Abstract
Dysregulation of glucose homeostasis contributes to insulin resistance and type 2 diabetes. Whilst exercise stimulated activation of AMP-activated protein kinase (AMPK), an important energy sensor, has been highlighted for its potential to promote insulin-stimulated glucose uptake, the underlying mechanisms for this remain largely unknown. Here we found that AMPK positively regulates the activation of Rab5, a small GTPase which is involved in regulating Glut4 translocation, in both myoblasts and skeletal muscles. We further verified that TBC1D17, identified as a potential interacting partner of Rab5 in our recent study, is a novel GTPase activating protein (GAP) of Rab5. TBC1D17-Rab5 axis regulates transport of Glut1, Glut4, and transferrin receptor. TBC1D17 interacts with Rab5 or AMPK via its TBC domain or N-terminal 1–306 region (N-Ter), respectively. Moreover, AMPK phosphorylates the Ser 168 residue of TBC1D17 which matches the predicted AMPK consensus motif. N-Ter of TBC1D17 acts as an inhibitory region by directly interacting with the TBC domain. Ser168 phosphorylation promotes intra-molecular interaction and therefore enhances the auto-inhibition of TBC1D17. Our findings reveal that TBC1D17 acts as a molecular bridge that links AMPK and Rab5 and delineate a previously unappreciated mechanism by which the activation of TBC/RabGAP is regulated.
Similar content being viewed by others
Introduction
Type 2 diabetes mellitus is an age-associated disease which affects more than 4 million people worldwide [1]. As a main feature of this disease, insulin resistance leads to dysregulation of glucose homeostasis [2,3,4]. Downregulation of glucose transporter 1 (Glut1) and impairment of insulin-responsive Glut4 translocation from specialized Glut4 storage vesicles (GSVs) to the cell surface membrane contribute to the development of insulin resistance [5,6,7,8]. In addition to insulin, exercise is another essential physiological stimulus for Glut4 translocation in skeletal muscles [9, 10]. The increased levels of AMP and ADP activate AMP-activated kinase (AMPK), the energy sensor in cells, which subsequently mediates the effect of exercise/muscle contraction on glucose transport [11,12,13,14]. Consequently, further understanding AMPK-mediated signaling could provide new insight into therapeutic targets for the treatment of insulin resistance associated with type 2 diabetes.
Rab5, a small GTPase, is an essential regulator for endosome fusion, transport, and biogenesis [15,16,17,18]. Accumulating evidences raise the notion that Rab5 is also critical for modulating metabolic events, including autophagy, and for the sorting of Glut4-containing vesicles [19,20,21,22]. This is supported by our in vivo studies and others reports showing that myogenic ablation of Rab5 suppresses AKT-mTOR activation [23] and that liver-specific knockdown of Rab5 results in metabolic abnormalities [24]. Ablation of APPL1 or PI3K-C2γ, two Rab5 effectors, consistently leads to insulin resistance in mice [25, 26]. However, the understanding of the upstream regulatory machinery for Rab5 activation and its involvement in modulating such metabolic factors remains elusive.
The activity of Rab family GTPases are specifically controlled by the Tre2-Bub2-Cdc16 (TBC) domain-containing GAPs (TBC/RabGAPs) [27]. Among the 40 TBC/RabGAP members, TBC1D1 and TBC1D4/AS160 are well characterized for their roles in negatively modulating Rab2, Rab8, Rab10, and Rab14 to inhibit GSVs trafficking [28,29,30]. AMPK can phosphorylate the two RabGAPs and suppress their activity [31, 32]. Given the presence of more than 40 members of the TBC/RabGAP family, other TBC/RabGAPs could also be involved in Glut4 transport. Here, we found that TBC1D17, previously identified to regulate Shiga toxin uptake and mitophagy [33, 34], is a novel GAP protein of Rab5. We show that AMPK-dependent phosphorylation strengthens the auto-inhibition of TBC1D17 by promoting its intra-molecular interaction. Our findings reveal a previously unappreciated AMPK-TBC1D17 axis that regulates the activation of Rab5 and glucose uptake.
Results
AMPK positively regulates Rab5 activation
To investigate the role of Rab5 in exercise-promoted Glut4 transport, we examined if AMPK regulates Rab5 activation by using the Rab5-binding domain (R5BD) of Rabaptin5, which specifically interacts with the active Rab5 [23]. Either AMPK activator aminoimidazole carboxamide ribonucleotide (AICAR) treatment or glucose-deprivation, both of which activate AMPK [35, 36], resulted in elevated levels of GTP-bound Rab5 in myoblasts (Fig. 1a, b). By contrast, AMPK inhibitor compound C inhibited Rab5 activation (Fig. 1c). Co-expression of AMPK catalytic α2 subunit, but not its kinase-dead mutant (K45R), increased Rab5 activation in myoblasts (Fig. 1d). We extended and validated our findings in mouse embryonic fibroblasts (MEFs) ablated both AMPKα1 and α2 isoforms (AMPK-DKO). Compared to wild-type MEFs, AMPK-DKO displayed much-reduced levels of active Rab5 (Fig. 1e).
a Immunoblot analysis of a Rab5 activation assay. Myoblasts transfected with HA-Rab5 were treated with DMSO or AICAR (2 mM) for 4 h. Lysates were incubated with GST-R5BD, or GST as a negative control, to pull-down GTP-bound Rab5. Data in the bar graphs represent the mean ± S.E. (error bars) values of the ratios of densities (active Rab5/total Rab5) for three independent experiments. **p < 0.01. b Myoblasts transfected with HA-Rab5 were deprived of glucose for 4 h. Lysates were incubated with GST control or GST-R5BD to pulldown GTP-bound Rab5. Data in the bar graphs represent the mean ± S.E. (error bars) values of the ratios of densities (active Rab5/total Rab5) for three independent experiments. *p < 0.05. c Myoblasts transfected with HA-Rab5 were treated with DMSO or Compound C (C. C) (20 µM) for 6 h. Lysates were incubated with GST control or GST-R5BD to pulldown GTP-bound Rab5. Data in the bar graphs represent the mean ± S.E. (error bars) values of the ratios of densities (active Rab5/total Rab5) for three independent experiments. **p < 0.01. d HA-Rab5 was co-transfected with FLAG vector, FLAG-AMPKα2, or FLAG-AMPKα2 K45R mutant in myoblasts. Lysates were incubated with GST-R5BD to pulldown GTP-bound Rab5. Data in the bar graphs represent the mean ± S.E. (error bars) values of the ratios of densities (active Rab5/total Rab5) for three independent experiments. *p < 0.05. e Wild-type (WT) or AMPKα1 and AMPKα2 double-knockout (DKO) MEFs were transfected with HA-Rab5. Lysates were incubated with GST control or GST-R5BD to pulldown GTP-bound Rab5. Data in the bar graphs represent the mean ± S.E. (error bars) values of the ratios of densities (active Rab5/total Rab5) for three independent experiments. **p < 0.01. f Effect of AMPK inactivation on the subcellular localization of Rab5. Myoblasts transfected with GFP-Rab5 were treated with DMSO or Compound C (C. C) (20 µM) for 6 h followed by confocal microscopy analysis for GFP-Rab5. Bar: 10 μm. Ratio of peripheral GFP-Rab5 to total GFP-Rab5 fluorescence intensity was quantified as described in Materials and Methods. ****p < 0.0001. g GFP-Rab5 was co-transfected with FLAG vector or FLAG-AMPKα2 K45R mutant in myoblasts followed by confocal microscopy analysis to examine the subcellular localization of GFP-Rab5. Bar: 10 μm. Ratio of peripheral GFP-Rab5 to total GFP-Rab5 fluorescence intensity was quantified as described in Materials and Methods. ***p < 0.001. h Lysates of extensor digitorum longus (EDL) muscles from rested or exercised mice (n = 4) were incubated with GST control or GST-R5BD to pulldown endogenous GTP-bound Rab5. The levels of active Rab5 were quantified. *p < 0.05. GST glutathione S-transferase, WCL whole cell lysate, GFP green fluorescent protein.
Rab5-positive endosomes move bidirectionally alone microtubules between the peripheral areas of the cytoplasm and the perinuclear region [37]. In control myoblasts, punctated Rab5 endosomal structures display a perinuclear fraction and a peripheral fraction (Fig. 1f). The ratio of peripheral to total signal of GFP-Rab5 fluorescence intensity was quantified as previously described [38]. Inhibition of AMPK by either Compound C treatment or co-expression AMPK K45R mutant led to juxtanuclear accumulation of Rab5 (Fig. 1f, g). Co-expression of AMPK K45R, but not treatment of Compound C, reduced cell size and suppressed cell protrusion (Supplementary Fig. S1). We further assessed Rab5 activity in both extensor digitorium longus (EDL) and tibialis anterior (TA) muscles from rested mice or after treadmill running. Exercise promoted the activation of Rab5, corresponding with the elevation of AMPK phosphorylation in exercised muscles (Fig. 1h, Supplementary Fig. S2). These findings demonstrate that AMPK positively regulates Rab5 activity.
