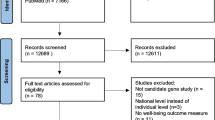
Abstract
Wellbeing is an important aspect of mental health that is moderately heritable. Specific wellbeing-related variants have been identified via GWAS meta-analysis of individual questionnaire items. However, a multi-item within-subject index score has potential to capture greater heritability, enabling improved delineation of genetic and phenotypic relationships across traits and exposures that are not possible on aggregate-data. This research employed data from the UK Biobank resource, and a wellbeing index score was derived from indices of happiness and satisfaction with family/friendship/finances/health, using principal component analysis. GWAS was performed in Caucasian participants (N = 129,237) using the derived wellbeing index, followed by polygenic profiling (independent sample; N = 23,703). The wellbeing index, its subcomponents, and negative indicators of mental health were compared via phenotypic and genetic correlations, and relationships with psychiatric disorders examined. Lastly, the impact of childhood maltreatment on wellbeing was investigated. Five independent genome-wide significant loci for wellbeing were identified. The wellbeing index had SNP-heritability of ~8.6%, and stronger phenotypic and genetic correlations with its subcomponents (0.55–0.77) than mental health phenotypes (−0.21 to −0.39). The wellbeing score was lower in participants reporting various psychiatric disorders compared to the total sample. Childhood maltreatment exposure was also associated with reduced wellbeing, and a moderate genetic correlation (rg = ~−0.56) suggests an overlap in heritability of maltreatment with wellbeing. Thus, wellbeing is negatively associated with both psychiatric disorders and childhood maltreatment. Although notable limitations, biases and assumptions are discussed, this within-cohort study aids the delineation of relationships between a quantitative wellbeing index and indices of mental health and early maltreatment.
Similar content being viewed by others
Introduction
Wellbeing is an important aspect of mental health, and is defined by the core concepts of happiness, life satisfaction and optimal psychological functioning [1]. This concept of wellbeing is consistent with the World Health Organisation definition of health: “a state of complete physical, mental and social wellbeing and not merely the absence of disease or infirmity”.
Wellbeing is defined by two core concepts: (1) subjective wellbeing (also referred to as ‘hedonia’), consisting of positive affect and satisfaction with life, and (2) psychological wellbeing (also referred to as ‘eudaimonia’), consisting of attributes that enable realisation of human potential including autonomy, mastery and life purpose [2]. Various questionnaires measure wellbeing [3, 4], but questionnaire items usually load onto a common wellbeing factor [5]. Indeed, we showed that a composite measure encapsulating elements of both subjective and psychological wellbeing, is more heritable and stable longitudinally than wellbeing indices derived from single-item questions [6].
Wellbeing is moderately heritable, with estimates from twin studies ranging from 17 to 67% [reviewed in Bartels [7]], leading to efforts to elucidate the specific genetic signatures (particularly Single Nucleotide Polymorphisms (SNPs)) underlying this trait [8, 9]. However, SNP-heritability estimated from genome-wide association studies (GWAS) are only ~4–6.4% [8, 9], thus new approaches are required to explain a greater proportion of trait heritability.
To improve power for gene discovery, analytic approaches have included meta-analyses of independent GWAS from different cohorts of the component traits which underlie subjective wellbeing [8]. As wellbeing is genetically and phenotypically correlated to negative mental health constructs including neuroticism and depressive symptoms [8, 10], other analytic approaches have used multivariate GWAS meta-analysis to include additional correlated phenotypes, as part of a ‘wellbeing spectrum’ [10, 11] that spans correlated traits such as loneliness and self-rated health [12]. However, broadening the spectrum to include negative health outcomes as a continuum may diminish the detection of unique genetic signals specifically relating to wellbeing. For example, previous studies have shown negative genetic correlations between subjective wellbeing and anxiety, depression, schizophrenia and bipolar disorder (ranging from −0.73 to −0.22) [13], suggesting different proportions of common and unique genetic factors across the spectrum of negative health outcomes. By separating the constructs and quantifying the influence of psychiatric disorders on wellbeing, one can subsequently examine the impact of environmental exposures known to predict higher risk for certain disorders. Furthermore, as no phenotype is derivable using meta-analytic genetic approaches, one cannot use this method to examine the phenotypic properties of wellbeing – this includes examining phenotypic correlations, quantifying the influence of psychiatric disorders, and environmental exposures. Therefore, a wellbeing score composed of multiple relevant unique components would facilitate detailed exploration of both genetic and phenotypic attributes of wellbeing.
One potentially important environmental influence on wellbeing is childhood maltreatment, for which a negative impact on wellbeing has been demonstrated in adulthood [14, 15]. Furthermore, recent evidence suggests that there is a genetic contribution to childhood maltreatment through gene–environment correlations [16, 17], suggesting that childhood maltreatment may influence wellbeing through both genetic and environmental processes, a relationship that has not hitherto been explored.
Here, building on our previous work [6], we hypothesise that a continuous wellbeing index – derived from multiple measures of wellbeing available in the UK Biobank – is more heritable than single-item measures, and can be used to explore both phenotypic and genetic correlations with related clinical features and environmental exposures. The availability of genotype data from UK Biobank participants provides the opportunity to use various genetic techniques, including gene discovery, genetic correlation analyses and exploration of predictive power of polygenic scores. Herein, we use five approaches to evaluate the wellbeing index. First, phenotypic correlations between the wellbeing index, its subcomponents and related negative mental health constructs are measured. Second, we perform discovery GWAS for the wellbeing index and functional analysis of identified genetic signals. Third, genetic correlations of wellbeing-related phenotypes are compared, head-to-head within the same sample. We also examine genetic correlations with several psychiatric illnesses and quantify the effect of specific psychiatric diagnoses on the wellbeing index. Fourth, we assess the predictive power of wellbeing index polygenic scores in an independent UK Biobank-derived sample. Finally, we evaluate the effect of childhood maltreatment exposure on the wellbeing index and examine their genetic correlations.
