Abstract
Psychiatric disorders are highly prevalent and display considerable clinical and genetic overlap. Dopaminergic and serotonergic neurotransmission have been shown to play an important role in many psychiatric disorders. Here we aim to assess the genetic contribution of these systems to eight psychiatric disorders (attention-deficit hyperactivity disorder (ADHD), anorexia nervosa (ANO), autism spectrum disorder (ASD), bipolar disorder (BIP), major depression (MD), obsessive-compulsive disorder (OCD), schizophrenia (SCZ) and Tourette’s syndrome (TS)) using publicly available GWAS analyses performed by the Psychiatric Genomics Consortium that include more than 160,000 cases and 275,000 controls. To do so, we elaborated four different gene sets: two ‘wide’ selections for dopamine (DA) and for serotonin (SERT) using the Gene Ontology and KEGG pathways tools, and two’core’ selections for the same systems, manually curated. At the gene level, we found 67 genes from the DA and/or SERT gene sets significantly associated with one of the studied disorders, and 12 of them were associated with two different disorders. Gene-set analysis revealed significant associations for ADHD and ASD with the wide DA gene set, for BIP with the wide SERT gene set, and for MD with the core SERT set. Interestingly, interrogation of a cross-disorder GWAS meta-analysis of the eight psychiatric conditions displayed association with the wide DA gene set. To our knowledge, this is the first systematic examination of genes encoding proteins essential to the function of these two neurotransmitter systems in these disorders. Our results support a pleiotropic contribution of the dopaminergic and serotonergic systems in several psychiatric conditions.
Similar content being viewed by others
Introduction
Psychiatric disorders represent a major health problem, affecting 29.2% of the worldwide population at some point during their lifetime [1], and are associated with considerable distress and functional impairment [2]. They constitute a set of complex traits that result from the interaction of genetic and environmental risk factors, with heritability ranging from 40 to 80%, depending on the disorder, as estimated by twin studies [3].
Interestingly, psychiatric disorders display considerable clinical overlap among them [4,5,6], and the presence of comorbidity is associated with increased severity and difficulty of treatment. Recent studies have shown strong genetic correlations among psychiatric phenotypes [7, 8], and the latest genome-wide association study (GWAS) meta-analysis conducted on eight psychiatric disorders (attention-deficit hyperactivity disorder (ADHD), anorexia nervosa (ANO), autism spectrum disorder (ASD), bipolar disorder (BIP), major depression (MD), obsessive-compulsive disorder (OCD), schizophrenia (SCZ) and Tourette’s syndrome (TS)) found that 75% of the LD-independent associated regions (109 out of 146) were associated with more than one disorder [9]. These results suggest that the high levels of comorbidity found in psychiatric disorders may be explained, at least in part, by shared genetic risk factors, supporting the existence of a set of genes that confer relatively broad liability to psychiatric disorders [9].
Dopamine (DA) and serotonin (SERT) are two important neurotransmitters that participate in the regulation of a wide range of essential functions of the organism (e.g. motor control, cognition, motivation, regulation of emotions or reward), and have been related to the physiopathology and treatment of many psychiatric disorders.
Dopaminergic dysfunction has been described in ADHD, ASD, OCD, TS, SCZ, mood disorders, and substance use disorders (SUD) among others [10]. For example, the positive symptoms of SCZ seem to be associated with hyperdopaminergic neurotransmission, especially in the mesolimbic system, while the negative symptoms and cognitive deficits might be caused by hypodopaminergic activity in the mesocortical pathway [11]. Therefore, most of the antipsychotic treatments block the dopamine D2 receptor (e.g. chlorpromazine and haloperidol), and some of them are combined with serotonin 5-HT2A receptor antagonists (e.g. clozapine and risperidone). Moreover, several studies have reported a reduction of dopaminergic receptor density in several brain regions of ADHD patients [12], which is in agreement with the mechanism of action of the ADHD treatments, like methylphenidate or amphetamine, that enhance dopamine transmission in prefrontal cortex [13].