TBC1D17 is a novel GAP protein of Rab5
Rab GTPases are positively or negatively regulated by GEF or GAP proteins respectively [29]. We considered that AMPK might indirectly regulate Rab5 activation via some unknown Rab5 regulators. Interestingly, our recent screening analysis had revealed two TBC/RabGAP proteins, TBC1D15 and TBC1D17, as potential interacting partners for Rab5 [23]. Previous screening also revealed that Rab5 binds TBC1D15 and TBC1D17 [39]. We wondered if these two TBC/RabGAPs regulate Rab5 activation. Ectopically transfected TBC1D17, but not TBC1D15 or TBC1D23, significantly reduced the levels of GTP-bound Rab5 in myoblasts (Fig. 2a). Moreover, the catalytic inactive TBC1D17 R381A mutant did not alter Rab5 activity (Fig. 2b). siRNA-mediated TBC1D17 depletion consistently elevated the levels of active Rab5 in myoblasts (Fig. 2c). Interestingly, the TBC domain of TBC1D17 displayed a stronger inhibitory effect on Rab5 activity than full-length TBC1D17 (Fig. 2d), indicating TBC1D17 auto-inhibition, possibly via the N-terminal region.
a HA-Rab5 was co-transfected with FLAG vector, FLAG-TBC1D15, FLAG-TBC1D17, or FLAG-TBC1D23 in myoblasts. Lysates were incubated with GST control or GST-R5BD to pulldown GTP-bound Rab5. Data in the bar graphs represent the mean ± S.E. (error bars) values of the ratios of densities (active Rab5/total Rab5) for three independent experiments. *p < 0.05. ns not significant. b HA-Rab5 was co-transfected with FLAG vector, FLAG-TBC1D17, or FLAG-TBC1D17 R381A mutant in myoblasts. Lysates were incubated with GST control or GST-R5BD to pulldown GTP-bound Rab5. Data in the bar graphs represent the mean ± S.E. (error bars) values of the ratios of densities (active Rab5/total Rab5) for three independent experiments. **p < 0.01. ns not significant. c Lysates of myoblasts transfected with control siRNA or TBC1D17 siRNA were incubated with GST control or GST-R5BD to pulldown GTP-bound Rab5. Data in the bar graphs represent the mean ± S.E. (error bars) values of the ratios of densities (active Rab5/total Rab5) for three independent experiments. *p < 0.05. d HA-Rab5 was co-transfected with FLAG vector, FLAG-TBC1D17, or FLAG-TBC domain of TBC1D17 in myoblasts. Lysates were incubated with GST control or GST-R5BD to pulldown GTP-bound Rab5. Data in the bar graphs represent the mean ± S.E. (error bars) values of the ratios of densities (active Rab5/total Rab5) for three independent experiments. **p < 0.01. ***p < 0.001. ****p < 0.0001. e Effect of TBC1D17 or TBC1D17 R381A mutant on GTP hydrolysis by Rab5 measured in GAP assays. Data in the bar graphs represent the mean ± S.E. (error bars) values for three independent experiments. *p < 0.05.**p < 0.01. f Time-dependent action of TBC1D17 or TBC1D17 R381A mutant on GTP hydrolysis by Rab5. Similar results were obtained in three different experiments. g Effect of TBC1D17 on the subcellular localization of Rab5. Myoblasts were co-transfected with GFP-Rab5 and FLAG vector, FLAG-TBC1D17, or FLAG-TBC1D17 R381A mutant followed by confocal microscopy analysis to examine the subcellular localization of Rab5. Bar: 10 μm. Ratio of peripheral GFP-Rab5 to total GFP-Rab5 fluorescence intensity was quantified as described in Materials and Methods. ***p < 0.001. ns not significant. h Effect of TBC GAP domain of TBC1D17 on the subcellular localization of Rab5. Myoblasts were co-transfected with GFP-Rab5 and FLAG vector, FLAG-TBC GAP domain, or FLAG-TBC R381A mutant followed by confocal microscopy analysis to examine the subcellular localization of Rab5. Bar: 10 μm. Ratio of peripheral GFP-Rab5 to total GFP-Rab5 fluorescence intensity was quantified as described in Materials and Methods. ****p < 0.0001. ns not significant. GST glutathione S-transferase, WCL whole cell lysate.
We further utilized an in vitro GAP assay to directly measure the ability of TBC1D17 to accelerate GTP hydrolysis by Rab5. TBC1D17, but not its R381A mutant, markedly enhanced GTP hydrolysis by Rab5 (Fig. 2e) in a time-dependent manner (Fig. 2f). We also examined the effect of TBC1D17, or its TBC domain, on Rab5 subcellular localization. Both TBC1D17 and its TBC domain, but not their R381A mutants, markedly promoted the perinuclear localization of Rab5 (Fig. 2g, h). Expression of the TBC domain, but not full-length TBC1D17, altered the size and shape of myoblasts (Supplementary Fig. S3). We concluded that TBC1D17 acts as a novel GAP for Rab5.
TBC1D17 physically associates with Rab5
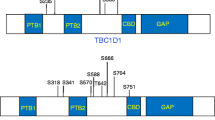
Though many TBC/RabGAPs directly bind to their substrates [27], the TBC1D17-Rab8 association is mediated by Optineurin [40]. To determine whether TBC1D17 directly binds Rab5, we purified recombinant FLAG-TBC1D17 and GST-Rab5 proteins for a pull-down assay. GST-Rab5, but not GST protein alone, directly interacted with TBC1D17 (Fig. 3a). To identify the regions within TBC1D17 that mediate its association to Rab5, a series of TBC1D17 mutants were generated and tested for their ability to interact with Rab5 in 293T cells (Fig. 3b). The TBC domain (307–542), but not the N terminus (1–306; N-Ter) or the C-terminal Proline-rich region (543–646), interacted with Rab5 (Fig. 3c).
a FLAG-TBC1D17 was transfected into 293T cell and immunoprecipitated with anti-FLAG-agarose beads followed by eluting with FLAG peptide. Control GST protein or GST-Rab5 fusion protein was incubated with purified FLAG-TBC1D17 for a direct interaction assay and then western blotted with FLAG antibody (top panel). The amounts of GST and GST-Rab5 used in this experiment were indicated by Coomassie blue staining (bottom panel). b Schematic diagram of TBC1D17 and its mutants. N-Ter N-terminal region, TBC Tre2/Bub2/Cdc16 domain, PR proline-rich region. c Lysates of 293T cells transfected with HA-Rab5 and FLAG-tagged TBC1D17 mutants as shown in (b) were immunoprecipitated with anti-FLAG beads and then western blotted with FLAG or HA antibodies. d Cell lysates of 293T cells transfected with FLAG-TBC1D17 were incubated with immobilized GST-Rab5 loaded with GDP or GTP-γS for GST Pull-down assay. The beads were washed and analyzed by western blot for FLAG (TBC1D17). The amounts of GST proteins were indicated by Coomassie blue staining. e HA-TBC1D17 and FLAG vector, FLAG-Rab5 (WT), FLAG-Rab5 Q79L (CA), or FLAG-Rab5 S34N (DN) were co-transfected into 293T cells for 24 h followed by co-immunoprecipitation analysis. f HA-Rab5 and FLAG-TBC1D17 or FLAG-TBC1D17 R381A mutant were co-transfected into 293T cells followed by co-immunoprecipitation analysis. GST glutathione S-transferase, WCL whole cell lysate, IP immunoprecipitation, CA constitutively active, DN dominant negative.
To determine if the Rab5-TBC1D17 interaction is dependent on the activation of Rab5, GST-Rab5 was unloaded or loaded with GTPγS (a nonhydrolyzable GTP analogue) or GDP, and incubated with lysates expressing TBC1D17. TBC1D17 preferentially bound to GDP-loaded GST-Rab5 (Fig. 3d). This is corresponded to the immunoprecipitation assay showing that TBC1D17 favorably interacted with dominant negative form of Rab5 (S34N) (Fig. 3e). Moreover, compared to wild-type TBC1D17, the inactive R381A mutant displayed weaker binding to Rab5 (Fig. 3f). These data indicated that the TBC1D17-Rab5 association is dependent on their respective activation statuses.
TBC1D17 is a novel regulator for glucose uptake
We observed that Rab5 colocalized with Glut4 vesicles while the Rab5 DN mutant induced perinuclear retention of Glut4 and reduced cell size in myoblasts (Fig. 4a, Supplementary Fig. S4). We further used an HA-Glut4-mCherry construct [41, 42] containing an intracellular mCherry tag fused to the COOH terminus of GLUT4 and an HA epitope in the first extracellular loop that is exposed on the cell surface when GLUT4 is inserted into the plasma membrane, to measure the membrane translocation of Glut4 in myoblasts as previously described [43]. The Rab5 DN mutant blocked Glut4 translocation (Supplementary Fig. S5a). Due to the low endogenous Glut4 Levels in myoblasts [44], we further measured glucose uptake and Glut4 translocation using differentiated C2C12 myotubes, which contained elevated Glut4 levels (Supplementary Fig. S5b) [45, 46], and displayed both insulin- and AICAR-dependent glucose uptake, based on the uptake of 2-deoxy-D-glucose (2-DG) (Supplementary Fig. S5c). Knockdown of Glut4 significantly impaired insulin-stimulated glucose uptake (Supplementary Fig. S5d). Next, we prepared the plasma membrane (PM) fraction and the post-PM fractions (supernatants, clear of PM) of serum-starved and insulin-stimulated myotubes [47]. By measuring the relative amounts of the Glut4 in the PM and the post-PM fractions we verified that AICAR enhanced insulin-stimulated Glut4 translocation in C2C12 myotubes (Supplementary Fig. S5e). The Rab5 DN mutant inhibited glucose uptake (Fig. 4b) and Glut4 translocation in C2C12 myotubes (Fig. 4c, Supplementary Fig. S5f), validating the positive role of Rab5 in regulating Glut4 translocation.
a C2C12 myoblasts transiently expressing GFP-Glut4 and FLAG vector, FLAG-Rab5, or FLAG-Rab5 S34N mutant were processed for immunostaining and confocal microscopy analysis to examine the subcellular localization of Glut4. Bar: 10 μm. b Myotubes expressing FLAG vector or FLAG-Rab5 S34N mutant were serum deprived for 4 h including incubation periods with insulin for 20 min prior to 2-DG uptake assay as described in Materials and Methods. Illustrations represent the means ± S.E. of six independent experiments performed in triplicate; **p < 0.01. ***p < 0.001. The expression levels of FLAG-Rab5 S34N mutant were determined by western blot analysis. c Myotubes expressing FLAG vector or FLAG-Rab5 S34N mutant were serum deprived for 4 h including incubation periods with insulin for 20 min prior to plasma membrane (PM) fraction as described in Materials and Methods. PM fractions were subjected to western blot analysis for examining the expression levels of Glut4. Illustrations represent the means ± S.E. of three independent experiments; **p < 0.01. d Lysates of indicated mouse tissues/organs were subjected to western blot analysis for examining the expression levels of TBC1D17. e GFP-Glut4 and HA-Rab5 were co-transfected with FLAG vector, FLAG-TBC1D17, or FLAG-TBC domain into C2C12 myoblasts in the indicated combination for 24 h followed by immunofluorescence analysis. Nuclei were visualized by Hoechst staining. Bar: 10 μm. f HA-Glut4-mCherry were co-transfected with GFP vector, GFP-TBC1D17, or GFP-TBC1D17 R381A mutant into C2C12 myoblasts for 24 h and processed for cell-surface HA-GLUT4-mCherry levels as described in Materials and Methods. Shown are mean ± SE fold changes from three experiments. ***P < 0.001. ****P < 0.0001. Bar: 10 μm. g Myotubes expressing FLAG vector, FLAG-TBC1D17, or FLAG-TBC1D17 R381A mutant were serum deprived for 4 h including incubation periods with insulin for 20 min prior to plasma membrane fraction. PM fractions were subjected to western blot analysis for Glut4. Illustrations represent the means ± S.E. of three independent experiments; *p < 0.05. **p < 0.01. ns not significant. The expression levels of FLAG-tagged proteins were determined by western blot analysis. h Myotubes expressing FLAG vector, FLAG-TBC1D17, or FLAG-TBC1D17 R381A mutant were serum deprived for 4 h including incubation periods with insulin for 20 min prior to 2-DG uptake assay as described in Materials and Methods. Illustrations represent the means ± S.E. of six independent experiments performed in triplicate; *p < 0.05. ***p < 0.001. ns not significant. GFP green fluorescent protein, PM plasma membrane. The expression levels of FLAG-tagged proteins were determined by western blot analysis.