Materials and methods
Participants
The UK Biobank is a population-based cohort with >500,000 participants from the United Kingdom, aged 40–69 years when recruited at baseline between 2006 and 2010, of predominantly British-Angloceltic ancestry [18]. Potentially eligible participants were identified from population-based registers and invited to participate. Due to the broad scope of the resource, the baseline questionnaire was designed to include questionnaire items with reliability and validity that relate to outcomes of public health importance. The UK Biobank received approval by the National Health Service National Research Ethics Service (11/NW/0382) and participants provided signed informed consent. The current project was approved by UK Biobank Data Access Committee (58534), with approval by University of New South Wales Human Research Advisory Panel (HC200191) for data analysis.
Defining wellbeing
We reviewed all UK Biobank questionnaire items and selected five wellbeing-related items from the baseline questionnaire, including general happiness and satisfaction with family, friendship, health and financial situation (Data-Fields:4526, :4559, :4570, :4581, :4548, respectively). Responses were on a 6-point Likert scale, related to general life experiences (not a defined timescale) and were reverse-scored. Principal component analysis was performed in SPSSv25, and a factor score created, herein referred to as the “wellbeing index”. To evaluate goodness of model fit, we employed confirmatory factor analysis in the validation dataset using R package lavvan. The internal consistency of the wellbeing index was tested using Cronbach’s Alpha. Additional details and analysis workflow are in Supplementary Methods (Fig. S1).
Discovery and confirmatory samples
To define the discovery sample, we used UK Biobank genetic data (March-2018 release), comprising 488,000 individuals genotyped using Affymetrix UK BiLEVE Axiom or UK Biobank Axiom arrays. Further information on the cohort, genotyping, imputation, and quality control (QC), is available elsewhere [18, 19]. Briefly, participants were removed who withdrew consent, reported non-Caucasian ancestry (Data-Field:22006), had >10% genotype missingness or QC failure (Data-Field:22051), were on genomic analysis exclusion list (Data-Field:22010), had gender mismatch (self-report vs. genotype-derived), sex chromosome aneuploidy or heterozygosity outliers (Data-Field:22027).
Participants meeting genotype QC criteria, with complete phenotype data for the five core wellbeing questions, were N = 129,237. Analysis of other mental health indicators was limited to this sample, but due to missingness of some phenotypes, sample size varies slightly.
An independent sample from UK Biobank was defined (N = 23,703; see Supplementary Methods) for confirmatory factor analysis, and to evaluate predictive power of polygenic scores from discovery GWAS.
Negative mental health indicators
Four negative mental health indicators were selected as outcome comparators for the wellbeing index score. These included measures of loneliness, neuroticism, depressive symptoms and whether the participant had ever seen a psychiatrist or GP for nerves, anxiety, tension or depression (Data-Fields provided in Table S1). Among these measures, loneliness and neuroticism ask about current experiences whereas the depressive symptoms and “seen a psychiatrist or GP” ask about lifetime experiences. Variable coding is described in Supplementary Methods.
Childhood maltreatment
Five items related to childhood maltreatment were identified, including emotional or physical neglect, and physical, emotional, or sexual abuse [Data-Field:20487-20490], and responses were on a 5-point Likert scale (see Supplemental Material). These exposures are collectively referred to as “maltreatment”, henceforth. First, the impact of each type of childhood maltreatment on the wellbeing index was examined as categorical variables, relating to the frequency of maltreatment experience. Then to evaluate the cumulative impact of maltreatment exposure, each item was dichotomised as “exposed” or “not-exposed” [see Table S2 for details], and a sum-score was created by summing across four maltreatment types [ranging from 0 to 4; excluding physical neglect due to identified coding issue (see Supplemental Methods for details)]. There were 45,723 participants with non-missing childhood maltreatment sum-score and wellbeing index.
Mean differences in wellbeing index were examined using Kruskal-Wallis (multi-category), and Wilcoxon (two-category) tests. Simple and multiple linear regression models, adjusting for age, age-squared, sex and Townsend Deprivation Index [Data-Field:189], examined how childhood maltreatment (or exposure sum-score) impacts wellbeing. The regression standardised estimate was considered the magnitude of effect of the dependant variable.
Psychiatric illnesses
The question “Have you been diagnosed with one or more of the following mental health problems by a professional, even if you don’t have it currently?” (Data-Field:20544) was used as the indicator of a lifetime psychiatric diagnosis. “Prefer not to answer” responses were coded as missing. As participants could select ≥1 of 16 listed diagnoses, representation in each mental health category was not mutually exclusive.
Phenotypic correlations (r p )
The correlation (v0.6.0) R package was employed to examine phenotypic correlations (rp), employing different types of correlations based on data type: Spearman’s rank for continuous/ordinal variables, Tetrachoric for binary, and Point-biserial for continuous/ordinal versus binary. Analyses were limited to participants with no missing phenotype data to enable head-to-head comparisons using the same sample size across all measures (N = 103,373; due to missing data in negative mental health phenotypes). The significance threshold for rp was set at p < 0.001, using a Bonferroni correction for 45 tests (α = 0.05/45).
Genetic association and SNP-heritability
To perform GWAS, we employed BOLT-LMM v2.3.4 [20]. Analysis employed imputed genotypes in BGENv.1.2 format (v3) and incorporated a Bayesian linear mixed effects model (LMM), accounting for population structure and sample relatedness using a genetic relatedness matrix, plus relevant covariates (sex, age, age-squared, genotype array, 20 PCs). BOLT-LMM automatically filters SNPs and individuals with >10% missing, and further filtering of SNPs with MAF < 0.01, INFO < 0.8 and Hardy-Weinberg Equilibrium p < 1 × 10−6, left ~8,068,119 SNPs for analysis. Results from the standard infinitesimal mixed-model association were employed for downstream analysis.
Functional Mapping and Annotation of GWAS (FUMA) [21] was employed to link candidate SNPs to genes and perform functional analysis (see Supplementary Methods).
SNP-heritability calculations employed Linkage Disequilibrium Score Regression (LDSC) [22], utilising GWAS summary statistics for each target phenotype derived from the same discovery sample. We used the LDSC method to estimate heritability and genetic correlations because it is a widely adopted method employed by most previous studies, hence ensuring more comparable associations. However, we acknowledge that this method has several assumptions that could limit the accuracy of estimates [22, 23], which are discussed later.
