).
Statistical analyses
Data were from at least three independent experiments unless otherwise specified. Variations within each group were estimated and were all statistically compared. Differences between groups were calculated by Student’s t-tests. Univariate analyses were performed by Chi-squared tests. Survival data were compared by log-rank tests. Correlations were analyzed by linear regression. All the statistical tests were two-tailed. P values < 0.05 were considered statistically significant. Data were presented as mean or mean ± standard deviation (SD).
Results
Elevated m6A is mainly regulated by its “writer” METTL3 in gastric cancer
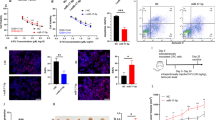
To explore the features of m6A in gastric cancer, we first compared the levels of m6A on total RNAs from 12 pairs of cancerous and adjacent tissues. The m6A levels were significantly elevated in tumor tissues compared with their adjacent tissues (Fig. 1a). To determine the key regulators, we performed differential expression analysis on the m6A “writers”, “erasers”, and “readers” from gastric cancer and normal tissues in The Cancer Genome Atlas Stomach Adenocarcinoma (TCGA-STAD) database. The “writers” and “readers” were all overexpressed in gastric cancer, whereas “erasers” were expressed similarly in tumors and controls (Fig. 1b). Based on this, we then focused on the most differentially expressed “writer”, METTL3, for further study.
a m6A levels of total RNA derived from 12 pairs of cancerous and noncancerous tissues. b Differential expression (gastric cancer vs. normal tissue) of the m6A regulators in the TCGA-STAD database. The colors indicate the functional subgroups of the proteins. c Expression of METTL3 mRNA in 12 pairs of cancerous and noncancerous tissues. d Correlation between METTL3 mRNA levels (y-axis) and percentage of m6A content in total RNA (x-axis) in our clinical cohorts (n = 24). e Genetic changes of METTL3 in gastric cohorts from online databases. Original data were extracted from cBioPortal. f Representative IHC staining of METTL3 in GC tissues (100×). One representative sample of staining intensity (-), (+), (++), and (+++) were shown. Scale bars represent 50 μm. g Clinicopathologic features of patients with METTL3-low (n = 36) and high tumors (n = 51). h, i Kaplan–Meier plots of RFS (n = 58) and OS (n = 80) of patients with METTL3-low and high tumors according to IHC staining intensity. Data were derived from a gastric cancer cohort followed-up by us. j, k Kaplan–Meier plots of RFS (n = 641) and OS (n = 876) of patients with METTL3-low and high tumors according to mRNA levels. Data were interrogated from the GEO datasets GSE14210, GSE15459, GSE22377, GSE29272, GSE38749, GSE51105, and GSE62254 with the probe 209265_s_at. Data are presented as mean ± SD. *P < 0.05; **P < 0.01.
METTL3 RNA level was significantly higher in cancerous tissues than that in adjacent tissues (Fig. 1c). In addition, METTL3 RNA positively correlated with total RNA m6A levels in our cohort (Fig. 1d). When mutations were considered in the previously published cohorts, genetic changes in METTL3 only accounted for 2.12% of total patients (Fig. 1e).
METTL3 overexpression indicates adverse pathological features and poor outcome in gastric cancer
To determine the clinical relevance of METTL3, we tested METTL3 expression in gastric cancer samples of 87 patients who had received radical or palliative gastrectomy (Fig. 1f). METTL3 was mainly distributed in the nucleus of tumor cells. According to the staining intensity, 12 negative (−), 24 weak positive (+), 32 medium positive (++), and 19 strong positive cases (+++) were observed. METTL3 expression significantly correlated with AJCC staging (the eighth edition, P = 0.0056), lymph node metastasis (P = 0.0251), and vascular invasion (P = 0.0346), but did not correlate with gender, age, differentiation, and tumor invasion (Table 1, Fig. 1g).
Patients with METTL3 overexpression (++/+++) had higher risks of recurrence (hazard ratio (HR) = 2.615, 95% confidence interval (CI) = 1.934–5.623, Fig. 1h) and death (HR = 1.712, 95% CI = 1.042–2.799, Fig. 1i), with significantly shorter median recurrence-free survival (mRFS, 9.3 vs. 18.7 months, P = 0.0002, Fig. 1h) and mOS (19.6 vs. 41.1 months, P = 0.0349, Fig. 1i) than those with low METTL3 expression (−/+). In addition, the prognostic value of METTL3 was also confirmed by the online tool Kaplan–Meier plotter. In this database, the METTL3-high patients had higher risks of recurrence (HR = 1.474, 95% CI = 1.211–1.955, Fig. 1j) and death (HR = 1.455, 95% CI = 1.238–1.841, Fig. 1k), with shorter mRFS (9.7 vs. 19.8 months, P = 0.0004, Fig. 1j) and mOS (19.8 vs. 33.2 months, P < 0.0001, Fig. 1k) than the METTL3-low gastric cancer patients.