Interestingly, TBC1D17 is highly expressed in both liver and skeletal muscle (Fig. 4d) (both being notably insulin sensitive), suggesting its involvement in regulating glucose signaling. Accumulating evidence demonstrates that both TBC1D1 and TBC1D4 regulate the abundance of Glut4 [48,49,50,51]. However, ectopic expression of either TBC1D17 or Rab5 had little effect on the expression levels of Glut4 (Supplementary Fig. S6a). Consistently, Glut4 abundance remains intact in TBC1D17- or Rab5-depleted myotubes (Supplementary Fig. S6b,c). Given the negative effects of TBC1D17 on Rab5 activation, we postulated that TBC1D17 might act as an inhibitor for glucose uptake. This was supported by the finding that the colocalization between Rab5 and Glut4 or Glut1 vesicles was impaired by TBC1D17 or its TBC domain (Fig. 4e; Supplementary Fig. S7a, b, c). Indeed, both TBC1D17 or its TBC GAP domain, but not their inactive R381A mutants, led to juxtanuclear accumulation of Glut4 in C2C12 myoblasts (Supplementary Fig. S7d, e). Moreover, TBC1D17 reduced the cell surface levels of HA-Glut4-mCherry in myoblasts (Fig. 4f) and suppressed Glut4 plasma membrane translocation and glucose uptake in differentiated C2C12 myotubes, while the R381A mutant has little effects (Fig. 4g,h).
Since Rab5 plays pivotal roles in regulating general endocytic recycling, we examined if TBC1D17 also modulates transporting of Glut1 and transferrin receptor (TfR). Co-expressing of Rab5 DN mutant or TBC1D17, but not TBC1D17 R381A mutant, induced perinuclear localization of TfR (Supplementary Fig. S8a,b). We incubated myoblasts with fluorescently labeled transferrin to label TfR, which is taken up by receptor-mediated endocytosis and recycled back to the plasma membrane. Consistently, Rab5 DN mutant and TBC1D17 inhibited recycling of TfR (Supplementary Fig. S8c,d). Both membrane fraction assay (Supplementary Fig. S8e,f) and immunostaining (Supplementary Fig. S8g,h) demonstrated that Rab5 DN mutant or TBC1D17, but not R381A mutant, reduced the plasma membrane localization of Glut1. These findings suggest that TBC1D17-Rab5 axis is involved in modulating divergent cellular functions.
AMPK interacts with and phosphorylates TBC1D17
Both AMPKα1 and α2 subunits associated with TBC1D17 through the N-terminal kinase domain (Fig. 5a, b, c). We aimed to determine whether AMPK regulates Rab5 activity in a TBC1D17-dependent manner. The reduction of GTP-bound Rab5 levels by TBC1D17 was prevented by the glucose metabolism inhibitor 2-deoxy-D-glucose (2-DG)-induced AMPK activation (Fig. 5d, lane 3 versus 4), while Compound C treatment restored the reduction of Rab5 activation (Fig. 5d, lane 4 versus 5). These findings indicated that AMPK modulates Rab5 activation through TBC1D17.
a Lysates of 293T cells transfected with HA-TBC1D17 and FLAG vector, FLAG-AMPKα1, or FLAG-AMPKα2 were immunoprecipitated with anti-FLAG beads and then western blotted with FLAG or HA antibodies. b Schematic diagram of AMPKα2 and its mutants. KD kinase domain, AID auto-inhibition sequence, CTD C-terminal regulatory domain. c Lysates of 293T cells transfected with HA-TBC1D17 and FLAG-tagged AMPK mutants, as shown in (b), were immunoprecipitated with anti-FLAG beads and then western blotted with FLAG or HA antibodies. d C2C12 myoblasts expressing HA-Rab5 and FLAG vector or FLAG-TBC1D17 were treated with 2-DG (20 mM) in the presence or absence of compound C (C. C) (20 µM). Lysates were incubated with GST control or GST-R5BD to pulldown GTP-bound Rab5. Data in the bar graphs represent the mean ± S.E. (error bars) values of the ratios of densities (active Rab5/total Rab5) for three independent experiments. *p < 0.05. e Lysates of 293T cells transfected with HA-AMPKα2 and FLAG vector, FLAG-1–306 of TBC1D17 (N-Ter), or FLAG-TBC domain were immunoprecipitated with anti-FLAG beads and then western blotted with FLAG or HA antibodies. f The amino acid sequence of TBC1D17 was analyzed using GPS 3.0 to search for potential AMPK phosphorylation site (http://gps.biocuckoo.org/online.php). The alignment of the sequence surrounding S168 in TBC1D17 is shown with the analogous sites in selective AMPK substrate proteins. g 293T cells were transfected with control FLAG vector or FLAG-TBC1D17 and treated with AICAR (2 mM). TBC1D17 was immunoprecipitated using FLAG antibody and the precipitate was analyzed on a protein blot using AMPK phosphorylation-specific substrate antibody (p-Sub/AMPK) and FLAG antibody. Densitometric quantification of p-Sub/AMPK levels from immunoblots are shown as the means ± S.E. from three independent experiments. **p < 0.01. h 293T cells were transfected with FLAG vector or FLAG-TBC1D17 followed by glucose deprivation. TBC1D17 was immunoprecipitated and was analyzed using p-Sub/AMPK antibody. Densitometric quantification of p-Sub/AMPK levels from immunoblots are shown as the means ± S.E. from three independent experiments. **p < 0.01. i 293T cells were co-transfected with FLAG vector or FLAG-TBC1D17 and HA-AMPKα2 or HA-AMPKα2 K45R mutant. TBC1D17 was immunoprecipitated and was analyzed using p-Sub/AMPK antibody. Densitometric quantification of p-Sub/AMPK levels from immunoblots are shown as the means ± S.E. from three independent experiments. **p < 0.01, ***p < 0.001. j In vitro phosphorylation of TBC1D17 by AMPK. Wild-type or mutant AMPK protein was purified from 293T cells expressing FLAG-AMPKα2 or FLAG-AMPKα2 K45R. Phosphorylation of GST-TBC1D17 was determined by immunoblotting with p-Sub/AMPK antibody. Densitometric quantification of p-Sub/AMPK levels from immunoblots are shown as the means ± S.E. from three independent experiments. *p < 0.05. **p < 0.01. ns not significant. GST glutathione S-transferase, WCL whole cell lysate, IP immunoprecipitation.
AMPK interacts with the 1–306 region (N-Ter), but not the TBC domain, of TBC1D17 (Fig. 5e). Bioinformatic analysis revealed a serine 168 residue matching the predicted AMPK consensus motif [52, 53] in the N-Ter of TBC1D17 (Fig. 5f; Supplementary Fig. S9a), but not in the homologous region of TBC1D15 (Supplementary Fig. S9b). We therefore proposed that AMPK might phosphorylate TBC1D17 to regulate its GAP activity towards Rab5. Using an AMPK phospho-substrate specific antibody (p-Sub/AMPK), we found that both AICAR treatment and glucose deprivation elevated TBC1D17 phosphorylation (Fig. 5g, h). Moreover, co-expression of wild-type AMPK, but not the K45R mutant, increased TBC1D17 phosphorylation (Fig. 5i). An in vitro kinase assay using purified AMPK and recombinant GST-TBC1D17 further validated phosphorylation of TBC1D17 by AMPK (Fig. 5j). All results indicated that TBC1D17 is a direct AMPK substrate.
AMPK-mediated Ser168 phosphorylation attenuates the GAP activity of TBC1D17
To recognize the site(s) on TBC1D17 phosphorylated by AMPK, a mutant was constructed where the Ser 168 residue was changed to alanine. TBC1D17 phosphorylation upon glucose deprivation was abolished by the serine-to-alanine replacement at the Ser168 residue (Fig. 6a). An in vitro kinase assay verified that AMPK directly phosphorylates TBC1D17 on Ser 168 (Fig. 6b). We further generated a phospho-TBC1D17 (Ser 168) specific antibody, which only recognized the phosphorylated peptide (Fig. 6c). Using this antibody, we showed that co-expressing wild-type AMPK, but not the K45R mutant, elevated Ser168 phosphorylation levels (Fig. 6d). Co-expression of AMPK or AICAR treatment consistently increased Ser 168 phosphorylation of wild-type TBC1D17, but not of the S168A mutant (Fig. 6e,f). Collectively, this indicates that Ser168 is a main energy stress response phosphorylation site by AMPK.