Genetic correlations (r g)
LDSC [22] was used to estimate genetic correlations (rg) between the wellbeing index, its five subcomponents, four negative indicators of mental health, and childhood maltreatment using GWAS summary statistics generated herein.
Published summary statistics of additional wellbeing-related traits were used to estimate rg with the derived wellbeing index phenotype, including: positive affect, life satisfaction, wellbeing spectrum, [10] subjective wellbeing [8], conscientiousness [24], extraversion [25], neuroticism [26], depressive symptoms [8], loneliness [27] and body mass index (BMI) [28].
In addition, rg with major psychiatric conditions were examined using published disease-specific summary statistics, including: major depressive disorder (MDD) [29], bipolar disorder (BIP) [30], schizophrenia (SCZ) [31], attention deficit/hyperactivity disorder (ADHD) [32], autism spectrum disorder (ASD) [33], obsessive-compulsive disorder (OCD) [34] and post-traumatic stress disorder (PTSD) [35]. We included HapMap3 SNPs with MAF > 0.01 and excluded the MHC region for rg estimations. The significance threshold for rg after Bonferroni correction for 72 tests (α = 0.05/72) was p < 6.94 × 10−4.
Polygenic scores
Polygenic scores (PGS) were constructed in the independent confirmatory cohort using PRS-CS software [36], employing summary statistics of the wellbeing index GWAS. We tested the predictive power of PGS in the confirmatory sample for the wellbeing index, its subcomponents, and negative mental health indicators using regression models, including relevant covariates (age, age-squared, sex, genotyping array, 10 PCs) in all models. An incremental R2, defined as the difference between the R2 from the full-model and the model without PGS as a predictor, was reported as predictive power. For binary variables (loneliness, depressive symptoms and “seen GP or psychiatrist”) a logistic regression was used instead of linear regression, and Nagelkerke R2 calculated using the R package fmsb (v0.7.0). A 95% confidence interval around the R2 was calculated using the R package psychometric (v2.2).
Results
Participant demographics
The discovery cohort comprised 129,237 participants (57.9% female) who were of British-white ancestry and had no missing phenotypes for the five core wellbeing questions. The age range was 40-70 years (mean ± SD = 57.3 ± 8.0), and 31.9% reported having college education. The data-field IDs and description of study variables in the discovery sample is presented in Table S1.
The confirmatory sample comprised 23,703 participants (51.3% female), who were genetically unrelated to the discovery sample but with identical inclusion criteria, and were ~7 years older (mean ± SD = 64.5 ± 7.7, range 46–82 years) than discovery cohort participants.
The wellbeing index and phenotypic correlations
Principal component analysis in the discovery sample indicated the first component explained 47.48% of the variance of the five questions (eigenvalue = 2.374) (Fig. S2), and was extracted using regression method as the wellbeing index score. The wellbeing index was approximately normally distributed with an extended left tail (skewness = −0.373, kurtosis=4.119) (Fig. S3A). Females had a slightly higher mean score than males (0.029 vs −0.034 respectively; Wilcoxon p < 2.2 × 10−16; Fig. S3B). The wellbeing index had a good internal consistency with Cronbach’s Alpha of 0.71. Confirmatory factor analysis in the independent sample showed a good model fit, loading onto a single factor (CFI = 0.98, TLI = 0.97, RMSEA = 0.047, SRMR = 0.034).
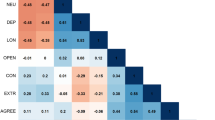
The wellbeing index had the strongest rp with the subcomponent happiness (rp = 0.77), and the weakest with health satisfaction (rp = 0.55). From the negative mental health indicators, the wellbeing index had the strongest rp with neuroticism (rp = −0.39) and weakest with depressive symptoms (rp = −0.22) (Fig. 1).
All correlations reported here are statistically significant after Bonferroni correction for multiple testing, although many have low (r = 0.3–0.5) to moderate (r = 0.5–0.7) effect size. Manhattan plots for each GWAS are provided in Figure S6.
GWAS findings for wellbeing index
Forty-two SNPs exceeded genome-wide significance for association from wellbeing index GWAS (p < 5 × 10–8) (Fig. 2), which represented five independent loci (Table S3 and Fig. S4).
A Manhattan plot representing association at 8,068,119 SNPs across the genome, with chromosome and base pair position is on x-axis and negative logarithm of the p-value from infinitesimal model is on y-axis. The red line indicates the genome-wide significance threshold of p < 5×10−8. B Quantile-quantile plot showing inflation of observed associations over that expected under null (λGC = 1.2005, mean χ2 = 1.2171, LD score regression intercept=1.005, Total Observed scale h2 = 0.0857 ± 0.005).
Candidate genes at each locus were mapped in FUMA using positional (n = 44 genes), eQTL (n = 27) and chromatin interaction mapping (n = 62) modules (total n = 101) (Table S4). The GWAS catalogue in FUMA identified signal overlaps amongst SNPs in linkage disequilibrium with wellbeing index and related traits; on chromosome 2, FSHR was associated with hedonic wellbeing [9] and both loci on chromosome 6 – the first associated with many related conditions including positive-affect, wellbeing spectrum [9], and depression [8, 29], and the second centred around TRIM26 associated with autism and schizophrenia [37, 38] (Table S5).
Tissue-specific enrichment of wellbeing index GWAS genes in MAGMA (implemented in FUMA) [39] showed significant enrichment in brain tissues, particularly cerebellum and frontal cortex (Fig. S5). Manhattan plots for GWAS of wellbeing subcomponents and related traits are provided (Fig. S6a–j).
Heritability and genetic correlations
For wellbeing and its subcomponents, the wellbeing index had the highest SNP-heritability (h2 = 8.6%, SE = 0.005), although it was not significantly different from health satisfaction (h2 = 7.8%, SE = 0.005). Other subcomponents had similar SNP-heritability (h2 = 6.2–5.5%, SE = 0.005) (Fig. S7).
Generally, the rg between traits were stronger than their rp (Fig. 1), and rg and rp were strongly correlated (r = 0.97). Amongst the subcomponents (Fig. S6a–e), wellbeing index had the strongest relationship with happiness (rg = 0.91, SE = 0.013). Across the negative mental health indicators (Fig. S6f-j), wellbeing index had the strongest relationship with loneliness (rg = −0.71, SE = 0.041).