METTL3 promotes gastric cancer cell proliferation and tumor growth
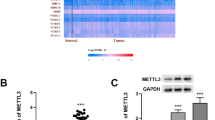
To investigate the biological role of METTL3 in gastric cancer, we established METTL3-downregulated (Fig. 2a and supplementary Fig. S1a) and -upregulated cells (Fig. 2b) using lentivirus containing shRNA and plasmids containing METTL3, respectively. Compared to those in control cells, the m6A levels of total RNA were significantly decreased in METTL3-downregulated cells (P = 0.0024 and 0.0015, Fig. 2c and supplementary Fig. S1b) and elevated in METTL3-upregulated cells (P = 0.0082 and 0.0002, Fig. 2d).
a METTL3 expression in HGC-27 cells transfected with lentivirus encoding shRNA against METTL3 (shM3) and control (shCTL). b METTL3 levels in MKN-45 and AGS cells transfected with overexpressing plasmid (OE) or empty vector (EV). c, d Percentage of m6A content in total RNA in METTL3-reducing HGC-27 and METTL3-overexpressing MKN-45 and AGS cells. e Proliferation curves of METTL3-reducing HGC-27 cells. f Colony-formation assays of METTL3-reducing HGC-27 cells. Representative plates are shown on the left, and quantification on the right. g Proliferation of METTL3-overexpressing MKN-45 and AGS cells. h Colony-formation assays of METTL3-overexpressing MKN-45 and AGS cells. n = 3 for each group in c–h. i METTL3 expression in xenograft tumors derived from stable cell lines transfected with lentivirus encoding METTL3 (lvM3, MKN-45 cells), shM3 (HGC-27 cells), and their corresponding controls (lvCTL and shCTL). j Kaplan–Meier plots of palpable tumor-free survival in METTL3-high (n = 6) and control (n = 6) MKN-45 cell xerographs. k Tumor-harboring mice (top) and xenograft tumors (bottom) 3 weeks post-implantation of METTL3-overexpressing and control MKN-45 cells. l Diameter (left) and weight (right) of METTL3-high and control MKN-45 tumors. Each dot represents one sample. m Ki67 levels of METTL3-high and control MKN-45 tumors. n Kaplan–Meier plots of palpable tumor-free survival in METTL3-low (n = 6) and control (n = 6) HGC-27 xerographs. o Tumor-harboring mice (top) and xenograft tumors (bottom) 35 days post-inoculation of METTL3-reducing and control HGC-27 cells. p Diameter (left) and weight (right) of METTL3-low and control HGC-27 tumors. Each dot represents one sample. q Ki67 levels of METTL3-low and control HGC-27 tumors. Data are presented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
In cell models, METTL3 downregulation impeded cell proliferation (all P < 0.05 from day 2, Fig. 2e and supplementary Fig. S1c) and colony formation (P = 0.0004 and 0.0010, Fig. 2f and supplementary Fig. S1d). Accordingly, METTL3 overexpression stimulated cell proliferation (all P < 0.05 from day 3, Fig. 2g) and colony-forming efficiencies (P = 0.0002 and 0.0055, Fig. 2h).
Subsequently, subcutaneous xenograft models were established for in vivo studies. The median times to form palpable tumors were 11.0 and 13.5 days in METTL3-overexpressed and control cells, respectively (P = 0.0055, Fig. 2i, j). Three weeks after subcutaneous injection, tumors from METTL3-overexpressing cells were remarkably larger (P = 0.0028, Fig. 2k, l) and heavier (P = 0.0027, Fig. 2l) than tumors from control cells. In addition, Ki67 was also overexpressed in METTL3-high tumors (P < 0.0001, Fig. 2m). In mice with METTL3-low tumors, the median time to form touchable tumors was significantly longer than that in control mice (24.0 vs. 22.0 days, P = 0.0055, Fig. 2i, n). Further, METTL3-low tumors were significantly smaller (P = 0.0369, Fig. 2o, p) and had lower weights (P = 0.0421, Fig. 2p) and Ki67 expression (P = 0.0005, Fig. 2q) than the control group.
METTL3 promotes gastric cancer cell migration, invasion, and peritoneal metastasis
In wound-healing assays, METTL3 downregulation reduced migration distances remarkably (P = 0.0023 and 0.0005, Fig. 3a and supplementary Fig. S1e), while METTL3 overexpression increased the migration distances (P < 0.0001 and =0.0003, Fig. 3b). In Transwell assays, the METTL3-low cells showed decreased migration (P = 0.0199 and 0.0061, Fig. 3c and supplementary Fig. S1f) and invasion (P = 0.0029 and 0.0003, Fig. 3d and supplementary Fig. S1g) compared to the control cells. Accordingly, METTL3 overexpression significantly augmented cell migration (P = 0.0001 and 0.0010, Fig. 3e) and invasion (P < 0.0001 and =0.0028, Fig. 3f) in two cell models.