Regarding the serotonergic system, there is strong evidence supporting its role in MD and in other mood disorders based on available pharmacological treatments. The serotonin hypothesis of depression postulates that reduced serotonin signaling is a risk factor in its etiology [14, 15], which is supported by the most effective antidepressant treatment, based on the use of serotonin selective reuptake inhibitors (SSRIs), which inhibit the serotonin transporter increasing the extracellular levels of the neurotransmitter [16]. However, the causal mechanisms connecting low serotonin signaling and depression are still unknown. The role of the serotonergic system in other psychiatric disorders like anxiety, SCZ, ADHD, or ASD is still unclear and further studies are needed [17]. Even so, all these psychiatric conditions seem to be related to serotonin dysfunction, and many psychotropic drugs interfere more or less directly with this system [18].
The contribution of these neurotransmitter systems to several psychiatric conditions is well known, as shown by the fact that several standard pharmacological approaches target these systems. However, drugs used nowadays that target the monoaminergic system are very unspecific. Genetic studies might help to identify more specific drug targets that share efficacy but add specificity. Dopaminergic and serotonergic neurotransmission have been widely investigated through candidate-gene association studies in many psychiatric disorders, which have mainly assessed genetic variants in core genes encoding DA and SERT transporters, receptors, and metabolizers [19,20,21,22]. One of the most studied genes is SLC6A3, encoding the dopaminergic transporter (DAT), with multiple variants mapping at this locus, including rare variants, copy number variations (CNVs), variable number of tandem repeats (VNTRs), and single nucleotide polymorphisms (SNPs) that have been found associated with several psychiatric conditions [23]. Although genetic variants in the serotonin transporter (SERT), encoded by SLC6A4, have also been extensively studied in psychiatric behavioral genetics, these different studies failed to obtain consistent results [24]. Interestingly, the outcome of GWAS evidenced that the classical serotonergic and dopaminergic candidate genes, like transporters (SLC6A3 and SLC6A4) and receptors are not significantly associated with any psychiatric disorder (https://www.ebi.ac.uk/gwas/home), with one exception, DRD2, that was found associated with both MD and SCZ [25,26,27].
In this study, we systematically explored the contribution of common variants in genes involved in dopaminergic and serotonergic neurotransmission in eight psychiatric disorders studied individually and in combination, using GWAS data from the Psychiatric Genomics Consortium (PGC).
Materials and methods
DA and SERT gene selection
We first obtained two core gene sets through manual curation (DA core with 12 genes, and SERT core with 23 genes), containing the well-known dopaminergic or serotonergic genes (neurotransmitter receptors: DRD1-5, HTR1A-B, HTR1D-F, HTR2A-C, HTR3A-E, HTR4, HTR5A, HTR6, HTR7; transporters: DAT1/SLC6A3, 5HTT/SLC6A4; and enzymes involved in their anabolism or catabolism: DDC, TH, TPH1, TPH2, DBH, COMT, MAOA, MAOB) (Fig. 1A).
A Strategy for the selection of the serotonin (SERT) and dopamine (DA) gene sets using Gene Ontology and KEGG’s pathways for the wide sets and manual curation for the core sets. All genes of the core gene sets are also included in their corresponding wide gene set. B Description of samples used in this study. The area of the circles is proportional to the sample size of the disorder.