Using published summary statistics, the rg between wellbeing index and other positively and negatively related phenotypes showed significant correlation for all tested traits after multiple-testing correction. Except for OCD and PTSD, all psychiatric conditions had a significant negative relationship with wellbeing index – the strongest effect was for MDD (rg = −0.55, SE = 0.03) (Fig. 3).
Positively and negatively correlated phenotypes and major psychiatric illnesses were examined using Linkage Disequilibrium Score Regression (LDSC). Published wellbeing-related summary statistics from independent studies included: positive affect, life satisfaction, wellbeing spectrum [10], subjective wellbeing [8], conscientiousness [24], extraversion [25], neuroticism [26], depressive symptoms [8], loneliness [27] and body mass index (BMI) [28]. Published disease-specific summary statistics from independent studies included: major depressive disorder (MDD) [29], bipolar disorder (BIP) [30], schizophrenia (SCZ) [31], attention deficit/hyperactivity disorder (ADHD) [32], autism spectrum disorder (ASD) [33], obsessive-compulsive disorder (OCD) [34] and post-traumatic stress disorder (PTSD) [35]. Error bars represent 95% confidence intervals (95% CI). Green bars indicate positive rg, orange bars indicate negative rg and grey bars indicate traits with non-significant genetic correlation after Bonferroni correction (p > 6.94 × 10−4).
Polygenic score analysis
In the replication sample, the wellbeing index PGS explained a small but significant portion of variance in all traits examined, with highest predictive power for the wellbeing index score itself (R2 = 0.01, p = 5.68 × 10−56), and the lowest for financial satisfaction (R2 = 0.003, p = 2.77 × 10−17). Amongst negative mental health indices, the best predictive power of wellbeing PGS was for loneliness (Nagelkerke R2 = 0.004, p = 2.93 × 10−14; Fig. 4).
The y-axis shows the incremental R2% and for binary variables (Loneliness, Depressive symptoms, and Seen GP or Psychiatrist) Nagelkerke R2. Error bars are lower and upper bound of 95% confidence intervals. While variance explained was <1%, P-values were all highly significant (p = 3.55 × 10−14 – 5.68 × 10−56).
Childhood maltreatment and wellbeing index
For all types of childhood maltreatment, the wellbeing index showed a stepwise reduction with increased frequency of maltreatment exposure (Fig. S8) – noting the potential data coding issue identified impacting physical neglect (Data-Field:20491) which indicated misinterpretation of the “never” response category, resulting in this variable’s exclusion (see Supplemental Methods).
Among participants reporting any childhood maltreatment, 43% reported multiple exposure types. The effect was dose dependant: by accumulation of maltreatment types, the mean wellbeing score decreased by about 0.2 for each additional maltreatment (Fig. 5). All types of childhood maltreatment exposure were significant in linear regression models predicting wellbeing. In simple models (with one trauma type as the predictor), the highest effect estimate was for the maltreatment sum score (β = −0.206, SE = 0.005), although not significantly different from emotional neglect (β = −0.200, SE = 0.010) (Fig. S9, panel A). In multiple linear model (with all traumas as predictors), emotional neglect and abuse had larger effects than physical or sexual abuse (Fig. S9, panel B). In people with exposure to a single maltreatment type, the wellbeing index was most significantly influenced by emotional neglect (mean = −0.373; Fig. S10).
The x-axis shows the childhood maltreatment sum score which represents number of childhood traumas in each group (0 = no trauma, 4 = experienced all four types of traumas). The y-axis is the wellbeing index z-score. The mean of wellbeing index z-score for each group is shown above each violin plot. The mean difference in wellbeing index between all categories was examined using Kruskal–Wallis test (p < 2.2 × 10−16). The pairwise mean difference between two adjacent groups employed the Wilcoxon test and the p-values are presented. The interquartile range is represented by vertical black lines inside the violin plots, and the dotted horizontal line is the median wellbeing index score in the sample (n = 45,723). The number of participants in each group is shown at the bottom of each category.
Childhood maltreatment GWAS and genetic correlations
Three loci exceeded genome-wide significance for association with childhood maltreatment: chromosome 7 (rs1015511, p = 1.7 × 10−12), chromosome 15 (rs4702, p = 4.3 × 10−12) and chromosome 16 (rs2043596, p = 2.4 × 10−8) (Fig. S6J). There was a significant negative rg between childhood maltreatment sum score and wellbeing index (rg = −0.56, p = 8.21 × 10−54) and its components (rg = −0.60 to −0.37), and a positive rg with negative indicators of mental health (Fig. S11). The strongest negative rg was for family satisfaction (rg = −0.60, p = 2.61 × 10−46) and the strongest positive rg for depressive symptoms (rg = 0.57, p = 1.61 × 10−37).
Psychiatric diagnoses and wellbeing
The mean wellbeing index z-score was negative for 16 self-reported psychiatric conditions (−1.66 to −0.22) (Fig. S12). Some psychiatric diagnoses were rare in the population, precluding formal statistical analysis. The most frequent psychiatric conditions – “depression” (N = 9,944; 7.7%) and “Anxiety, nerves or generalised anxiety disorder” (N = 6,508; 5%) were associated with lower mean wellbeing index z-score of −0.355 and −0.289, respectively. Of mental illness groups with >200 participants (>0.2% population frequency), the largest mean differences were with over-eating/binge-eating, social-anxiety/social-phobia, OCD, and bipolar-mania.
Discussion
A wellbeing index score was constructed and evaluated phenotypically and genetically in the UK Biobank. The wellbeing index had moderate to high correlation with its subcomponents (rg = 0.65–0.91, rp = 0.55–0.77) and moderate to low negative correlation with adverse mental health indicators (rg = −0.71 to −0.50, rp = −0.39 to −0.22). The wellbeing GWAS had five genome-wide significant associated loci, with a relatively high SNP-heritability. Despite very small estimated effects on phenotypic variance, the wellbeing index PGS significantly predicted the wellbeing score and correlated items in an independent sample. Moreover, both childhood maltreatment and psychiatric illness were associated with lower wellbeing.