a, b Wound-healing assays of METTL3-reducing (shM3) HGC-27, METTL3-overexpressing (OE) MKN-45, and AGS, and their corresponding control (shCTL and EV) cells. c, d Migration and invasion assays of METTL3-reducing HGC-27 cells. e, f Migration and invasion assays of METTL3-overexpressing MKN-45 and AGS cells. Representative images on the left, and quantification bars on the right in a–f (n = 3 for each group). g Mice with peritoneal implant nodes derived from METTL3-overexpressing (lvM3; n = 4) and control (lvCTL; n = 4) MKN-45 cells. h, i Curves of abdominal circumferences and body weights of the peritoneal metastasis models. j Representative pictures of the peritoneal implants from stable METTL3-overexpressing and control MKN-45 cells. k Number (left) and weight (right) of implanted nodes in peritonea of mouse models. Each dot represents one sample. l Representative pictures of METTL3 expression in peritoneal nodes by IHC. Data are presented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Next, we established a gastric cancer peritoneal metastasis model by intraperitoneal injection of tumor cells (Fig. 3g). The abdominal circumferences of the mice bearing METTL3-high cells increased rapidly and became larger than those of the mice bearing control cells from the second week (all P < 0.05, Fig. 3h). However, body weights were not significantly different between the two groups (Fig. 3i). Most mice with METTL3-high cells showed palpable masses on the abdominal wall, which was not observed in the control group (Fig. 3g). When dissected four weeks later, the mice bearing METTL3-high cells possessed more and larger peritoneal-implanted nodules (both P < 0.0001, Fig. 3j, k). They were mainly distributed in the mesentery and omentum (Fig. 3j), while few grew on the surface of the liver or spleen. METTL3 overexpression in implanted peritoneal nodes was confirmed by IHC (Fig. 3l).
METTL3 promotes tumor progression by facilitating biogenesis of miR-17-92 cluster
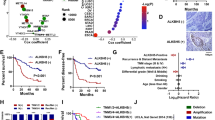
To decipher the mechanisms of METTL3 in tumor growth and metastasis, we performed correlation analysis between METTL3 mRNA and all the other mRNA, miRNA, and long ncRNA in the TCGA-STAD database. Among the top 7 METTL3-correlated miRNAs, 6 (miR-17, 19b-1, 20a, 92a-1, 19a, and 18a) were derived from one miRNA cluster, the miR-17-92 cluster, which shared a primary miRNA, the pri-miR-17-92 (the transcript of MIR17HG, Fig. 4a, b). Meanwhile, none of these miRNAs targeted METTL3 mRNA, as predicted by miRDB. By cluster enrichment analysis of miRNA with software TAM, 76 METTL3-correlated miRNAs (correlation coefficients >0.2 and FDR < 4 × 10−6) were also significantly enriched in the miR-17-92 cluster (P < 0.0001, Fig. 4c). In terms of function, these METTL3-correlated miRNAs were enriched in several cancer-associated pathways, such as cell proliferation, immunity, AKT pathway, onco-miRNA, angiogenesis, apoptosis, and cell cycle (Fig. 4d).
a Correlation between METTL3 mRNA and all miRNAs in the TCGA-STAD database. Pearson correlation coefficients were transformed into Z-scores (y-axis) and ranked in descending order (x-axis). Blue and red dots represent negatively and positively METTL3-correlated miRNAs, respectively. Green dots represent the members of the miR-17-92 cluster. b Correlations between METTL3 (y-axis) and members of the miR-17-92 cluster (x-axis). Data were derived from the TCGA-STAD database. c MiRNA cluster enrichment results of the top METTL3-correlated miRNA (n = 76). The y-axis indicates the miRNA clusters that miRNAs were enriched in, and the x-axis indicates the numbers of METTL3-correlated miRNAs enriched in this cluster. Dot sizes indicate percentage of the METTL3-correlated miRNA in total miRNAs of the cluster, and colors indicate the P values of the enrichment. d Top 10 functional terms from METTL3-correlated miRNA enrichment. e, f Relative expressions of the miR-17-92 cluster and pri-miR-17-92 in METTL3-overexpressing (OE and lvM3) and control (EV and lvCTL) MKN-45 cells in vitro and in vivo. MiR-mean indicates mean value of the miR-17-92 cluster. g, h Relative expression of the miR-17-92 cluster and pri-miR-17-92 in METTL3-reducing (shM3) and control (shCTL) HGC-27 cells in vitro and in vivo. n = 3 for each group in e–h. i Relative expression of miR-17-92 cluster in fresh cancerous (T1–T12) and noncancerous tissues (P1–P12). Relative expression was indicated by the heatmap (left). Fold change (cancerous vs. noncancerous tissues) and statistical significance were shown on the right. j Correlation among METTL3 mRNA and miRNA miR-17-92 clusters. Correlation coefficients indicated by the heatmap. k Correlation between METTL3 mRNA (y-axis) and miR-17-92 cluster (x-axis) shown with scatter plots. l Expression of miR-17-92 cluster in HGC-27 cells expressing shM3 rescued by miniMIR17HG and empty vector (EV) transfection. m, n Proliferation and migration assays of HGC-27 cells expressing shM3 rescued by miniMIR17HG expression. The indicated significances were for comparisons between METTL3-reducing cells and those transfected with miniMIR17HG. Data are presented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We then examined the expression levels of the miR-17-92 cluster and pri-miR-17-92 in cells and xenograft tumors. Both in vitro or in vivo, METTL3 overexpression significantly increased levels of all six miRNAs (all P < 0.05, Fig. 4e) and reduced the level of pri-miR-17-92 (P = 0.0229 in vitro and P = 0.0009 in vivo, Fig. 4f), while METTL3 downregulation reduced all miRNAs (all P < 0.05, Fig. 4g) and increased the level of pri-miR-17-92 (P = 0.0078 in vitro and P = 0.0009 in vivo, Fig. 4h).