For a wide selection of genes encoding proteins involved in dopamine and serotonin neurotransmission and signaling pathways we used two main databases, GO and KEGG. We queried the GO database (Gene Ontology Consortium, http://www.geneontology.org/, September 2016) by the search terms “dopamine” or “serotonin/serotonergic”. The resulting list of GO terms (89 for dopamine and 78 for serotonin/serotonergic) was filtered to keep only those of “biological process” ontology source and with presence of the search term as part of the GO identifier. Then, we examined the hierarchical tree structure of the GO database to filter out the most specific terms (child terms) that were contained within broader ones (parental terms). The final GO terms selection included: “dopamine transport” (GO:0015872), “dopamine receptor signaling pathway” (GO:0007212), “dopamine receptor binding” (GO:0050780), “synaptic transmission, dopaminergic” (GO:0001963), “dopaminergic neuron axon guidance” (GO:0036514), “dopaminergic neuron differentiation” (GO:0071542), “response to dopamine” (GO:1903350), “dopamine metabolic process” (GO:0042417), “serotonin receptor signaling pathway” (GO:0007210), “serotonin transport” (GO:0006837), “serotonin metabolic process” (GO:0042428) and “serotonin production involved in inflammatory response” (GO:0002351). For the exploration of KEGG pathways (https://www.genome.jp/kegg/pathway.html, September 2016) we followed a similar procedure, using the terms “dopamine” or “serotonin” as keywords, and only those pathways more tightly related to neurotransmitters’ function were kept: “dopaminergic synapse” (hsa04728) and “tyrosine metabolism” (hsa00350), and “serotonergic synapse” (hsa04726) and “tryptophan metabolism” (hsa00380). The human entries obtained were then filtered for more inclusive and non-disease pathways.
We elaborated the dopaminergic (DA wide, 275 genes) and serotonergic (SERT wide, 176 genes) gene sets with human genes belonging to the final GO categories plus the selected KEGG pathways (Supplementary Table S1). Although GO and KEGG are based on experimentally supported annotations, we confirmed that these two lists contained the corresponding core genes. We also checked the expression of the included genes in the human brain using the GTEx database (https://gtexportal.org), and 95% of them were found to be expressed in brain tissues (Supplementary Table S2).
As the summary statistics used in our analyses did not include the X chromosome, we filtered out those genes from our gene sets. In the case of DA genes, 10 genes were removed: two core genes (MAOA and MAOB) and 8 other genes from the wide set (ATP7A, AGTR2, FLNA, GPR50, GRIA3, HPRT1, PPP2R3B, and VEGFD). And 8 genes were excluded for SERT: three core genes (MAOA, MAOB, and HTR2C) and 5 other genes from the wide set (ATP7A, ARAF, ASMT, CACNA1F, and GPM6B) (Fig. 1A).
Finally, four gene lists were obtained: (i) DA core (10 genes); (ii) DA wide (265 genes); (iii) SERT core (20 genes); and (iv) SERT wide (168 genes) (Fig. 1A).
Gene symbols were converted to NCBI Entrez gene ID with the DAVID Gene ID Conversion Tool (https://david.ncifcrf.gov/conversion.jsp) for downstream analysis.
Data used
We inspected the four DA and SERT gene sets in European samples of eight psychiatric disorders and in the meta-analysis of all of them (cross-disorder meta-analysis; CD-MA) [9] using publicly available GWAS summary statistics of attention-deficit hyperactivity disorder (ADHD) [28], anorexia nervosa (ANO) [29], autism spectrum disorder (ASD) [30], bipolar disorder (BIP) [31], major depression (MD) [32], obsessive-compulsive disorder (OCD) [33], schizophrenia (SCZ) [26] and Tourette’s syndrome (TS) [34] (Fig. 1B).
All of them were downloaded from the Psychiatric Genomics Consortium (PGC) webpage (https://www.med.unc.edu/pgc/results-and-downloads/). The MD and CD-MA summary statistics do not include 23andMe data used by the PGC in the MD GWAS.
Gene-based and gene-set association analyses
Gene-based and gene-set association analyses of each phenotype were performed with MAGMA 1.06 [35] using the summary statistics from each individual GWAS meta-analysis and also from the cross-disorder GWAS, which is a meta-analysis of all eight conditions. For the gene-based analysis the SNP-wise mean model was used, in which the test statistic used was the sum of -log(SNP p value) for SNPs located within the transcribed region (defined on NCBI 37.3 gene definitions), with a 0 Kb window around genes. MAGMA accounts for gene size, number of SNPs in a gene and linkage disequilibrium between markers, using as a reference panel the European ancestry samples from the 1000 Genomes Project, phase 3 [36]. The resulting p values were corrected for multiple testing using False Discovery Rate (5% FDR).