Strengths and weaknesses
While previous meta-analyses of GWAS summary statistics for correlated phenotypes has significantly advanced our understanding of wellbeing genetics [8, 10, 11], these methods have limitations. First, employing genetic-based methods on aggregated data ignores clinical and genetic heterogeneity, and precludes detailed analysis of corresponding phenotypic environmental modifiers. Second, the SNP-heritability of the resulting phenotype is usually reduced due to heterogeneity in the phenotypes (e.g., SNP-heritability of “wellbeing spectrum” derived from multi-trait meta-analysis is only ~0.02 [10]). Herein, we used the same measures for all participants, making it possible to precisely examine rp and rg relationships. We demonstrated that SNP-heritability of the wellbeing index score was slightly higher than previously reported wellbeing SNP-heritability (h2 = 8.6% vs. ~4.0–6.4%) [8,9,10]. Thus, as covariance is greater due to elevated SNP-heritability, the wellbeing index improves reliability for genetic correlation analysis. However, while the present wellbeing index slightly increased SNP-heritability estimates compared to those previously reported, most of the causal variants for well-being were not discovered, and SNP-heritability remains much smaller than the heritability estimates derived from twin studies (phenotypic heritability of ~36% [7] vs. SNP-heritability of 8.6%). This ‘missing heritability’ highlights the inability of conventional genetic methods to uncover the complete variance of causal variants for complex traits such as wellbeing, which are likely the result of an intricate interaction of genes and environmental factors in the presence of heterogeneity, pleiotropy, and epistasis. The challenge to close the gap between twin and SNP-heritability estimates likely requires alternative analytic approaches and carefully phenotyped cohorts, with more delicate methods such as machine learning and incorporation of non-linear models of gene-gene and gene-environment effects [40, 41], which seem promising avenues that may provide better understanding of networks involving complex interrelated phenotypes [42].
Although mental illness can influence wellbeing, we defined wellbeing in the entire population – including those with a mental illness – to ensure that findings were closely representative of a general population, within inherent cohort selection biases [43]. While the mean wellbeing index in participants reporting a psychiatric condition was lower than the general population, some individuals in each mental illness group had a positive z-score, indicating that even within a background of mental illness an individual’s wellbeing can be positive – supporting the notion that wellbeing is not simply the absence of mental illness [44,45,46]. However, these results should be interpreted with caution, as psychiatric conditions herein were self-reported, not mutually exclusive (due to psychiatric comorbidities), and could have been current or historical, with variable impacts on current wellbeing. Finally, we cannot conclude causation or infer direction of effect between psychiatric illness and wellbeing, as effects are likely bidirectional. However, the negative rg between wellbeing index and major psychiatric illnesses implies that relationships might be partly due to overlapping genes or gene-environmental correlations influencing both outcomes.
While the wellbeing index was derived from the baseline questionnaire, childhood maltreatment data was collected in the follow-up mental health assessment, 6–10 years later. We did not consider this a methodological concern given that the trauma questionnaire asked retrospectively about childhood events. Notably, we observed discrepancies relating to response endorsement for physical neglect – this potential issue has not, to our knowledge, been reported previously – hence we recommend caution with the use of Data-Field:20491, which was excluded herein. The stepwise decline in mean wellbeing index with increasing frequency of each individual type of maltreatment, as well as maltreatment accumulation, is consistent with existing evidence indicating that frequency and accumulation of different trauma types are key factors that can influence the way childhood maltreatment affects adults [15, 47, 48]. Furthermore, the largest effect on wellbeing index was for emotional rather than physical and sexual maltreatments – consistent with a recent report [49]. Collectively, emotional maltreatment appears to have profound consequences on wellbeing and mental health, potentially due to adverse developmental consequences in understanding and controlling emotions and cognitive development.
A strong correlation of rg and rp was found across all phenotypes (r = 0.97), with rg consistently higher than rp (e.g., loneliness and neuroticism rg = 0.79, rp = 0.50) – consistent with comparisons of genotype/phenotype correlations of other traits [50]. We note that the nature of rp and rg are somewhat distinct, with the former comparing individual-level data (and different variable types), and the latter comparing summary statistics across a population. Therefore, these findings should be interpreted with caution. A stronger rg for wellbeing-related phenotypes has previously been reported [12], and a recent study observed that even phenotypically uncorrelated mental health profiles can be genetically correlated, suggesting either a genetic overlap that is distinct from clinical overlap, or unique environmental factors impact the phenotype in the presence of pleiotropy [51]. The stronger rg between two traits may indicate that although shared genetic variants influence both phenotypes, environmental factors may impact them differently [as discussed [50]]. Amongst negative mental health indicators, loneliness had the strongest negative rg with the wellbeing index, as well as its subcomponents. This further highlights the importance of loneliness in wellbeing [12], which despite a moderate to weak rp, shows strong rg [52]. Interestingly, loneliness had the strongest rg with the financial satisfaction subcomponent, despite its stronger rp with happiness, family- and friendship-satisfaction. The rg between wellbeing index and depressive symptoms or neuroticism (−0.67 or 0.55, respectively) were weaker than previously reported relationships with subjective wellbeing (−0.81 or −0.75, respectively) [8]. Factors influencing these estimates include different phenotype measurement, cohort size, and SNPs employed (e.g., the present study excluded the Major Histocompatibility Complex (MHC) region for rg estimates). Furthermore, the genetic correlation between the wellbeing index and Okbay’s subjective wellbeing is 0.77, indicating the two scores are not identical; possibly due to inclusion of more elements of life satisfaction than affect in our score, which would diminish apparent relationships with neuroticism and depressive symptoms. One of the strengths of our study is using the same cohort and measures for different phenotypes, which makes genetic and phenotypic correlations more comparable – though we acknowledge that consistent reporting biases (e.g. relating to social desirability, or emotional state at the time of reporting) may influence associations derived from a single cohort.