The correlation between METTL3 and the miRNA-17-92 cluster was also confirmed in our clinical samples. Levels of miR-17, 18, 19a, 19b-1, and 20a, as well as the average level of all six miRNAs (miR-mean), were significantly elevated in tumors compared with those in adjacent tissues (P < 0.05 for all, Fig. 4i). The expression of each member of the miRNA-17-92 cluster was positively correlated with the others (Fig. 4j). In addition, most members (except miR-92a-1) and miR-mean correlated positively with the level of METTL3 mRNA with statistical significance (P < 0.05 for all, Fig. 4j, k).
To verify whether METTL3 exerts the onco-promoting role by facilitating pri-miR-17-92 maturation, we constructed a miniMIR17HG plasmid to force expression of 6 miRNAs for rescue studies14. This construct contains all miRNAs from the miR-17-92 cluster but no flanking sequences and can be rapidly processed into mature miRNAs. Transfection of miniMIR17HG in METTL3-knocked-down cells significantly raised all miRNAs (all P < 0.01) to similar levels of control cells (Fig. 4l). In addition, miniMIR17HG partially rescued cell proliferation (P < 0.05 from day 2, Fig. 4m) and cell migrations (P = 0.0019, Fig. 4n).
METTL3 facilitates pri-miR-17-92 processing by m6A modification on its A879 locus
To explore the mechanisms by which METTL3 regulates pri-miR-17-92 processing, we examined its function on m6A regulation of pri-miR-17-92 and pri-miR-17-92-DGCR8 binding by RNA RIP. METTL3 overexpression significantly increased the m6A modification on pri-miR-17-92 (P < 0.0001, Fig. 5a). Besides, METTL3 remarkably facilitated DGCR8 binding to pri-miR-17-92 (P = 0.0077, Fig. 5a). Consistently, METTL3 downregulation repressed m6A modification on pri-miR-17-92 (P = 0.0021) and DGCR8-pri-miR-17-92 binding (P < 0.0001, Fig. 5b).
a Results of m6A-RIP and DGCR8-RIP in METTL3-overexpressing (lvM3) and control (lvCTL) MKN-45 cells. b Results of m6A-RIP and DGCR8-RIP in METTL3-reducing (shM3) and control (shCTL) HGC-27 cells. Input RNA and RNA pulled down by specific (anti-m6A or anti-DGCR8) antibodies (Sab) or polyclonal immunoglobulin G (IgG) were quantified by RT-PCR, and their ratios to corresponding inputs were shown in a, b. c Schematic diagram of the wild type pri-miR-17-92 (WT), miniMIR17HG, and pri-miR-17-92 with point mutations. “GGAC” motifs were not displayed in the diagram when mutated into “GGCC”. d Fold change (control vs. METTL3-reducing HGC-27 cells) of miR-19a, 19b, 20a, and their mean values when cells were transfected with vectors encoding pri-miR-17-92-WT or those with point mutations. Statistical significance is for comparisons between cells transfected with pri-miR-17-92-WT and those with mutations. e m6A-RIP in METTL3-reducing and control HGC-27 cells transfected with pri-miR-17-92-WT, A879, or empty vector (EV). RNAs pulled down by anti-m6A or IgG were quantified by RT-PCR, and their ratios to inputs are displayed. Statistical significance is for comparisons between the m6A-RIP data from cells transfected with WT and A879C. f, g Proliferation and migration assays of HGC-27 cells overexpressing pri-miR-17-92-WT, A879C, and EV. The indicated significances were for comparisons between cells overexpressing pri-miR-17-92-WT and A879C. n = 3 for each group. Data are presented as mean ± SD. nsP > 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To identify the specific sites of m6A on pri-miR-17-92 by METTL3, we analyzed its sequence and found five “GGAC” motifs that were potentially recognized by METTL3 (Fig. 5c). Five plasmids with adenine to cytosine mutation in these motifs were constructed and transfected into cells (Fig. 5c). Compared to the wild type plasmid, A879C mutation hindered miRNA biogenesis significantly, while the others did not (Fig. 5d). Also, A879C mutation remarkably reduced the precipitated pri-miR-17-92 by anti-m6A antibody in control cells with normal expression of METTL3 (P = 0.0004), but this difference was not observed in METTL3-knocked-down cells (Fig. 5e). In another aspect, the wild type pri-miR-17-92 with m6A in control cells was 4.3 times as much as that in METTL3-low cells (Fig. 5f). In contrast, the difference of pri-miR-17-92-A879C with m6A was only 1.5 times in these two cell lines, indicating a key role of A879 in the METTL3-mediated pri-miR-17-92 processing (Fig. 5f).
In cell proliferation assays, cells transfected with wild type pri-miR-17-92 showed superior proliferation ability to both pri-miR-17-92-A879C-expressing and control cells from day 1 to 4 (all P < 0.05 from day 2, Fig. 5g). In addition, cells transfected with pri-miR-17-92-A879C also showed inferior migrating ability compared to those transfected with wild type pri-miR-17-92 (P = 0.0082, Fig. 5h).