Based on the gene-based p values we analysed the four sets of genes described above (DA wide, DA core, SERT wide, and SERT core). MAGMA applies a competitive test to analyse whether the genes of a gene set are more strongly associated with the trait than other genes, while correcting for a series of confounding effects such as gene length and size of the gene set. In our analyses, only genes on autosomes were included. Multiple testing corrections were performed for each disorder separately. As the Bonferroni correction is quite conservative for gene sets strongly overlapped, we used an empirical multiple testing correction implemented in MAGMA, based on a permutation procedure.
Gene-based Manhattan plot
Results of the gene-based analysis of the cross-disorder meta-analysis were plotted in R using the package “qqman” [37].
MetaXcan analyses
We considered all the SNPs located in each DA and SERT gene to infer whether the genetically determined expression of each of those genes correlates with the phenotypes considered in the study. These analyses were carried out on MetaXcan [38] using the summary statistics of each disorder. Prediction models were constructed considering SNPs located within ± 1 Mb from the transcription start site (TSS) of each gene and were trained with RNA-Seq data of 13 GTEx (release V7) brain tissues and whole blood [39]. The SNP covariance matrices were generated using the 1000 Genomes Project Phase 3 [36] EUR genotypes of the prediction model SNPs. For each brain tissue, the threshold for significance was calculated using the Bonferroni correction for multiple testing.
Results
Dopaminergic and serotonergic neurotransmission have been pointed out as involved in many psychiatric disorders. In the present study, we aimed to assess the genetic contribution of these two monoaminergic systems to eight different psychiatric disorders (ADHD, ANO, ASD, BIP, MD, OCD, SCZ, and TS) as well as to the meta-analysis of all of them (CD-MA). For that purpose, we elaborated four different gene sets (DA core, 10 genes; DA wide, 265 genes; SERT core, 20 genes; and SERT wide, 168 genes; Fig. 1A and Supplementary Table S1) that were subsequently interrogated in the summary statistics from nine case-control GWAS meta-analyses (Fig. 1B). Importantly, we found some overlap between the wide datasets, with about 20% of genes in the DA gene sets present also in SERT, and 32% the other way round. One gene from the wide DA gene set (KIF5C) and two from the wide SERT gene set (OR11H7 and OR10J6P) were not present in any of the GWAS datasets inspected.
DA and SERT genes associated with psychiatric disorders
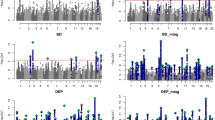
At the gene-wide level, we found 67 genes from DA and/or SERT gene sets significantly associated (5% FDR) with at least one of the studied disorders, and 309 genes were not associated with any of them (Supplementary Table S3). Interestingly, twelve of these genes were associated with two different disorders, eight of them with SCZ and BIP (Fig. 2 and Supplementary Table S3). Eleven out of these twelve genes also showed a significant association with the CD-MA phenotype, DRD2 from the DA core gene set among them. Interestingly, five out of these twelve genes (CACNA1C, CACNA1D, GNAS, GRIN2A and ITR3) belong to both the DA and SERT gene sets, highlighting the importance of those genes that are involved in both monoamine pathways (Supplementary Table S3). Then, we plotted the gene-based results of the cross-disorder meta-analysis in a Manhattan plot and highlighted the DA core and SERT core genes, to visualize the performance of these genes in the combination of all the psychiatric conditions considered in the study (Fig. 3A). As shown in the figure, only the DRD2 gene overcomes the Bonferroni correction for multiple testing, and HTR6 surpasses 5% FDR. Then, we repeated the analysis by highlighting the DA and SERT wide genes and found 10 genes with associations that overcome the Bonferroni correction and 32 additional genes overcoming 5% FDR (Fig. 3B) and Supplementary Table S3). Among them, CACNA1C, present in both DA and SERT gene lists, was the one showing the strongest association in the cross-disorder meta-analysis (raw P = 8.9E-14, Padj = 7.8E-11).