The wellbeing index GWAS revealed five independent genome-wide significant loci; three consistent with previous reports [9, 10]. The chromosome 2 locus was previously associated with hedonic wellbeing [10]. Tagging SNP rs373377070 (max-p = 2.7 × 10−9) lies in an intron of FSHR (Follicle Stimulating Hormone Receptor), and influences its expression (GTeX v8; p = 1.1 × 10−11 [53]). Knockout mice (fshr−/−) display enhanced anxiety- and depression-like behaviours, and modulate gene expression in mood-mediating brain regions [54]. Two additional significant wellbeing-associated loci lie in the MHC/HLA region on chromosome 6, which is robustly associated with psychiatric conditions [30, 55]. One of these loci, tagged by rs3131073, was previously associated with positive affect and wellbeing spectrum [9, 10]. This suggests potentially pleiotropic effects of the MHC region on wellbeing and psychiatric illnesses. Two new loci identified on chromosomes 11 and 14 had minimal support from adjacent SNPs and may be spurious associations. Associations with related traits, including childhood maltreatment [16], are consistent with previously reported loci. Tissue-specific expression of mapped wellbeing-related genes using MAGMA showed significant enrichment of expression in brain tissues, consistent with previous reports [9]. Furthermore, the wellbeing index PGS significantly predicted the wellbeing index score and related phenotypes in the independent sample, albeit accounting for a small percentage of variance. Together, these data support the reliability of results reported herein, and the validity of the wellbeing index phenotype.
Limitations and assumptions
One design limitation of our index score is that only subjective wellbeing indicators were included. The single item that related to psychological wellbeing was excluded due to the time-lag between baseline and follow-up assessments. Additionally, job satisfaction was excluded due to significant data missingness – representing individuals who were unemployed or retired at the time of assessment – which may have skewed the index score and limited generalisability, despite job satisfaction being an important component of wellbeing. Another caveat was the weak correlation (~0.25) of financial- and health-satisfaction to other items in the factor score. However, given their importance on subjective wellbeing [12, 56] and to maximise variable number to generate a quantitative index, we retained financial- and health-satisfaction as components in factor analysis. This led to moderate variance explained (47.5%) by the wellbeing index. Despite these limitations, the factor score had acceptable model fit in the confirmatory factor analysis and is a good proxy for subjective wellbeing.
Our study inherits the limitations of UK Biobank data. All the measures used in this study were self-report, with limited evidence of their validity, and reliability over time. The large sample sizes available through the UK Biobank enable highly significant associations to be derived from weak effect sizes, and caution should be taken in their interpretation. One should consider participant age (40–70 years), recall and reporting bias, and under-representation of psychiatric illnesses when interpreting results [43]. We cannot exclude misreports, longitudinal changes, and self-report bias [57] that could potentially influence our results. The confirmatory sample was ~6 years older than the discovery sample, and childhood maltreatment data came from follow-up assessments of participants who are better educated, with higher socioeconomic status and healthier than the baseline cohort and the general population [58]. Notably, our analyses were restricted to European-ancestry participants, which limits generalisability to other populations.
Finally, genetic correlations were estimated from common SNP variants identified via GWAS, and thus may underestimate pleiotropic contributions of other variant classes (e.g. rare SNVs or CNVs), and non-additive effects. Furthermore, as the SNP-heritability of the traits examined were small, only a small fraction of the causal SNPs for each phenotype contributed to the genetic correlations reported herein. While the LDSR method assumes that this small fraction is a random selection of all causal variants [22], recent studies have shown that these assumptions might not hold in similar complex traits to wellbeing such as human temperament [59]. LDSR also assumes that each phenotype is measured in ways that are specific, reliable and valid, and are well-matched across studies [23], which may not always be true and therefore may bias interpretation. Therefore, genetic correlations should be interpreted with caution, acknowledging the limitations and assumptions of the LDSR method.
Conclusions and recommendations
Using a quantitative index of subjective wellbeing – which encompasses elements of general happiness and satisfaction with family, friendship, health, and financial situation – we explored the genetic and phenotypic relationships between positive and negative aspects of mental health and their correlation with mental illness and childhood maltreatment. The wellbeing index GWAS suggested that improving the measure by using multiple indicators of wellbeing could be a valid method to increase the explained genetic variance (SNP heritability) of wellbeing; although we note that unrealistic underlying models that are fundamental to PGS and LDSR that assume independent additive effects on traits in isolation from environmental exposures may contribute more substantially to ‘missing heritability’ and limit clinical utility. However, future studies will benefit from a combination of denser phenotyping, improved statistical genetic methods that are based on realistic assumptions (i.e. that reflect networks of genotype-phenotype effects across complex interrelated traits), and probably integration of technologies such as artificial intelligence and machine learning to advance the field. The negative association between childhood maltreatment and wellbeing, especially relating to emotional domains, highlights the importance of early life environment on wellbeing later in life. Most studies focus on the association of childhood maltreatment and psychopathologies; however, our study suggests that we might also need to measure the positive spectrum of mental health in relation to the childhood environment. Furthermore, our work may help to delineate the possible negative effects of maltreatment (i.e. on reduced wellbeing) before they become clinically significant. This is useful to know because indices of wellbeing can enable targeted provision of early support for people with an increased risk of developing psychiatric illnesses, which will segue into mental illness prevention. Finally, reduced wellbeing in the context of psychiatric illness, alongside a negative genetic correlation between wellbeing and psychiatric outcomes, demonstrates an interdependent relationship that can be influenced both by genetic and environmental factors, although directionality and causality remain to be determined in future studies. Determining the direction of effects and causality is possible using longitudinal studies with appropriate experimental design or genetic methods such as Mendelian randomisation. A better understanding of causality can then directly inform population-based interventions at societal or policy levels, to reduce the incidence of mental disorders and improve health and quality of life.
Data availability
The UKB Data described in the manuscript is available to all researchers and can be accessed upon approval of the UK Biobank (https://www.ukbiobank.ac.uk/enable-your-research/apply-for-access). We will return the derived data fields following UKB policy; in due course, they will be available through the UK Biobank Access Management System.
Code availability
Analytic codes used in this study are stored in the UNSW data archive at www.dataarchive.unsw.edu.au, under Research Data Management Plan reference number H0237934, and are available on request.
References
Ryan RM, Deci EL. On happiness and human potentials: a review of research on hedonic and eudaimonic well-being. Annu Rev Psychol. 2001;52:141–66.
Keyes CL, Shmotkin D, Ryff CD. Optimizing well-being: the empirical encounter of two traditions. J Pers Soc Psychol. 2002;82:1007–22.