METTL3 inhibits PTEN/TMEM127 expression and activates AKT/mTOR pathway by facilitating biogenesis of miR-17-92 cluster
To determine the downstream targets of the miR-17-92 cluster in gastric cancer progression, we performed miRNA cluster enrichment analysis and found the members of the miR-17-92 cluster were significantly enriched in several cancer-related terms, including cell proliferation, AKT pathway, angiogenesis, onco-miRNAs, and apoptosis (Fig. 6a). Also, we obtained 2056 target genes of the miR-17-92 cluster predicted by miRDB and 525 mRNAs that negatively correlated with METTL3 with a correlation coefficient < −0.18 in the TCGA-STAD database. The two gene pools shared a total of 98 genes, which were enriched in PI3K/mTOR, p53, TGF-β, and MAPK pathways by KEGG analysis (Fig. 6b). Thus, we chose the members that were clustered in the PI3K/mTOR pathway, PTEN and TMEM127, for further study.
a Top 10 terms from miRNA functional enrichment of the miR-17-92 cluster. b KEGG analysis of the METTL3- and miR-17-92 cluster-regulated genes. (Upper) Number of potential target genes of the miR-17-92 cluster, METTL3-correlated genes, and their intersections. (Lower) Table with the KEGG results (pathways and involved genes) of the intersectional genes. c Correlation between METTL3 (y-axis) and PTEN or TMEM127 (x-axis) in the TCGA-STAD database. d Correlation between METTL3 (y-axis) and PTEN or TMEM127 (x-axis) in our clinical cohorts. e Genetic change of PTEN and TMEM127 in gastric cohorts from online databases. Original data were extracted from cBioPortal. FDR, false discovery rate adjusted P value.
In the TCGA-STAD databases, both PTEN and TMEM127 were negatively correlated with METTL3 expression with statistical significance (both P < 0.0001, Fig. 6c). These negative correlations between METTL3 mRNA and PTEN/TMEM127 mRNA were confirmed in our clinical samples by quantitative RT-PCR (Fig. 6d). Nonsynonymous mutation and copy number variations (CNVs) of PTEN occurred in about 10.6% of gastric patients (Fig. 6e). Among them, all CNVs of PTEN were deletion instead of amplification (Fig. 6e). In contrast, nonsynonymous mutations and CNVs of TMEM127 only occurred in 1.6% of the patients, and all CNVs were amplification (Fig. 6e).
By RT-PCR, METTL3 overexpression significantly reduced mRNA levels for PTEN (P < 0.0001) and TMEM127 (P = 0.0019, Fig. 7a), while METTL3 downregulation elevated their mRNA levels (P = 0.0002 and 0.0083, Fig. 7b). Furthermore, forced expression of miniMIR17HG in METTL3-reducing cells reversed (of PTEN, P = 0.0124) or diminished (of TMEM127, P = 0.0929) the differences (Fig. 7b). By western blot and IHC, both PTEN and TMEM127 were remarkably reduced in METTL3-high cells, subcutaneous xenografts, and peritoneal implants (Fig. 7c, g). Accordingly, when METTL3 was reduced, PTEN and TMEM127 were significantly elevated in cells and subcutaneous xenografts (Fig. 7h–j). What is more, the overexpression of PTEN and TMEM127 caused by METTL3-knockdown was reversed by miniMIR17HG (Fig. 7k).
a, b Relative mRNA of PTEN and TMEM127 in METTL3-overexpressing MKN-45 (lvM3), METTL3-reducing HGC-27 (shM3), and control cells (lvCTL and shCTL) by RT-PCR. c Protein levels in METTL3-overexpressing and control MKN-45 cells by western blot. d, e Protein levels in METTL3-high (n = 3) and control (n = 3) xenograft MKN-45 tumors by western blot or IHC. Quantification is derived from data of three mice. f, g Protein levels in METTL3-high (n = 3) and control (n = 3) peritoneal implants by western blot and IHC. h Protein levels in METTL3-reducing and control HGC-27 cells by western blot. i, j Protein levels in METTL3-low (n = 3) and control (n = 3) xenograft HGC-27 tumors by western blot and IHC. k Protein levels in control, METTL3-reducing, and METTL3-reducing/miR-17-92 cluster-expressing HGC-27 cells by western blot. Data are presented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We continued to investigate whether METTL3/miR-17-92 cluster influenced the AKT/mTOR pathway activation. In METTL3-high cells, subcutaneous xenografts, and peritoneal metastatic nodes, the phosphorylation levels of AKT/mTOR pathway-related proteins, including AKT, S6K, S6, and 4E-BP1, were all upregulated (Fig. 7c, d, f), which indicated the activation of the AKT/mTOR pathway. In the METTL3-reducing cells and xenograft tumors, phosphorylation levels of AKT, S6K, S6, and 4E-BP1 were significantly decreased (Fig. 7h, i). Further, forced expression of miniMIR17HG counteracted the AKT/mTOR pathway inactivation by METTL3-knocked-down (Fig. 7k).