Venn diagrams of the significantly associated DA and SERT genes across the studied disorders (5% False Discovery Rate). Gene names are only provided for those genes significantly associated with more than one disorder (overlapping genes). All the genes overlapping across phenotypes are significantly associated with the phenotype in the cross-disorder meta-analysis, except for GNAS. The area of circles is proportional to the number of significantly associated genes in each disorder. DA dopaminergic gene set, SERT serotonergic gene set, ADHD Attention-Deficit/Hyperactivity Disorder, ASD Autism Spectrum Disorder, BIP Bipolar Disorder, MD Major Depression, SCZ Schizophrenia.
Represented (A) DA core and SERT core genes, and (B) DA wide and SERT wide gene sets. Colored dots correspond to DA genes in pink, SERT genes in blue and overlapping genes in purple. Continuous line: threshold for Bonferroni significance (P = 2.8E-06). Discontinuous line: threshold for 5% False Discovery Rate (P = 4.9E-03).
Gene expression correlates in brain regions in psychiatric disorders
In addition, to identify genes which expression correlates with the phenotypes included in the study, we performed MetaXcan analysis, considering only those phenotypes that show significant association with DA or SERT genes (ADHD, ASD, MD, SCZ, and CD-MA). Taking into account the genes associated at gene level with a given disorder, we found that the expression of CTNNB1 significantly correlates with ADHD in the amygdala (P = 8.9E-04, Z = −3.3246), ATF6B with SCZ in the spinal cord (P = 1.6E-06, Z = 4.7925) and DNM1 and ITPR3 with the phenotype in the CD-MA in the cortex and caudate, respectively (P = 2.6E-04, Z = 3.6516 and P = 1.8E-05, Z = −4.2860) (Table 1 and Supplementary Tables S4–S16). Interestingly, the expression of the DNM1 gene in the cortex also correlates with ADHD (P = 8.5E-03, Z = 2.6308), ASD (P = 5.9E-03, Z = 2.7524), MD (P = 3.0E-03, Z = 2.9630) and SCZ (P = 0.0312, Z = 2.1549) in the same direction, and this gene was found nominally associated with all these disorders in the gene-based analysis (raw PADHD = 2.5E-04, raw PASD = 3.6E-03, raw PMD = 1.5E-03 and raw PSCZ = 0.0156). It is important to note that for most DA and SERT genes (ranging from 77% to 93.6% depending on the tissue and disorder) we could not retrieve information from MetaXcan because it only provides data about those gene models where the predictive power is good enough. For that reason, we decided to perform the analysis also on whole blood as the expression models are better for this tissue and we could retrieve information for about 30% of DA/SERT genes. This analysis highlighted additional genes (ARNTL, FZD3, PPP1CB, PPP2R3C and PRMT5) for SCZ and BIP, and supported some of the results obtained in brain tissues (Table 1 and Supplementary Table S17).
DA and SERT gene sets associated with psychiatric disorders
Finally, we performed a gene-set analysis that revealed interesting significant associations. The DA wide gene set was significantly associated with ADHD (Pperm = 0.023) and ASD (Pperm = 0.025), and the SERT wide gene set with BIP (Pperm = 0.015). Besides, the SERT core (Pperm = 0.018) gene set was significantly associated with MD (Fig. 4 and Supplementary Table S18). Interrogation of the cross-disorder GWAS meta-analysis displayed association with the DA wide gene set (Pperm = 0.044). It should be mentioned that the DA wide gene set was nominally associated also with BIP, TS, and SCZ, in line with the results obtained in the cross-disorder analysis for this neurotransmitter system (Fig. 4). No significant results were found for ANO, OCD, and TS in either the gene-based or in the gene-set analyses after multiple testing correction, maybe due to limited sample sizes (number of patients below 5000; Fig. 1B).