a C2C12 myoblasts transfected with FLAG-TBC1D17 or FLAG-TBC1D17 S168A mutant were deprived of glucose for 4 h followed by immunoprecipitation and western blot using p-Sub/AMPK antibody and FLAG antibody. Densitometric quantification of p-Sub/AMPK levels from immunoblots are shown as the means ± S.E. from three independent experiments. **p < 0.01. b In vitro phosphorylation of TBC1D17 or TBC1D17 S168A by AMPK. AMPK protein was purified from 293T cells expressing FLAG-AMPKα2. Phosphorylation of GST-TBC1D17 or TBC1D17 S168A was determined by immunoblotting with p-Sub/AMPK antibody. Densitometric quantification of p-Sub/AMPK levels from immunoblots are shown as the means ± S.E. from three independent experiments. **p < 0.01. c Characterization of the anti-phospho-TBC1D17(Ser168) specific antibody. Different amounts of phospho-S168 peptide or unmodified peptide were spotted on nitrocellulose membranes, and the specificity of the antibody against the phosphorylated Ser 168 residue was determined by dot blot assay. d C2C12 myoblasts were co-transfected with FLAG-TBC1D17 and HA-AMPKα2 subunit or HA-AMPK α2 K45R mutant for 24 h followed by immunoprecipitation and western blot using pTBC1D17(Ser168) antibody and FLAG antibody. The ratios of densities (pTBC1D17 Ser168/total TBC1D17) from three independent experiments were quantified. ***p < 0.001. e C2C12 myoblasts were co-transfected with HA-AMPKα2 subunit and FLAG-TBC1D17 or FLAG-TBC1D17 S168A mutant for 24 h followed by immunoprecipitation and western blot using pTBC1D17(Ser168) antibody and FLAG antibody. The ratios of densities (pTBC1D17 Ser168/total TBC1D17) from three independent experiments were quantified. ***p < 0.001. f C2C12 myoblasts transfected with FLAG-TBC1D17 or FLAG-TBC1D17 S168A mutant were treated with AICAR for 4 h followed by immunoprecipitation and western blot using pTBC1D17(Ser168) antibody and FLAG antibody. The ratios of densities (pTBC1D17 Ser168/total TBC1D17) from three independent experiments were quantified. **p < 0.01. g 293T cells were co-transfected with HA-Rab5 and control FLAG vector, FLAG-TBC1D17 (WT) or FLAG-TBC1D17 S168A mutant (S168A) and treated with AICAR (2 mM). Lysates were incubated with GST control or GST-R5BD to pulldown GTP-bound Rab5. Data in the bar graphs represent the mean ± S.E. (error bars) values of the ratios of densities (active Rab5/total Rab5) for three independent experiments. *p < 0.05. **p < 0.01. ****p < 0.0001. h 293T cells were transfected with FLAG-TBC1D17 or FLAG-TBC1D17 S168A mutant and treated with AICAR (2 mM) for 4 h. Effect of purified TBC1D17 or TBC1D17 S168A mutant on GTP hydrolysis by Rab5 was measured in GAP assays. Data in the bar graphs represent the mean ± S.E. (error bars) values for three independent experiments. *p < 0.05.**p < 0.01. ns not significant. i Myotubes expressing FLAG vector or FLAG-Rab5 were serum deprived for 4 h including incubation periods with C.C for 2 h and insulin for 20 min prior to 2-DG uptake assay. Illustrations represent the means ± S.E. of six independent experiments performed in triplicate; *p < 0.05. **p < 0.01. ***p < 0.001. The expression levels of FLAG-Rab5 were determined by western blot analysis. j C2C12 myoblasts were co-transfected with GFP-Glut4 and control FLAG vector, FLAG-TBC1D17 or FLAG-TBC1D17 S168A mutant and treated with DMSO or AICAR (2 mM). Ratio of peripheral GFP-Glut4 to total GFP-Glut4 fluorescence intensity was quantified as described in Materials and Methods. *p < 0.05. **p < 0.01. k Myotubes expressing FLAG vector, FLAG-TBC1D17, or FLAG-TBC1D17 S168A mutant were serum deprived for 4 h including incubation periods with AICAR for 2 h and insulin for 20 min prior to plasma membrane fraction. PM fractions were subjected to western blot analysis for Glut4. Illustrations represent the means ± S.E. of three independent experiments; *p < 0.05. ns not significant. The expression levels of FLAG-tagged proteins were determined by western blot analysis. l Myotubes expressing FLAG vector, FLAG-TBC1D17, or FLAG-TBC1D17 S168A mutant were serum deprived for 4 h including incubation periods with AICAR for 2 h and insulin for 20 min prior to 2-DG uptake assay. Illustrations represent the means ± S.E. of six independent experiments performed in triplicate; *p < 0.05. **p < 0.01. ***p < 0.001. ****p < 0.0001. GST glutathione S-transferase, WCL whole cell lysate, IP immunoprecipitation, GFP green fluorescence protein, PM plasma membrane.
We next investigated whether the phosphorylation of TBC1D17 at Ser168 modulates its GAP activity toward Rab5. GAP activity of TBC1D17 was inhibited by AICAR treatment as evidenced by significantly elevated GTP-bound Rab5 in myoblasts (Fig. 6g; lane 4 versus 5). By contrast, AICAR failed to suppress GAP activity of the TBC1D17 S168A mutant (Fig. 6g; lane 5 versus 6). We further performed in vitro GAP assays using FLAG-TBC1D17 or S168A mutant purified from DMSO- or AICAR-treated cells. AICAR treatment inhibited GAP activity of wild-type TBC1D17, but not of its S168A mutant (Fig. 6h). Given the above findings, we reasoned that AMPK-mediated phosphorylation of TBC1D17 at Ser168 reduced its GAP activity towards Rab5.
We further proposed that AMPK-dependent phosphorylation modulates the functions of the TBC1D17-Rab5 axis in Glut4 translocation and glucose uptake, as supported by the finding that Compound C-mediated AMPK inhibition suppressed Rab5-dependent glucose uptake in C2C12 myotubes (Fig. 6i). Activation of AMPK by AICAR treatment also diminished TBC1D17-mediated Glut4 perinuclear accumulation (Fig. 6j). Conversely, TBC1D17 S168A, which could not be phosphorylated by AMPK, still induced juxtanuclear accumulation of Glut4 in the presence of AICAR (Fig. 6j). Glucose deprivation could attenuate Glut4 accumulation in myoblasts expressing TBC1D17 wild type, but not in those expressing the TBC1D17 S168A mutant (Supplementary Fig. S10a). Moreover, AICAR treatment elevated the cell surface levels of HA-Glut4-mCherry in myoblasts expressing GFP-TBC1D17, but not TBC1D17 S168A mutant (Supplementary Fig. S10b). We further verified AICAR promotion of Glut4 located in the PM fraction in myotubes expressing TBC1D17, in contrast to those expressing the TBC1D17 S168A mutant (Fig. 6k). AICAR could suppress the negative effect of TBC1D17 wild type, but not the TBC1D17 S168A mutant, on glucose uptake in C2C12 myotubes (Fig. 6l). We thus concluded that AMPK-dependent Ser168 phosphorylation inhibits the GAP activity of TBC1D17.
The N terminus of TBC1D17 plays auto-inhibitory roles by interacting with the TBC GAP domain
We next explored the mechanism by which the Ser168 phosphorylation modulates the GAP activity of TBC1D17. Given that the TBC domain displayed stronger GAP activity than the full-length TBC1D17 (Fig. 2d), we proposed that auto-inhibition of TBC1D17 must occur and be strengthen by AMPK-mediated phosphorylation. To verify these hypotheses, we first investigated if the N-terminal 1–306 region (N-Ter) of TBC1D17 exerts an inhibitory effect on the TBC domain. When co-expressed with N-Ter, the GAP activity of TBC domain was reduced, as demonstrated by the R5BD assay (Fig. 7a; lane 4 versus 3). Similarly, the interaction between TBC domain and its substrate, Rab5, was impaired by co-expressing N-Ter (Fig. 7b; lane 3 versus 2). Moreover, expression of TBC domain led to juxtanuclear localization of Rab5 and altered cell size and shape, while co-expressing of N-Ter restored the cell peripheral fraction of Rab5 and rescued cell size and shape (Fig. 7c, Supplementary Fig. S11). These results support the hypothesis that the N-Ter acts as an inhibitory region to the TBC domain.
a HA vector or HA-TBC domain was co-transfected with FLAG vector or FLAG-N-Ter region in the indicated combination in 293T cells. Lysates were incubated with GST control or GST-R5BD to pulldown GTP-bound Rab5. Data in the bar graphs represent the mean ± S.E. (error bars) values of the ratios of densities (active Rab5/total Rab5) for three independent experiments. *p < 0.05. **p < 0.01. b FLAG vector, FLAG-TBC domain, or HA-N-Ter were co-transfected with HA-Rab5 into 293T cells in the indicated combination followed by immunoprecipitation analysis. c GFP-Rab5 were co-transfected with FLAG vector, FLAG-TBC domain, or HA-N-Ter into C2C12 myoblasts for 24 h followed by immunofluorescence analysis. Nuclei were visualized by Hoechst staining. Bar: 20 μm. Ratio of peripheral GFP-Rab5 to total GFP-Rab5 fluorescence intensity was quantified. ****p < 0.0001. d HA-N-Ter was co-transfected with FLAG vector, FLAG-TBC domain, or FLAG-N-Ter in 293T cells followed by immunoprecipitation analysis. e Control GST protein or GST-N-Ter fusion protein was incubated with purified FLAG-TBC for direct Pull-down assay and then western blotted with FLAG antibody (top panel). The amounts of GST proteins were indicated by the Coomassie blue staining (bottom panel). f HA-TBC was co-transfected with FLAG vector or FLAG-N-Ter in the absence or presence of AICAR (2 mM) for 4 h followed by immunoprecipitation analysis. g HA-TBC was co-transfected with FLAG vector or FLAG-N-Ter in the absence or presence of Compound C (20 µM) for 6 h followed by immunoprecipitation analysis.
Intra-molecular interaction between functional domains and inhibitory elements acts as a common mechanism for auto-inhibition [54, 55]. We examined if N-Ter physically interacts with the TBC domain. Robust co-immunoprecipitation of the TBC domain with the N-Ter region (Fig. 7d) was noted with GST-N-Ter directly interacting with the purified recombinant TBC domain (Fig. 7e). Importantly, manipulating AMPK activation by AICAR or Compound C treatment led to enhancement or reduction of TBC domain-N-Ter region interaction, respectively (Fig. 7f, g), indicating pivotal roles for AMPK in modulating the intra-molecular interaction of TBC1D17.