Gastric cancer with METTL3 overexpression is more sensitive to everolimus
Due to the remarkable impact of the METTL3/miR-17-92 cluster on the AKT/mTOR pathway, we explored whether everolimus, an mTOR inhibitor, could inhibit the onco-promoting role of METTL3. Cell viability was measured in cell models with different levels of METTL3 after treatment with solvent or different concentrations of everolimus. Without everolimus, METTL3 overexpression increased cell viability (P < 0.05 from day 1, Fig. 8a), whereas METTL3 downregulation decreased cell viability (P < 0.05 from day 1, Fig. 8b), compared to their corresponding controls. Addition of everolimus remarkably suppressed cell viability, regardless of its concentrations or METTL3 expression (Fig. 8a, b). However, METTL3-overexpressing cells showed more sensitivity to everolimus in a dose-dependent manner than the control cells (P < 0.05 at day 2 and 3, Fig. 8a). In accordance, the METTL3-low cells showed lower sensitivity to everolimus in both concentrations than the control cells (P < 0.05 at day 2 and 3, Fig. 8b).
a, b Cell viability assays of METTL3-overexpressing MKN-45 (lvM3), METTL3-reducing HGC-27 (shM3), and control (lvCTL and shCTL) cells, with (5 or 50 µg/mL) or without everolimus (E). c Tumor volume of mice carrying METTL3-high or control tumors and fed with everolimus or solvent. Mice were pre-inoculated with MKN-45 tumor cells, administrated with everolimus or solvent daily from day 0 to 16, then sacrificed at day 17. d Photos of xenograft tumors when mice were sacrificed. e Tumor inhibition rate of everolimus in METTL3-high and control tumors. Tumor inhibition rate = 1 − the tumor weight with everolimus/the corresponding tumor weight with solvent. f Overview of METTL3/m6A-mediated miRNA cluster biogenesis and AKT/mTOR pathway activation in gastric cancer development. n = 3 for each group. Data are presented as mean ± SD. *P < 0.05.
With in vivo models, everolimus was effective in reducing tumor volumes in both METTL3-overexpressing or control tumors (Fig. 8c). The tumor inhibition rate of everolimus in METTL3-high tumors was significantly higher than that in control tumors (89.92% vs. 73.26%, P = 0.0465, Fig. 8c–e), suggesting that METTL3-high tumors could be inhibited by everolimus more effectively.
Discussion
Due to its heterogeneity and specificity, gastric cancer has been gradually shown to lack common driven mutations or CNVs that are commonly seen in lung, breast, or colorectal cancer. Consequently, it is difficult to copy the success of the translational therapies based on genetic alterations in certain types of cancers, such as the EGFR or ALK-driven non-small cell lung cancer. Increasing scholars believe that cancer is an epigenetic disease, and disrupted and unstable epigenomes exist widely among different types of tumors with heterogeneity and therapeutic resistance15,16. m6A is the most abundant internal modification in eukaryotic RNA and may represent a critical mechanism for regulating malignant behaviors of tumors17. Our data show that the m6A level was upregulated in gastric cancers and suppressing m6A by METTL3-knockdown hindered tumor growth and metastasis in vitro and in vivo. Based on these discoveries, we believe that abnormal m6A modification is an important epigenetic feature of gastric cancer and a potential therapeutic target for further study.
As the predominant “writer” of m6A, METTL3 is dysregulated and plays dual roles in cancers, coupled with different substrates and cell types18. It directly regulates transcription, translation, and RNA maturation of a broad range of oncogene and onco-suppressors, many of which are undiscovered, and the role of METTL3 cancers depends on orchestration of multiple effects18. In non-tumor cells, METTL3 methylated primary miRNAs, such as pri-let-7e and pri-miR221, and facilitated their maturation19. Recent studies reported that METTL3 accelerated the maturation of pri-miR221/22220 and pri-miR-2521 in bladder and pancreatic cancers. So far, there has been no study reported the regulating function of METTL3 on ncRNA in gastric cancer. As we found, one of the most prominent substrates of METTL3 is a non-coding primary miRNA, pri-miR-17-92, a 6-tandem stem hairpin-containing polycistronic transcript of the gene MIR17HG that encodes a miRNA cluster composed of 6 onco-miRNAs22. Through m6A modification, METTL3 facilitated pri-miR-17-92 to bind to DGCR8, which recognizes the stem-flanking junctions and orients DROSHA to cleave primary miRNA into precursor miRNA23,24. Our study revealed that the over-m6A modification on pri-miR-17-92, instead of its overexpression, caused upregulation of the miR-17-92 cluster and gastric cancer progression. To our knowledge, this is the first report of METTL3-mediated-m6A-dependent maturation of ncRNA and its downstream signal pathway and biological effect in gastric cancer. Most importantly, we mapped pri-miR-17-92 and found its A879 was the dominant site responsible for the m6A-mediated pri-miR-17-92 maturation because A879C mutation significantly abolished the METTL3-mediated m6A modification, DGCR8 binding, and miR-17-92 cluster formation. Therefore, the pri-miR-17-92 A879 represented the key mediator of the m6A-mediated pri-miR-17-92 maturation and was likely a highly precise target for intervention as an epigenetically modifiable position.