Diener E, Emmons RA, Larsen RJ, Griffin S. The Satisfaction With Life Scale. J Pers Assess. 1985;49:71–5.
World Health Organization. Development of the World Health Organization WHOQOL-BREF quality of life assessment. Psychol Med. 1998;28:551–8.
Gatt JM, Burton KLO, Schofield PR, Bryant RA, Williams LM. The heritability of mental health and wellbeing defined using COMPAS-W, a new composite measure of wellbeing. Psychiatry Res. 2014;219:204–13.
Jamshidi J, Williams LM, Schofield PR, Park HRP, Montalto A, Chilver MR, et al. Diverse phenotypic measurements of wellbeing: Heritability, temporal stability and the variance explained by polygenic scores. Genes Brain Behav. 2020;19:e12694.
Bartels M. Genetics of wellbeing and its components satisfaction with life, happiness, and quality of life: a review and meta-analysis of heritability studies. Behav Genet. 2015;45:137–56.
Okbay A, Baselmans BM, De Neve JE, Turley P, Nivard MG, Fontana MA, et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet. 2016;48:624–33.
Baselmans BML, Bartels M. A genetic perspective on the relationship between eudaimonic -and hedonic well-being. Sci Rep. 2018;8:14610.
Baselmans BML, Jansen R, Ip HF, van Dongen J, Abdellaoui A, van de Weijer MP, et al. Multivariate genome-wide analyses of the well-being spectrum. Nat Genet. 2019;51:445–51.
Turley P, Walters RK, Maghzian O, Okbay A, Lee JJ, Fontana MA, et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet. 2018;50:229–37.
Baselmans BML, van de Weijer MP, Abdellaoui A, Vink JM, Hottenga JJ, Willemsen G, et al. A genetic investigation of the well-being spectrum. Behav Genet. 2019;49:286–97.
Brainstorm Consortium, Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J et al. Analysis of shared heritability in common disorders of the brain. Science. 2018;360:eaap8757.
Herrenkohl TI, Klika JB, Herrenkohl RC, Russo MJ, Dee T. A prospective investigation of the relationship between child maltreatment and indicators of adult psychological well-being. Violence Vict. 2012;27:764–76.
Edwards VJ, Holden GW, Felitti VJ, Anda RF. Relationship between multiple forms of childhood maltreatment and adult mental health in community respondents: results from the adverse childhood experiences study. Am J Psychiatry. 2003;160:1453–60.
Dalvie S, Maihofer AX, Coleman JRI, Bradley B, Breen G, Brick LA, et al. Genomic influences on self-reported childhood maltreatment. Transl Psychiatry. 2020;10:38.
Warrier V, Kwong ASF, Luo M, Dalvie S, Croft J, Sallis HM et al. Gene–environment correlations and causal effects of childhood maltreatment on physical and mental health: a genetically informed approach. The Lancet Psychiatry. 2021;8:373–86.
Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779.
Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203–9.
Loh PR, Kichaev G, Gazal S, Schoech AP, Price AL. Mixed-model association for biobank-scale datasets. Nat Genet. 2018;50:906–8.
Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826.
Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics Consortium et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5.
Ni G, Moser G, Wray NR, Lee SH, Ripke S, Neale BM, et al. Estimation of genetic correlation via linkage disequilibrium score regression and genomic restricted maximum likelihood. Am J Hum Genet. 2018;102:1185–94.
de Moor MH, Costa PT, Terracciano A, Krueger RF, de Geus EJ, Toshiko T, et al. Meta-analysis of genome-wide association studies for personality. Mol Psychiatry. 2012;17:337–49.
van den Berg SM, de Moor MH, Verweij KJ, Krueger RF, Luciano M, Arias Vasquez A, et al. Meta-analysis of genome-wide association studies for extraversion: findings from the genetics of personality consortium. Behav Genet. 2016;46:170–82.
Nagel M, Jansen PR, Stringer S, Watanabe K, de Leeuw CA, Bryois J, et al. Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat Genet. 2018;50:920–7.
Day FR, Ong KK, Perry JRB. Elucidating the genetic basis of social interaction and isolation. Nat Commun. 2018;9:2457.
Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, et al. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum Mol Genet. 2018;27:3641–9.
Howard DM, Adams MJ, Clarke TK, Hafferty JD, Gibson J, Shirali M, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22:343–52.
Mullins N, Forstner AJ, O’Connell KS, Coombes B, Coleman JRI, Qiao Z et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet. 2021;53:817–29.
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014;511:421–7.
Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51:63–75.
Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51:431–44.
International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF-GC) and OCD CollaborativeGenetics Association Studies (OCGAS). Revealing the complex genetic architecture of obsessive-compulsive disorder using metaanalysis.Mol Psychiatry. 2018;23:1181–8.
Duncan LE, Ratanatharathorn A, Aiello AE, Almli LM, Amstadter AB, Ashley-Koch AE, et al. Largest GWAS of PTSD (N=20 070) yields genetic overlap with schizophrenia and sex differences in heritability. Mol Psychiatry. 2018;23:666–73.
Ge T, Chen CY, Ni Y, Feng YA, Smoller JW. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun. 2019;10:1776.
Ikeda M, Takahashi A, Kamatani Y, Momozawa Y, Saito T, Kondo K, et al. Genome-wide association study detected novel susceptibility genes for schizophrenia and shared trans-populations/diseases genetic effect. Schizophr Bull. 2019;45:824–34.
Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium. Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol Autism. 2017;8:21.
de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219.
Zwir I, Arnedo J, Del-Val C, Pulkki-Råback L, Konte B, Yang SS, et al. Uncovering the complex genetics of human character. Mol Psychiatry. 2018;25:2295–312.
Zwir I, Mishra P, Del-Val C, Gu CC, de Erausquin GA, Lehtimäki T, et al. Uncovering the complex genetics of human personality: response from authors on the PGMRA Model. Mol Psychiatry. 2019;25:2210–3.
Zwir I, Del-Val C, Arnedo J, Pulkki-Raback L, Konte B, Yang SS, et al. Three genetic-environmental networks for human personality. Mol Psychiatry. 2021;26:3858–75.