Ser 168 phosphorylation enhance TBC1D17 auto-inhibition by promoting its intra-molecular interaction
To investigate whether Ser168 phosphorylation modulates the N-Ter-TBC interaction and thereby strengthens auto-inhibition, we compared the TBC-binding activity of the N-Ter and its S168A mutant. The N-Ter S168A mutant showed a substantial reduction in TBC-binding activity (Fig. 8a). Furthermore, glucose deprivation elevated the association of TBC domain to wild-type N-Ter, but not to the N-Ter S168A mutant (Fig. 8b; lane 3 versus 5), indicating that the Ser 168 phosphorylation modulates the TBC-N-Ter association. Notably, the TBC domain reduced the active Rab5 levels (Fig. 8c, lane 3 versus lane 2), while wild-type N-Ter robustly restored Rab5 activation (Fig. 8c, lane 5 versus lane 3). In contrast, the N-Ter S168A mutant displayed reduced interaction ability (Fig. 8a) to, and loss of the inhibitory effect (Fig. 8c, lane 4 versus lane 5) upon, TBC domain. Only the wild-type N-Ter region, but not the S168A mutant, rescued the peripheral localization of Rab5 in TBC domain-expressing myoblasts (Supplementary Fig. S12a). These results suggest that AMPK-mediated Ser 168 phosphorylation impairs GAP activity of TBC1D17 by enhancing the association between N-Ter and the TBC domain. We also evaluated the effect of TBC1D17 auto-inhibition on Glut4 translocation. Co-expressing N-Ter, but not the N-Ter S168A mutant, suppressed the TBC domain-induced juxtanuclear accumulation of GFP-Glut4 in myoblasts (Fig. 8d). The inhibitory effects of TBC domain on Glut4 plasma membrane localization and cell size were similarly suppressed by N-Ter, but not the N-Ter S168A mutant, in C2C12 myotubes (Fig. 8e; Supplementary Fig. S12b). These findings favor a model that AMPK-mediates Ser168 phosphorylation promoting the auto-inhibition of TBC1D17 to activate Rab5 (Fig. 8f).
a HA-TBC was co-transfected with FLAG vector, FLAG-N-Ter wild-type, or FLAG-N-Ter S168A in 293T cells followed by immunoprecipitation analysis. b HA-TBC was co-transfected with FLAG vector, FLAG-N-Ter wild-type, or FLAG-N-Ter S168A in 293T cells followed by glucose deprivation and immunoprecipitation analysis. c HA vector, HA-N-Ter, or HA-N-Ter S168A was co-transfected with FLAG vector or FLAG-TBC domain in the indicated combination in 293T cells. Lysates were incubated with GST control or GST-R5BD to pulldown GTP-bound Rab5. Data in the bar graphs represent the mean ± S.E. (error bars) values of the ratios of densities (active Rab5/total Rab5) for three independent experiments. *p < 0.05. **p < 0.01. d GFP-Glut4 were co-transfected with FLAG vector, FLAG-TBC domain, HA vector, HA-N-Ter, or HA-N-Ter S168A mutant in the indicated combination for 24 h followed by immunofluorescence analysis. Nuclei were visualized by Hoechst staining. Bar: 20 μm. Ratio of peripheral GFP-Glut4 to total GFP-Glut4 fluorescence intensity was quantified as described in Materials and Methods. **p < 0.01. ***p < 0.001. ns not significant. e Myotubes co-expressing FLAG-TBC domain and HA-N-Ter or HA-N-Ter S168A mutant were serum deprived for 4 h including incubation periods with insulin for 20 min prior to plasma membrane fraction. PM fractions were subjected to western blot analysis for Glut4. Illustrations represent the means ± S.E. of three independent experiments; *p < 0.05. **p < 0.01. The expression levels of FLAG-tagged and HA-tagged proteins were determined by western blot analysis. f Schematic model depicting the regulatory mechanism for glucose uptake by AMPK-TBC1D17-Rab5 axis. In sedentary state, AMPK is inactivated while TBC1D17 inhibits Rab5 activation. During exercise, AMPK is activated and phosphorylates TBC1D17 on the Ser168 residue (Step 1). AMPK-mediated phosphorylation enhances the intra-molecular interaction of TBC1D17 to promote its auto-inhibition (Step 2). N-Ter-TBC domain association-mediated auto-inhibition thus suppresses the GAP activity of TBC1D17 and subsequently elevates Rab5 activation and enhances glucose uptake (Step 3).
Discussion
In addition to promoting insulin-stimulated glucose uptake [20, 22, 56], Rab5 is also involved in adiponectin-triggered Glut4 translocation [21, 26]. Moreover, Rab5 has been found to cause Glut4 retention in chronic hyperinsulinemia-induced cell models [57], indicating that Rab5 modulates Glut4 translocation in a context-dependent manner. Exercise-activated AMPK also drives Glut4 transport [5, 58]. Prior exercise can induce a subsequent elevation in insulin-stimulated glucose transport in the muscle, but the underlying mechanism of this remains elusive [59]. Here we found that AMPK inhibits the GAP activity of TBC1D17 towards Rab5, suggesting that TBC1D17 acts as a molecular bridge that links AMPK and Rab5-mediated glucose uptake (Fig. 8f). In addition, a previous report disclosed that insulin activates Rab5-dependent Glut4 translocation through stimulating GAPEX-5, a VPS9 domain-containing Rab5 GEF [22]. These two studies suggest that insulin and exercise signaling converge at the level of Rab5 to regulate glucose homeostasis, by activating Rab5 GEF (GAPEX-5) and suppressing Rab5 GAP (TBC1D17), respectively (Supplementary Fig. S13). Given the essential roles of Rab5 in general endocytic recycling, the TBC1D17-Rab5 axis is not limited to regulating Glut4 translocation. This notion is supported by our findings that both TBC1D17 and Rab5 DN mutant is also involved in regulating the positioning of TfR and Glut1 (Supplementary Fig. S8). It would also be of interest to explore the roles of AMPK-TBC1D17-Rab5 signaling in other physiological events.
Given the pivotal roles of Rab5 in regulating not only endosome trafficking and biogenesis [15, 16, 60], but also cell metabolism, migration, and the cell cycle [20, 23, 24], it is conceivable that the activity of Rab5 needs to be tightly regulated. A series of TBC/RabGAPs have been identified as upstream modulators for Rab5. As the first identified Rab5GAP, RN-tre regulates EGFR internalization and focal adhesion turnover [61]. Moreover, both RN-tre and TBC1D3 act on Rab5 to regulate macropinocytosis [62, 63]. Another Rab5 GAP, RabGAP5/RUTBC3, regulates endosome trafficking [33, 64]. In Caenorhabditis elegans, Rab5 is governed by TBC-2 during phagosome maturation [65]. Our recent screening revealed that both TBC1D15 and TBC1D17 were potential interaction partners for Rab5 [23], while only TBC1D17 elicits GAP activity toward Rab5 (Fig. 2). Thus, ours and others findings clearly demonstrate that the diverse cellular functions of Rab5 are modulated by distinct TBC/RabGAPs.
Most TBC/RabGAPs preferentially interact to GTP-bound Rabs [33, 66]. However, other interaction patterns between TBC/RabGAPs and their substrates do exit. For example, TBC1D5 co-immunoprecipitated only wild-type Rab7b, but not its CA or DN mutant [67]. Moreover, the wild-type and mutated Rab26 display similar binding affinities towards TBC1D6 [68]. Here, we show that TBC1D17 preferentially associates with the GDP-bound form of Rab5 (Fig. 3d) and thus demonstrate another unique interaction profile between TBC/RabGAP and Rab GTPase. Besides acting as a potent inhibitor of TBC GAP domain in cis (Fig. 7a), the N-Ter of TBC1D17 also modulates the TBC domain-Rab5 association (Fig.7b). This raises the possibility that the unique TBC1D17-Rab5 interaction pattern might be due to the presence of the N-Terminal region. This hypothesis awaits confirmation.
AMPK has long been recognized as a potential therapeutic target of metabolic diseases [32, 69]. As a commonly used AMPK agonist, metformin is a widely prescribed drug for Type 2 diabetes [70, 71]. AMPK regulates Glut4 translocation through both TBC1D1 and TBC1D4/As160 [11, 47, 72, 73]. Though recent indications show that AMPK regulates the association of TBC1D1 to IRAP [74], the molecular basis by which AMPK-mediated phosphorylation modulates TBC/RabGAPs remains largely unknown. Our study sheds light into this issue by pinpointing AMPK-mediated Ser168 phosphorylates (Fig. 5) acts as a regulatory signaling to TBC1D17 by strengthening the intra-molecular interaction of TBC1D17 (Fig. 8). Whether similar regulatory strategies are employed by AMPK to modulate TBC1D1 and TBC1D4 activation requires further exploration. Interestingly, compared with TBC1D1, TBC1D17 displays a different AMPK interaction pattern. TBC1D1 only interacts with AMPK α1, but not α2 [75], while TBC1D17 binds both isoforms (Fig. 5a). Moreover, TBC1D1 interacts with AMPK through two N-terminal PTB domains [75], which are not present in TBC1D17. Nevertheless, our study proposes that the TBC1D17-Rab5 axis might be a promising target applicable to both metabolic diseases and aging.
Materials and methods
Antibody and reagent
Antibodies from Cell Signaling Technology were as follows: AMPKα (2532), Phospho-AMPKα (Thr172) (2535S), Phospho-AMPK Substrate Motif (5759S), Rab5 (3547). Other antibodies were as follows: anti-FLAG epitope (Sigma-Aldrich, F7425), anti-HA epitope (Invitrogen, 715500), GFP (Santa Cruz Biotechnology, sc-9996), anti-Tubulin (HuaAn Biotechnology, M1305-2), anti-Actin (HuaAn Biotechnology, M1210-2), anti-TBC1D17 (Novusbio, NBP2-49546), anti-Glut1 (abcam, ab15309), anti-Glut4 (Santa Cruz Biotechnology, sc-53566). Chemical reagents used were as follows: AICAR (Topscience, T1477), Compound C (Topscience, T1977), 2-deoxy-D-Glucose (Cayman Chemical, 14325), Phalloidin (Sigma-Aldrich, P1951), Hoechst (Invitrogen, H1399), 488-CF-Dye-transferrin (Biotium, #00081).
Construction of expression plasmids
Full-length cDNAs of TBC1D17, TBC1D15, TBC1D23, Rab5, Glut1, Glut4, or mutant proteins were cloned into a hemagglutinin (HA)-tagged, GFP-tagged, or FLAG-tagged pXJ40 expression vector (E Manser, IMCB, Singapore). HA-Glut4-mCherry were a gift from Dr. Joshua J Zimmerberg (NICHD, NIH). For each construct, several clones were chosen and sequenced to their entirety in both directions to confirm their identity. All plasmids were purified using an Axygen miniprep kit for use in transfection experiments. The Escherichia coli strain DH5α was used as a host for propagation of the clones.
Cell culture and transfection
C2C12 myoblasts, 293T cells, and MEF cells were from American Type Culture Collection, USA. C2C12 with less than 15 passage numbers were used. Cell lines were authenticated by examination of morphology and growth characteristics and tested for mycoplasma contamination using GMyc-PCR Mycoplasma Test Kit (Yeasen biotech, 40601ES20). MycGuard™-1 Solution (Yeasen biotech, 40609ES60) was added in water trays of CO2 tissue culture incubator to prevent contamination. Cells were grown in DMEM supplemented with 10% (v/v) fetal bovine serum, 100 U/ml penicillin, 2 mM L-glutamine, and 100 mg/ml streptomycin (all from Hyclone Laboratories, Logan, UT). 293T cells were transfected with polyethylenimine (PEI), C2C12 myoblasts and MEF cells were transfected with lipofectamine 2000 (Invitrogen), and myotubes were transfected using the 4D-Nucleofector™ Y kit System (Lonza) according to manufacturer’s instructions (using solution AD2 and program FB-166).