The regulation of the miR-17-92 cluster remains largely unclear, other than that c-Myc directly binds to MIR17HG and promotes pri-miR-17-92 transcription25. Our findings uncovered another layer of regulation of this miRNA cluster by m6A-mediated processing. Widely accepted as an onco-miRNA26, the miR-17-92 cluster targets many onco-suppressing genes, such as SMAD2 and SMAD427, p2128, and TRAF329. In this study, we found that TMEM127 and PTEN were part of the targets. Furthermore, forced expression of the miR-17-92 cluster counteracted the effects of METTL3-knockdown in PTEN/TMEM127 regulation, cell proliferation, and metastasis. TMEM127 is a negative regulator of the mTOR pathway and a tumor suppressor located on chromosome 2q1130. So far, the function of TMEM127 in gastric cancer and its association with the miR-17-92 cluster have not been reported. In this study, we did not observe frequent mutations or deletions of TMEM127. Instead, TMEM127, along with PTEN and the mTOR pathway, was efficiently regulated by the miR-17-92 cluster as its target. Therefore, we suspect that METTL3/miR-17-92 cluster activates mTOR pathways by targeting PTEN and TMEM127 in gastric cancer progression. Of note, several other signaling pathways, including p53, TGF-beta, and MAPK signaling pathways, were probably also involved, which are of value for further investigation in future studies.
Currently, drugs targeting epigenetic changes, such as DNA methyltransferase inhibitors decitabine, have become standard treatments in hematological malignancies31. Up to now, the small molecule inhibitor against METTL3 is not available. Based on the above discovery, the mTOR inhibitor everolimus was chosen to interfere with METTL3/miR-17-92 cluster/TMEM127 or PTEN/mTOR signaling pathway in gastric cancer. We found that everolimus indeed reversed the METTL3-induced proliferation in a dose-dependent manner. This effect was remarkably pronounced when METTL3 was highly expressed. The high sensitivity to everolimus in METTL3-high cells could be because the mTOR pathway was greatly activated by METTL3 in these cells. These data confirmed that the oncogenic role of METTL3 relies on mTOR activation in another aspect. Everolimus has shown promising efficacy in patients with previously treated advanced gastric cancer in a phase II study32. However, phase III studies33,34,35 failed to repeat the positive results, except for that the progression-free survival was significantly prolonged. Our findings provided evidence that the METTL3 level might be a potential predictor for treatment efficacy of everolimus in gastric cancer and may help screen possible advantageous subgroups out from the previous failed clinical trials with everolimus-treated gastric cancer. These inferences are yet to be further studied and confirmed in clinical trials.
In conclusion, METTL3/m6A promotes gastric cancer growth and metastasis by facilitating pri-miR-17-92 processing into the oncogenic miRNA cluster and activating the AKT/mTOR pathway by targeting PTEN and TMEM127, which could be targeted by everolimus (Fig. 8f). These findings provide us with a novel insight into the role of METTL3 in the regulation of cancer development and a theoretical rationale for use of everolimus in the treatment of m6A/METTL3-high gastric cancer.
Data availability
Expression data of mRNA and miRNA in TCGA-STAD were downloaded from the GDC data portal (https://portal.gdc.cancer.gov). Survival data from GEO databases, including GSE14210, GSE15459, GSE22377, GSE29272, GSE38749, GSE51105, and GSE62254, were extracted from Kaplan–Meier Plotter (http://kmplot.com). Genomic alternation data were extracted from cBioPortal (http://www.cbioportal.org). All the other data and materials are available from the corresponding author upon reasonable request.
References
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA 68, 394–424 (2018).
Van Cutsem, E., Sagaert, X., Topal, B., Haustermans, K. & Prenen, H. Gastric cancer. Lancet 388, 2654–2664 (2016).
Thomassen, I. et al. Peritoneal carcinomatosis of gastric origin: a population-based study on incidence, survival and risk factors. Int. J. Cancer 134, 622–628 (2014).
Bang, Y. J. et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 376, 687–697 (2010).
Ohtsu, A. et al. Bevacizumab in combination with chemotherapy as first-line therapy in advanced gastric cancer: a randomized, double-blind, placebo-controlled phase III study. J. Clin. Oncol. 29, 3968–3976 (2011).
Fuchs, C. S. et al. Safety and efficacy of pembrolizumab monotherapy in patients with previously treated advanced gastric and gastroesophageal junction cancer: phase 2 clinical KEYNOTE-059 trial. JAMA Oncol. 4, e180013 (2018).
Kang, Y. K. et al. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 390, 2461–2471 (2017).
Zhao, B. S., Roundtree, I. A. & He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18, 31–42 (2017).
Yang, D. et al. N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 46, 3906–3920 (2018).
Liu, N. & Pan, T. N6-methyladenosine-encoded epitranscriptomics. Nat. Struct. Mol. Biol. 23, 98–102 (2016).
Wang, Q. et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut, https://doi.org/10.1136/gutjnl-2019-319639 (2019).
Yang, D. D. et al. METTL3 promotes the progression of gastric cancer via targeting the MYC pathway. Front Oncol. 10, 115 (2020).