Fry A, Littlejohns TJ, Sudlow C, Doherty N, Adamska L, Sprosen T, et al. Comparison of sociodemographic and health-related characteristics of UK biobank participants with those of the general population. Am J Epidemiol. 2017;186:1026–34.
Slade M. Mental illness and well-being: the central importance of positive psychology and recovery approaches. BMC Health Serv Res. 2010;10:26.
Kendler KS, Myers JM, Maes HH, Keyes CL. The relationship between the genetic and environmental influences on common internalizing psychiatric disorders and mental well-being. Behav Genet. 2011;41:641–50.
World Health Organization. Prevention of Mental Disorders: Effective Interventions and Policy Options. 2004.
Turner HA, Finkelhor D, Ormrod R. Poly-victimization in a national sample of children and youth. Am J Prev Med. 2010;38:323–30.
Chu DA, Williams LM, Harris AW, Bryant RA, Gatt JM. Early life trauma predicts self-reported levels of depressive and anxiety symptoms in nonclinical community adults: relative contributions of early life stressor types and adult trauma exposure. J Psychiatr Res. 2013;47:23–32.
Dye HL. Is emotional abuse as harmful as physical and/or sexual abuse? J Child Adolesc Trauma. 2020;13:399–407.
Sodini SM, Kemper KE, Wray NR, Trzaskowski M. Comparison of genotypic and phenotypic correlations: cheverud’s conjecture in humans. Genetics 2018;209:941–8.
Roelfs D, Alnaes D, Frei O, van der Meer D, Smeland OB, Andreassen OA, et al. Phenotypically independent profiles relevant to mental health are genetically correlated. Transl Psychiatry. 2021;11:202.
Abdellaoui A, Sanchez-Roige S, Sealock J, Treur JL, Dennis J, Fontanillas P, et al. Phenome-wide investigation of health outcomes associated with genetic predisposition to loneliness. Hum Mol Genet. 2019;28:3853–65.
GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5.
Bi WK, Luan SS, Wang J, Wu SS, Jin XC, Fu YL, et al. FSH signaling is involved in affective disorders. Biochem Biophys Res Commun. 2020;525:915–20.
Tamouza R, Krishnamoorthy R, Leboyer M. Understanding the genetic contribution of the human leukocyte antigen system to common major psychiatric disorders in a world pandemic context. Brain Behav Immun. 2021;91:731–9.
Ng W, Diener E. What matters to the rich and the poor? Subjective well-being, financial satisfaction, and postmaterialist needs across the world. J Pers Soc Psychol. 2014;107:326–38.
Xue A, Jiang L, Zhu Z, Wray NR, Visscher PM, Zeng J, et al. Genome-wide analyses of behavioural traits are subject to bias by misreports and longitudinal changes. Nat Commun. 2021;12:20211.
Davis KAS, Coleman JRI, Adams M, Allen N, Breen G, Cullen B, et al. Mental health in UK Biobank - development, implementation and results from an online questionnaire completed by 157 366 participants: a reanalysis. BJPsych Open. 2020;6:e18.
Cloninger CR, Cloninger KM, Zwir I, Keltikangas-Järvinen L. The complex genetics and biology of human temperament: a review of traditional concepts in relation to new molecular findings. Transl Psychiatry. 2019;9:290.
Acknowledgements
Analyses of these data was supported by the UNSW Scientia PhD Scholarship Scheme (JJ), and will form part of his PhD thesis dissertation. This research was funded in part by the Australian National Health and Medical Research Council (NHMRC) Project Grants 1122816 (JMG), 1066177 (JMF) and Investigator Grant 1176716 (PRS). JMF gratefully acknowledges the Janette Mary O’Neil Research Fellowship in support of this work. This research has been conducted using the UK Biobank Resource. The project was approved by UK Biobank Data Access Committee under Project ID #58534. This research was undertaken with the assistance of resources from the National Computational Infrastructure (NCI), which is supported by the Australian Government. The National Computational Merit Allocation Scheme & UNSW High Performance Computing Resource Allocation Scheme provided support of em5 project (Neuroscience Research Australia; PI A/Prof Janice M Fullerton).
We thank Martin Thompson (Research Technology Services, UNSW) and Andrew Cartwright (NeuRA) for assistance with the transfer of derived phenotype data to the Gadi HPC workspace, and access to UKB genotype data held at UNSW, which formed part of UKB approved project #37103 (PI Professor Wei Wen). We thank Dr Anbu Thalamuthu and Dr Karen Mather (Centre for Healthy Brain Ageing, UNSW) for providing early advice on managing and analysing UKB genotype data. For the purposes of Open Access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript (AAM) version arising from this submission.
Author information
Authors and Affiliations
Contributions
The study was conceptualised by JJ, PRS, JMG and JMF. Analysis methodology was developed by JJ, JMG and JMF, and administrative efforts and resources were provided by JMF, PRS and JMG. Formal analysis was conducted by JJ with supervision by JMF and JMG. The original draft was written by JJ, with review & editing by JJ, PRS, JMG and JMF, who all approved the final manuscript. Funding relevant to the current work was acquired by JJ (UNSW Scientia PhD scholarship) with the support of JMG and JMF, and was supported by grants acquired by JMG, PRS and JMF.
Corresponding author
Ethics declarations
Competing interests
JMG is a stockholder in MAP Biotech Pty Ltd. There are no other conflicts of interest to report, nor competing financial interests in relation to the work described.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jamshidi, J., Schofield, P.R., Gatt, J.M. et al. Phenotypic and genetic analysis of a wellbeing factor score in the UK Biobank and the impact of childhood maltreatment and psychiatric illness. Transl Psychiatry 12, 113 (2022). https://doi.org/10.1038/s41398-022-01874-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41398-022-01874-5
- Springer Nature Limited
This article is cited by
-
Principled distillation of UK Biobank phenotype data reveals underlying structure in human variation
Nature Human Behaviour (2024)
-
Time to Change How We Measure Quality of Life and Well-Being in Autism: A Systematic Review
Review Journal of Autism and Developmental Disorders (2024)
-
Genetic associations between alcohol phenotypes and life satisfaction: a genomic structural equation modelling approach
Scientific Reports (2023)