RNA interference
C2C12 myoblasts at 40% confluency were transfected with siRNA using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s protocol. Sequences of siRNAs were: TBC1D17 siRNA #1: 5′-CCCAGAUUCUCUUCAAGAA-3′; TBC1D17 siRNA #2: 5′- CCUGCCUGCCUUACACUUU-3′; Rab5 siRNA #1: 5′-GCCAAAUACUGGAAAUAAA-3′; Rab5 siRNA #2: 5′-GGUUCUUCGCUUUGUGAAA-3′; Glut4 siRNA: 5′-CCAACUGGACCUGUAACUU-3′; control siRNA: 5′-UUCUCCGAACGUGUCACGU-3′.
Immunoprecipitation studies and western blot analyses
Control cells or cells transfected with expression plasmids were lysed in lysis buffer (50 mM Tris, pH 7.3, 150 mM sodium chloride, 1% (w/v) sodium deoxycholate, 0.25 mM EDTA, 1% (v/v) Triton X-100, 0.1% sodium orthovanadate, 0.2% sodium fluoride, and protease Inhibitor cocktail (Selleck Chemicals, B14001)). Lysates were immunoprecipitated (IP) with anti-FLAG M2 beads (Bimake, B23102). Samples were analyzed by western blotting with the indicated antibodies.
R5BD Pulldown (Rab5 activation) assay
The Rab5-binding domain (R5BD, residues 739–862) of Rabaptin5 was cloned into the pGEX4T1 vector (GE Healthcare). The constructs were subsequently transformed in to E. coli. BL21 cells for expression and purified using glutathione-sepharose 4B beads (GE Healthcare Life Sciences). Subsequently, GST control or GST-R5BD beads were incubated with whole cell lysate at 4 °C for 2–3 h, finally proceeding to western blot using the anti-Rab5 antibody.
In vitro GAP assay
GST-Rab5 proteins bound to GST-Sepharose were loaded with 4 mM GTP (R0461, Thermo Scientific) at 25 °C for 1 h in a GTPase loading buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM EDTA, 1 mM dithiothreitol). The matrix was washed with the same buffer to remove unbound GTP. The kinetics of intrinsic and GAP-accelerated GTP hydrolysis were measured using the ATPase/GTPase Activity Assay Kit (MAK-113, Sigma-Aldrich) according to the manufacturer’s instructions.
In vitro AMPK kinase assay
FLAG-tagged AMPKα2 WT or K45R protein was transfected into 293T cells and purified using M2-FLAG beads. As previously described [76], in vitro kinase assays were performed for 30 min in a 50 µl reaction mixture containing 200 ng FLAG-AMPKα2 WT or K45R as kinase, 2 µg GST-TBC1D17 WT or S168A as substrates, with 1 mM ATP in the kinase buffer (10 mM MgCl2, 25 mM HEPES pH7.5, 2 mM DTT, 50 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA). The reactions were stopped by boiling in 10 µl 5X SDS sample buffer for 5 min and analyzed by western blot using anti-Phospho-(Ser/Thr) AMPK substrate antibody.
Direct binding assay
293T lysates transfected with FLAG-tagged expression plasmids were subjected to immunoprecipitation with anti-FLAG M2 affinity gel. The desired proteins were eluted from the beads using 200 ng/μl FLAG peptide. Subsequently GST control or GST fusion protein-beads were incubated with eluted FLAG-tagged proteins at 4 °C for 2–3 h in binding buffer (200 mM sodium chloride, 50 mM Tris, pH 7.3, 0.25 mM EDTA, 1% (w/v) sodium deoxycholate, 1% (v/v) Triton X-100, 0.2% sodium fluoride, 0.1% sodium orthovanadate, and a mixture of protease inhibitors from Roche Applied Science). The bound proteins were separated by SDS-PAGE for western blot using anti-FLAG antibody.
In vitro GTP/GDP loading and binding assay
A total of 10 μg of GST-Rab5 fusion protein were preloaded with 5 mM GTPγS or GDP in binding buffer (20 mM Tris-HCl, pH 7.5, 25 mM NaCl, 10 mM EDTA, and 0.1 mM DTT) at 30 °C for 20 min. The reaction was stopped with MgCl2 to a final concentration of 10 mM. The beads were incubated with cell lysates at 4 °C in lysis buffer (150 mM NaCl, 50 mM HEPES pH 7.4, 1.5 mM MgCl2, 10% glycerol, 5 mM EGTA, 1% Triton X-100, 5 mM sodium orthovanadate, and a mixture of protease inhibitors). Samples were analyzed by western blotting.
Preparation of antibody targeted to phosphorylated TBC1D17-Ser168
Phosphorylated TBC1D17-Ser168 antibody production was commercially customized by the Youke Bio Ltd. Company (Shanghai, China). Briefly, the phosphorylated peptide C-ALLRVL(pS)RYLL-NH2 and unphosphorylated peptide C-ALLRVLSRYLL-NH2 were synthesized and used to immunize rabbits. The phosphorylated peptide and unphosphorylated peptide were coupled to the affinity column and purified using the antibody against phosphorylated TBC1D17-Ser168 from the serum of immunized rabbits.
Glucose uptake assay
After 3 h of serum starvation, C2C12 myotubes cultured in 96-well plates were washed twice with Krebs‑Ringer‑Phosphate‑HEPES (KRPH) buffer (20 mM HEPES, 5 mM KH2PO4, 1 mM MgSO4, 1 mM CaCl2, 136 mM NaCl and 4.7 mM KCl, pH 7.4) and starved for glucose by incubation with KRPH buffer for 40 min. The myotubes were then stimulated with 100 nM insulin for 20 min and incubated with 10 μl of 10 mM 2-deoxyglucose at 37 °C for 60 min. To analyze glucose uptake, a Screen Quest™ Colorimetric Glucose Uptake Assay Kit (36503, AAT Bioquest) was used according to the manufacturer’s instructions. At the end of the reaction, the absorbance of the samples and standards were measured by reading the OD ratio at wavelengths of 570–610 nm on a Spectrophotometric multi-well plate reader (SynergyMx M5, Molecular Devices). Total cellular protein was determined by the Bradford method (Bio-Rad, 500–0205).
Plasma membrane isolation
Subcellular fractionation was carried out as previously described [47] with some modifications. C2C12 myotubes grown in 10-cm-dishes were lysed using 12 passes through a 22-gauge needle followed by 20 passes through a 27-gauge needle in 1 ml homogenization buffer containing 250 mM sucrose, 10 mM EDTA, and 20 mM HEPES (pH 7.4) supplemented with protease inhibitor cocktail and phosphatase inhibitors on ice. Homogenates were centrifuged (500 g, 10 min, 4 °C) to remove unbroken cells, and supernatants were centrifuged at 10,080 g for 12 min at 4 °C to obtain crude plasma membrane (PM) together with nucleus and mitochondria, and intracellular fraction (Post-PM) containing cytoplasm. The PM fraction was solubilized in detergent containing (1% v/v Triton) lysis buffer.
Cell-surface HA-Glut4-mCherry detection
Surface HA-GLUT4-mCherry was detected by immunofluorescence in C2C12 myoblasts plated on glass coverslips as previously described [43]. Briefly, myoblasts were serum starved for 4 h including preincubation with or without AICAR (2 mM, 2 h), and then either treated or untreated with 100 nM insulin for 15 min. Cells were then rinsed with PBS, fixed with 3% paraformaldehyde (PFA), quenched with 0.1 M glycine, and blocked with 5% nonfat milk (w/v) in PBS for 10 min. Surface HA-GLUT4-mCherry was detected by anti-HA (1:100) for 4 h. After five washes with PBS, cells were incubated with Alexa 633–conjugated anti-rabbit secondary antibody (1:500) in the dark for 2 h, rinsed with PBS for 3 times, and mounted on slides. Fluorescence images were acquired with a LSM 880 laser-scanning confocal microscope (Carl Zeiss, Germany). Cells were scanned along the z-axis, and single collapsed images (collapsed xy projection) were assembled from optical stacks taken at 0.5 µm intervals. The pixel intensity in each cell (≥25 cells/condition) was quantified using ImageJ software (National Institutes of Health, Bethesda, MD).
Immunofluorescence and direct fluorescence studies
As previously described [77], cells were seeded on coverslips in a 24-well plate and transfected with various expression constructs for 24–36 h and then stained for immunofluorescence detection using confocal fluorescence microscopy or directly visualized for cells expressing GFP or mCherry-tagged proteins. The images were collected using a ×63 oil immersion objective with appropriate laser excitation on a Zeiss LSM 880 confocal microscope. The detector gain was first optimized by sampling various regions of the coverslip and then fixed for each specified channel. Once set, the detector gain value was kept constant throughout the image acquisition process. Images were analyzed using the ZEN2 Software.
Transferrin recycling
As previously described [78], cells were serum starved in DMEM with 0.5% BSA for 2h at 37 °C, incubated with 25 µg/ml 488-CF-Dye-conjugated transferrin (Biotium, #00081) for 1 h at 37 °C in serum-free media, and chased in full media for up to 40 min.
Image quantification
The intensity of the GFP-Rab5, GFP-Glut1, or GFP-Glut4 fluorescence signal in the cell periphery were analyzed in the raw images of the Zeiss LSM 880 confocal microscope using ImageJ software. The periphery cytoplasmic area signal (avoiding the perinuclear area) of GFP-Rab5, GFP-Glut1, or GFP-Glut4 in myoblasts was selected and quantified relative to the total GFP signal as previously described [38]. Cell sizes were measured by ImageJ of more than 30 randomly selected cells from each experimental group. Cell shape change was determined by measuring the length/width ratio of randomly selected cells from each experimental group. The degree of colocalization was determined with Pearson’s coefficient (GFP-Glut4 to Rab5 or Glut1 to Rab5). Values were analyzed by GraphPad Prism software and are presented as mean ± SE.
Mice and animal care
10–12 weeks old male mice without stress or discomfort (such as stereotypic behaviors and hair loss) were employed to minimize physiological variability. In every experiment, eight mice were allocated randomly into two groups (four mice per group) and were sufficient to obtain statistical differences. Animal experiments were blinded using number codes till the final data analyses were performed. All mice used in this study had a C57BL/6J genetic background and were housed in a pathogen-free facility in the University Laboratory Animal Center. All animal experiment protocols were approved by the Review Committee of Zhejiang University School of Medicine and were in compliance with ethical regulations and institutional guidelines.