Zhang, D. et al. LIMD1 is a survival prognostic marker of gastric cancer and hinders tumor progression by suppressing activation of YAP1. Cancer Manag Res. 10, 4349–4361 (2018).
Olive, V. et al. A component of the mir-17-92 polycistronic oncomir promotes oncogene-dependent apoptosis. eLife 2, e00822 (2013).
Feinberg, A. P., Koldobskiy, M. A. & Gondor, A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat. Rev. Genet. 17, 284–299 (2016).
Heyn, H. et al. Epigenomic analysis detects aberrant super-enhancer DNA methylation in human cancer. Genome Biol. 17, 11 (2016).
Roundtree, I. A., Evans, M. E., Pan, T. & He, C. Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 (2017).
Zheng, W. et al. Multiple functions and mechanisms underlying the role of METTL3 in human cancers. Front Oncol. 9, 1403 (2019).
Alarcon, C. R., Lee, H., Goodarzi, H., Halberg, N. & Tavazoie, S. F. N6-methyladenosine marks primary microRNAs for processing. Nature 519, 482–485 (2015).
Han, J. et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer 18, 110 (2019).
Zhang, J. et al. Excessive miR-25-3p maturation via N(6)-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat. Commun. 10, 1858 (2019).
Ota, A. et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 64, 3087–3095 (2004).
Gregory, R. I. et al. The Microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240 (2004).
Denli, A. M., Tops, B. B., Plasterk, R. H., Ketting, R. F. & Hannon, G. J. Processing of primary microRNAs by the Microprocessor complex. Nature 432, 231–235 (2004).
O’Donnell, K. A., Wentzel, E. A., Zeller, K. I., Dang, C. V. & Mendell, J. T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435, 839–843 (2005).
Ventura, A. et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132, 875–886 (2008).
Mestdagh, P. et al. The miR-17-92 microRNA cluster regulates multiple components of the TGF-beta pathway in neuroblastoma. Mol. Cell 40, 762–773 (2010).
Fontana, L. et al. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PloS ONE 3, e2236 (2008).
Liu, F., Cheng, L., Xu, J., Guo, F. & Chen, W. miR-17-92 functions as an oncogene and modulates NF-kappaB signaling by targeting TRAF3 in MGC-803 human gastric cancer cells. Int. J. Oncol. 53, 2241–2257 (2018).
Qin, Y. et al. Germline mutations in TMEM127 confer susceptibility to pheochromocytoma. Nat. Genet. 42, 229–233 (2010).
Nie, J., Liu, L., Li, X. & Han, W. Decitabine, a new star in epigenetic therapy: the clinical application and biological mechanism in solid tumors. Cancer Lett. 354, 12–20 (2014).
Doi, T. et al. Multicenter phase II study of everolimus in patients with previously treated metastatic gastric cancer. J. Clin. Oncol. 28, 1904–1910 (2010).
Ohtsu, A. et al. Everolimus for previously treated advanced gastric cancer: results of the randomized, double-blind, phase III GRANITE-1 study. J. Clin. Oncol. 31, 3935–3943 (2013).
Lorenzen, S. et al. Phase III randomized, double-blind study of paclitaxel with and without everolimus in patients with advanced gastric or esophagogastric junction carcinoma who have progressed after therapy with a fluoropyrimidine/platinum-containing regimen (RADPAC). Int. J. Cancer, https://doi.org/10.1002/ijc.33025 (2020).
Shen, Y. C. et al. Phase II multicentered study of low-dose everolimus plus cisplatin and weekly 24-hour infusion of high-dose 5-fluorouracil and leucovorin as first-line treatment for patients with advanced gastric cancer. Oncology 87, 104–113 (2014).
Acknowledgements
The manuscript was edited by Alexandra H. Marshall (Marshall Medical Communications). This work was supported by the National Natural Science Foundation of China (81172487 to L.Liu, and 81500092 to S.Li.), Natural Science Foundation of Shandong Province (ZR201702180008 to L.Liu), and Chinese Postdoctoral Science Foundation (2019M652406 to S.Li.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by N. Barlev
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sun, Y., Li, S., Yu, W. et al. N6-methyladenosine-dependent pri-miR-17-92 maturation suppresses PTEN/TMEM127 and promotes sensitivity to everolimus in gastric cancer. Cell Death Dis 11, 836 (2020). https://doi.org/10.1038/s41419-020-03049-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-020-03049-w
- Springer Nature Limited
This article is cited by
-
The regulatory mechanism of m6A modification in gastric cancer
Discover Oncology (2024)
-
METTL14-dependent maturation of pri-miR-17 regulates mitochondrial homeostasis and induces chemoresistance in colorectal cancer
Cell Death & Disease (2023)
-
Role of N6-methyladenosine RNA modification in gastric cancer
Cell Death Discovery (2023)
-
Crosstalk Between m6A RNA Methylation and miRNA Biogenesis in Cancer: An Unholy Nexus
Molecular Biotechnology (2023)
-
METTL3 facilitates multiple myeloma tumorigenesis by enhancing YY1 stability and pri-microRNA-27 maturation in m6A-dependent manner
Cell Biology and Toxicology (2023)