Exercise experiments
Exercise experiments were performed on an Exer3/6 mouse treadmill (Columbus Instruments). As previously published [79], mice (10–12 weeks) underwent a 3-day period of treadmill running acclimatization that consisted of running for 15 min at a 0% incline and a speed of 13 m/min each day. On the day of acute exercise, mice allocated to perform treadmill running were subject to 5% incline and 10 min at 13 m/min, 10 min at 16 m/min, 50 min at 19 m/min, and then 20 min at 21 m/min. Mice were sacrificed following treadmill running and muscle tissues were harvested immediately after exercise.
Bioinformatics
To search for putative AMPK phosphorylation site, the peptide sequence of TBC1D17 was analyzed using GPS 3.0 (http://gps.biocuckoo.org/online.php). Multiple sequence alignment was performed using the Clustal Omega (http://www.clustal.org/omega/).
Statistical analysis
Statistical analyses were performed with GraphPad Prism 6 (GraphPad Software, Inc.). Values were presented as mean ± SE of three to six independent experiments as shown in each legend. Statistical significance was determined as indicated in the figure legends. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. The data distribution was first checked using a Shapiro–Wilk normality test, Kolmogorov–Smirnov test and D’Agostino & Pearson omnibus normality test. For comparison between two groups, if the data fitted a normal distribution, a two-tailed unpaired Student’s t-test was used when variances were similar by F test (p > 0.05). Whereas a two-tailed unpaired Student’s t-test with Welch’s correction was used when variances were different by F test (p < 0.05). If the data did not fit a normal distribution, a Mann–Whitney U-test was used. If the variation among three or more groups was minimal, ANOVA followed by Dunnett’s post-test or Tukey’s post hoc test was applied for comparison of multiple groups.
References
Atella V, Piano Mortari A, Kopinska J, Belotti F, Lapi F, Cricelli C, et al. Trends in age-related disease burden and healthcare utilization. Aging Cell. 2019;18:e12861.
Boucher J, Kleinridders A, Kahn CR. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb Perspect Biol. 2014;6:a009191.
Bogan JS. Regulation of glucose transporter translocation in health and diabetes. Annu Rev Biochem. 2012;81:507–32.
Gong XM, Li YF, Luo J, Wang JQ, Wei J, Wang JQ, et al. Gpnmb secreted from liver promotes lipogenesis in white adipose tissue and aggravates obesity and insulin resistance. Nat Metab. 2019;1:570–83.
Jaldin-Fincati JR, Pavarotti M, Frendo-Cumbo S, Bilan PJ, Klip A. Update on GLUT4 vesicle traffic: a cornerstone of insulin action. Trends Endocrinol Metab. 2017;28:597–611.
Klip A, McGraw TE, James DE. Thirty sweet years of GLUT4. J Biol Chem. 2019;294:11369–81.
Bryant NJ, Gould GW. Insulin stimulated GLUT4 translocation—size is not everything! Curr Opin Cell Biol. 2020;65:28–34.
Harcourt BE, Penfold SA, Forbes JM. Coming full circle in diabetes mellitus: from complications to initiation. Nat Rev Endocrinol. 2013;9:113–23.
Richter EA, Hargreaves M. Exercise, GLUT4, and skeletal muscle glucose uptake. Physiol Rev. 2013;93:993–1017.
Hoffman NJ, Parker BL, Chaudhuri R, Fisher-Wellman KH, Kleinert M, Humphrey SJ, et al. Global phosphoproteomic analysis of human skeletal muscle reveals a network of exercise-regulated kinases and AMPK substrates. Cell Metab. 2015;22:922–35.
Cartee GD. Roles of TBC1D1 and TBC1D4 in insulin- and exercise-stimulated glucose transport of skeletal muscle. Diabetologia. 2015;58:19–30.
Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–62.
Lin SC, Hardie DG. AMPK: sensing glucose as well as cellular energy status. Cell Metab. 2018;27:299–313.
Gonzalez A, Hall MN, Lin SC, Hardie DG. AMPK and TOR: The Yin and Yang of Cellular Nutrient Sensing and Growth Control. Cell Metab. 2020;31:472–92.
Pfeffer SR. Rab GTPases: master regulators that establish the secretory and endocytic pathways. Mol Biol Cell. 2017;28:712–15.
Zhen Y, Stenmark H. Cellular functions of Rab GTPases at a glance. J Cell Sci. 2015;128:3171–6.
Zeigerer A, Gilleron J, Bogorad RL, Marsico G, Nonaka H, Seifert S, et al. Rab5 is necessary for the biogenesis of the endolysosomal system in vivo. Nature. 2012;485:465–70.
Langemeyer L, Frohlich F, Ungermann C. Rab GTPase function in endosome and lysosome biogenesis. Trends Cell Biol. 2018;28:957–70.
Zhou F, Wu ZL, Zhao MZ, Murtazina R, Cai J, Zhang A, et al. Rab5-dependent autophagosome closure by ESCRT. J Cell Biol. 2019;218:1908–27.
Su X, Lodhi IJ, Saltiel AR, Stahl PD. Insulin-stimulated interaction between insulin receptor substrate 1 and p85 alpha and activation of protein kinase B/Akt require Rab5. J Biol Chem. 2006;281:27982–90.
Mao X, Kikani CK, Riojas RA, Langlais P, Wang L, Ramos FJ, et al. APPL1 binds to adiponectin receptors and mediates adiponectin signalling and function. Nat Cell Biol. 2006;8: 516–23.
Lodhi IJ, Bridges D, Chiang SH, Zhang YL, Cheng A, Geletka LM, et al. Insulin stimulates phosphatidylinositol 3-phosphate production via the activation of Rab5. Mol Biol Cell. 2008;19:2718–28.
Cong XX, Gao XK, Rao XS, Wen J, Liu XC, Shi YP, et al. Rab5a activates IRS1 to coordinate IGF-AKT-mTOR signaling and myoblast differentiation during muscle regeneration. Cell Death Differ. 2020;27:2344–62.
Zeigerer A, Bogorad RL, Sharma K, Gilleron J, Seifert S, Sales S, et al. Regulation of liver metabolism by the endosomal GTPase Rab5. Cell Rep. 2015;11:884–92.
Braccini L, Ciraolo E, Campa CC, Perino A, Longo DL, Tibolla G, et al. PI3K-C2 gamma is a Rab5 effector selectively controlling endosomal Akt2 activation downstream of insulin signalling. Nat Commun. 2015;6:7400.
Ryu J, Galan AK, Xin X, Dong F, Abdul-Ghani MA, Zhou L, et al. APPL1 potentiates insulin sensitivity by facilitating the binding of IRS1/2 to the insulin receptor. Cell Rep. 2014;7:1227–38.
Fukuda M. TBC proteins: GAPs for mammalian small GTPase Rab? Biosci Rep. 2011;31:159–68.
Frasa MA, Koessmeier KT, Ahmadian MR, Braga VM. Illuminating the functional and structural repertoire of human TBC/RABGAPs. Nat Rev Mol Cell Biol. 2012;13:67–73.
Barr F, Lambright DG. Rab GEFs and GAPs. Curr Opin Cell Biol. 2010;22:461–70.
Mafakheri S, Chadt A, Al-Hasani H. Regulation of RabGAPs involved in insulin action. Biochem Soc Trans. 2018;46:683–90.
Chen L, Chen Q, Xie B, Quan C, Sheng Y, Zhu S, et al. Disruption of the AMPK-TBC1D1 nexus increases lipogenic gene expression and causes obesity in mice via promoting IGF1 secretion. Proc Natl Acad Sci USA. 2016;113:7219–24.
Kjobsted R, Hingst JR, Fentz J, Foretz M, Sanz MN, Pehmoller C, et al. AMPK in skeletal muscle function and metabolism. Faseb J. 2018;32:1741–77.
Fuchs E, Haas AK, Spooner RA, Yoshimura S, Lord JM, Barr FA. Specific Rab GTPase-activating proteins define the Shiga toxin and epidermal growth factor uptake pathways. J Cell Biol. 2007;177:1133–43.
Yamano K, Fogel AI, Wang C, van der Bliek AM, Youle RJ. Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. Elife. 2014;3:e01612.
Jeon SM, Chandel NS, Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485:661–5.
Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–75.
Neefjes J, Jongsma MML, Berlin I. Stop or go? endosome positioning in the establishment of compartment architecture, dynamics, and function. Trends Cell Biol. 2017;27:580–94.
Sun Y, Chiu TT, Foley KP, Bilan PJ, Klip A. Myosin Va mediates Rab8A-regulated GLUT4 vesicle exocytosis in insulin-stimulated muscle cells. Mol Biol Cell. 2014;25:1159–70.
Gillingham AK, Sinka R, Torres IL, Lilley KS, Munro S. Toward a comprehensive map of the effectors of rab GTPases. Dev Cell. 2014;31:358–73.
Vaibhava V, Nagabhushana A, Chalasani ML, Sudhakar C, Kumari A, Swarup G. Optineurin mediates a negative regulation of Rab8 by the GTPase-activating protein TBC1D17. J Cell Sci. 2012;125:5026–39.
Fazakerley DJ, Holman GD, Marley A, James DE, Stockli J, Coster AC. Kinetic evidence for unique regulation of GLUT4 trafficking by insulin and AMP-activated protein kinase activators in L6 myotubes. J Biol Chem. 2010;285:1653–60.
Lizunov VA, Stenkula KG, Lisinski I, Gavrilova O, Yver DR, Chadt A, et al. Insulin stimulates fusion, but not tethering, of GLUT4 vesicles in skeletal muscle of HA-GLUT4-GFP transgenic mice. Am J Physiol Endocrinol Metab. 2012;302:E950–60.
Sun Y, Jaldin-Fincati J, Liu Z, Bilan PJ, Klip A. A complex of Rab13 with MICAL-L2 and alpha-actinin-4 is essential for insulin-dependent GLUT4 exocytosis. Mol Biol Cell. 2016;27:75–89.
Tortorella LL, Pilch PF. C2C12 myocytes lack an insulin-responsive vesicular compartment despite dexamethasone-induced GLUT4 expression. Am J Physiol Endocrinol Metab. 2002;283:E514–24.
Mitsumoto Y, Burdett E, Grant A, Klip A. Differential expression of the GLUT1 and GLUT4 glucose transporters during differentiation of L6 muscle cells. Biochem Biophys Res Commun. 1991;175:652–9.
Sun Y, Bilan PJ, Liu Z, Klip A. Rab8A and Rab13 are activated by insulin and regulate GLUT4 translocation in muscle cells. Proc Natl Acad Sci USA. 2010;107:19909–14.
Chen S, Wasserman DH, MacKintosh C, Sakamoto K. Mice with AS160/TBC1D4-Thr649Ala knockin mutation are glucose intolerant with reduced insulin sensitivity and altered GLUT4 trafficking. Cell Metab. 2011;13:68–79.
Lansey MN, Walker NN, Hargett SR, Stevens JR, Keller SR. Deletion of Rab GAP AS160 modifies glucose uptake and GLUT4 translocation in primary skeletal muscles and adipocytes and impairs glucose homeostasis. Am J Physiol Endocrinol Metab. 2012;303:E1273–86.
Wang HY, Ducommun S, Quan C, Xie B, Li M, Wasserman DH, et al. AS160 deficiency causes whole-body insulin resistance via composite effects in multiple tissues. Biochem J. 2013;449:479–89.
Chadt A, Immisch A, de Wendt C, Springer C, Zhou Z, Stermann T, et al. "Deletion of both Rab-GTPase-activating proteins TBC1D1 and TBC1D4 in mice eliminates insulin- and AICAR-stimulated glucose transport [corrected]. Diabetes. 2015;64:746–59.
Hargett SR, Walker NN, Keller SR. Rab GAPs AS160 and Tbc1d1 play nonredundant roles in the regulation of glucose and energy homeostasis in mice. Am J Physiol Endocrinol Metab. 2016;310:E276–88.
Banko MR, Allen JJ, Schaffer BE, Wilker EW, Tsou P, White JL, et al. Chemical genetic screen for AMPKalpha2 substrates uncovers a network of proteins involved in mitosis. Mol Cell. 2011;44:878–92.
Schaffer BE, Levin RS, Hertz NT, Maures TJ, Schoof ML, Hollstein PE, et al. Identification of AMPK phosphorylation sites reveals a network of proteins involved in cell invasion and facilitates large-scale substrate prediction. Cell Metab. 2015;22:907–21.
Pufall MA, Graves BJ. Autoinhibitory domains: modular effectors of cellular regulation. Annu Rev Cell Dev Biol. 2002;18:421–62.
Wang JJ, Liu F, Yang F, Wang YZ, Qi X, Li Y, et al. Disruption of auto-inhibition underlies conformational signaling of ASIC1a to induce neuronal necroptosis. Nat Commun. 2020;11:475.
Huang J, Imamura T, Olefsky JM. Insulin can regulate GLUT4 internalization by signaling to Rab5 and the motor protein dynein. P Natl Acad Sci USA. 2001;98:13084–89.
Tessneer KL, Jackson RM, Griesel BA, Olson AL. Rab5 activity regulates GLUT4 sorting into insulin-responsive and non-insulin-responsive endosomal compartments: a potential mechanism for development of insulin resistance. Endocrinology. 2014;155:3315–28.
Kjobsted R, Treebak JT, Fentz J, Lantier L, Viollet B, Birk JB, et al. Prior AICAR stimulation increases insulin sensitivity in mouse skeletal muscle in an AMPK-dependent manner. Diabetes. 2015;64:2042–55.
Maarbjerg SJ, Sylow L, Richter EA. Current understanding of increased insulin sensitivity after exercise - emerging candidates. Acta Physiol. 2011;202:323–35.
Liu H, Wang S, Hang W, Gao J, Zhang W, Cheng Z, et al. LET-413/Erbin acts as a RAB-5 effector to promote RAB-10 activation during endocytic recycling. J Cell Biol. 2018;217:299–314.
Palamidessi A, Frittoli E, Ducano N, Offenhauser N, Sigismund S, Kajiho H, et al. The GTPase-activating protein RN-tre controls focal adhesion turnover and cell migration. Curr Biol. 2013;23:2355–64.
Lanzetti L, Palamidessi A, Areces L, Scita G, Di Fiore PP. Rab5 is a signalling GTPase involved in actin remodelling by receptor tyrosine kinases. Nature. 2004;429:309–14.
Frittoli E, Palamidessi A, Pizzigoni A, Lanzetti L, Garre M, Troglio F, et al. The primate-specific protein TBC1D3 is required for optimal macropinocytosis in a novel ARF6-dependent pathway. Mol Biol Cell. 2008;19:1304–16.
Haas AK, Fuchs E, Kopajtich R, Barr FA. A GTPase-activating protein controls Rab5 function in endocytic trafficking. Nat Cell Biol. 2005;7:887–U36.
Li W, Zou W, Zhao D, Yan J, Zhu Z, Lu J, et al. C. elegans Rab GTPase activating protein TBC-2 promotes cell corpse degradation by regulating the small GTPase RAB-5. Development. 2009;136:2445–55.
Haas AK, Fuchs E, Kopajtich R, Barr FA. A GTPase-activating protein controls Rab5 function in endocytic trafficking. Nat Cell Biol. 2005;7:887–93.
Borg Distefano M, Hofstad Haugen L, Wang Y, Perdreau-Dahl H, Kjos I, Jia D, et al. TBC1D5 controls the GTPase cycle of Rab7b. J Cell Sci. 2018;131:17.
Wei Z, Zhang M, Li C, Huang W, Fan Y, Guo J, et al. Specific TBC Domain-Containing Proteins Control the ER-Golgi-Plasma Membrane Trafficking of GPCRs. Cell Rep. 2019;28:554–66. e4
Han Y, Hu Z, Cui A, Liu Z, Ma F, Xue Y, et al. Post-translational regulation of lipogenesis via AMPK-dependent phosphorylation of insulin-induced gene. Nat Commun. 2019;10:623.
Burkewitz K, Zhang Y, Mair WB. AMPK at the nexus of energetics and aging. Cell Metab. 2014;20:10–25.
Zhang CS, Li M, Ma T, Zong Y, Cui J, Feng JW, et al. Metformin activates AMPK through the lysosomal pathway. Cell Metab. 2016;24:521–22.
Xie B, Chen Q, Chen L, Sheng Y, Wang HY, Chen S. The inactivation of RabGAP function of AS160 promotes lysosomal degradation of GLUT4 and causes postprandial hyperglycemia and hyperinsulinemia. Diabetes. 2016;65:3327–40.
Treebak JT, Taylor EB, Witczak CA, An D, Toyoda T, Koh HJ, et al. Identification of a novel phosphorylation site on TBC1D4 regulated by AMP-activated protein kinase in skeletal muscle. Am J Physiol Cell Physiol. 2010;298:C377–85.
Mafakheri S, Florke RR, Kanngiesser S, Hartwig S, Espelage L, De Wendt C, et al. AKT and AMP-activated protein kinase regulate TBC1D1 through phosphorylation and its interaction with the cytosolic tail of insulin-regulated aminopeptidase IRAP. J Biol Chem. 2018;293:17853–62.
Thomas EC, Hook SC, Gray A, Chadt A, Carling D, Al-Hasani H, et al. Isoform-specific AMPK association with TBC1D1 is reduced by a mutation associated with severe obesity. Biochem J. 2018;475:2969–83.
Yi C, Tong J, Lu P, Wang Y, Zhang J, Sun C, et al. Formation of a Snf1-Mec1-Atg1 module on mitochondria governs energy deprivation-induced autophagy by regulating mitochondrial respiration. Dev Cell. 2017;41:59–71. e4
He MY, Xu SB, Qu ZH, Guo YM, Liu XC, Cong XX, et al. Hsp90beta interacts with MDM2 to suppress p53-dependent senescence during skeletal muscle regeneration. Aging Cell. 2019;18:e13003.
Roy S, Leidal AM, Ye J, Ronen SM, Debnath J. Autophagy-dependent shuttling of TBC1D5 controls plasma membrane translocation of GLUT1 and glucose uptake. Mol Cell. 2017;67:84–95. e5
Laker RC, Drake JC, Wilson RJ, Lira VA, Lewellen BM, Ryall KA, et al. Ampk phosphorylation of Ulk1 is required for targeting of mitochondria to lysosomes in exercise-induced mitophagy. Nat Commun. 2017;8:548.
Acknowledgements
We thank Ms. Yanwei Li and Guifeng Xiao from the core facility platform of Zhejiang university school of medicine for their technical support. We thank Prof. Wei Liu for providing the AMPKα1/α2 double-knockout MEFs and Dr. Joshua J Zimmerberg (NICHD, NIH) for providing the HA-Glut4-mCherry plasmid.
Funding
This work was supported by the National Key R&D Program of China (2018YFA0800400), the National Natural Science Foundation of China (91954121, 81871760, 32070821, 81670740), grants from Science and Technology Commission of Zhejiang Province (LZ20H060001, LQ20H060006), China Postdoctoral Science Foundation (2019M662034), the International Science & Technology Cooperation Program of China (the Ministry of Science and Technology of PRC, 2015DFG32130), the Training Program of Hangzhou Key Discipline of Geriatrics, and the Fundamental Research Funds for the Central Universities.
Author information
Authors and Affiliations
Contributions
LLZ, YTZ, and PL designed and coordinated the study. YTZ, LLZ, PL, ZXM, CY, and YX wrote the paper. XSR, XXC, and XKG performed and analyzed the immunofluorescence assays and the RT-qPCR assays. XSR, XXC, XKG, YPS, LJS, CYN, and MJH performed and analyzed the protein-protein interaction and western blot experiments. XSR, JFW, and JL designed and constructed vectors for expression. All authors analyzed the results and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval
All animal experiment protocols were approved by the Review Committee of Zhejiang University School of Medicine and were in compliance with ethical regulations and institutional guidelines.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by A. Degterev
Supplementary information
Rights and permissions
About this article
Cite this article
Rao, X.S., Cong, X.X., Gao, X.K. et al. AMPK-mediated phosphorylation enhances the auto-inhibition of TBC1D17 to promote Rab5-dependent glucose uptake. Cell Death Differ 28, 3214–3234 (2021). https://doi.org/10.1038/s41418-021-00809-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-021-00809-9
- Springer Nature Limited
This article is cited by
-
VAPB-mediated ER-targeting stabilizes IRS-1 signalosomes to regulate insulin/IGF signaling
Cell Discovery (2023)
-
Mapping of the plant SnRK1 kinase signalling network reveals a key regulatory role for the class II T6P synthase-like proteins
Nature Plants (2022)
-
Phase separation of insulin receptor substrate 1 drives the formation of insulin/IGF-1 signalosomes
Cell Discovery (2022)