A small circle represents nominally significant p value, a big circle represents significant p value after the empirical multiple testing correction implemented in MAGMA. DA dopaminergic gene set, SERT serotonergic gene set, ADHD Attention-deficit/hyperactivity disorder, ANO Anorexia nervosa, ASD Autism spectrum disorder, BIP Bipolar disorder, MD Major depression, OCD Obsessive-compulsive disorder, TS Tourette’s syndrome SCZ Schizophrenia, CD-MA Cross- disorder.
Discussion
Dopaminergic (DA) and serotonergic (SERT) functions represent good candidates for psychiatric disorders, mainly on the basis of the effectiveness of several pharmacological treatments and their target molecules [10, 23, 40,41,42]. Common genetic variation of genes encoding key participants of these two neurotransmission systems (receptors, transporters, and enzymes) has been broadly studied in psychiatric disorders, mainly through candidate-gene association studies, with several positive findings [23, 43,44,45]. However, except for a few cases [25,26,27], these hits have not been replicated by GWAS, which raises questions on how alterations in these two neurotransmitter systems can actually be causally related to psychiatric conditions, but also on the value of genome-wide significance for functional variation [46].
To our knowledge, this is the first systematic study of the DA and SERT neurotransmitter systems in eight psychiatric disorders (ADHD, ANO, ASD, BIP, MD, OCD, SCZ, and TS) using data from GWAS meta-analyses that include large number of samples. Here, we assess the contribution of genetic variation not only of genes encoding the core participants of DA and SERT functions, but also of a comprehensive list of genes involved in DA and SERT neurotransmission obtained by a systematic database search.
Our results show a contribution to psychiatric disorders of both dopaminergic and serotonergic systems at genetic level. We found several DA and SERT genes associated with some psychiatric disorders, twelve of them with two conditions, and also in the cross-disorder meta-analysis, which underscores the relevance of genetic risk factors in these genes for psychiatric disorders. Among the associated genes we identified the DA core gene coding for the dopamine receptor D2, DRD2, which has been widely studied as a candidate for different psychiatric disorders with some positive findings [47,48,49,50,51], especially in SCZ (recently meta-analyzed by [52]). The association between DRD2 and SCZ observed through candidate-gene studies has subsequently been confirmed through GWAS [26]. Interestingly, a recent GWAS identified association of polymorphisms in DRD2 with depressive symptoms, neuroticism and different traits of the well-being spectrum [53, 54]. These findings are in agreement with ours, obtaining associations of DRD2 with SCZ and MD (Fig. 2 and Supplementary Table S3). Another interesting gene that encodes the calcium voltage-gated channel subunit alpha1 C (CACNA1C), present in both the DA and SERT wide gene lists and found associated by us with BIP and with SCZ (disorders with a very high genetic correlation, rg = 0.7 [9]), displayed the strongest association in the cross-disorder meta-analysis. Noteworthy, CACNA1C polymorphisms have been consistently associated with BIP and SCZ, among other psychiatric disorders [55, 56]. Mutations in this gene have been related to Timothy Syndrome, a monogenic disorder that affects multiple organs and presents with cognitive impairment, major developmental delays, and autism-like behaviors [57]. We also identified the potassium calcium-activated channel subfamily N member 2 (KCNN2) gene, from the SERT wide gene set, associated with BIP and with ASD, as previously described in two studies [30, 31].
Interestingly, two genes emerged from the analysis of gene expression using MetaXcan. First, the gene for inositol 1,4,5-trisphosphate receptor type 3 (ITPR3), found associated with ADHD, SCZ and the CD-MA at gene level in our study, which expression would be altered in the caudate of patients from the CD-MA and in whole blood from ADHD cases. The caudate nucleus is a crucial component of the ventral striatum, part of the basal ganglia that control motor, cognitive control, motivational and emotional processing [58], functions involved in the physiopathology of many psychiatric conditions. Several studies have found alterations both in the volume of the caudate nucleus and in its functional connections with other brain regions in many psychiatric disorders [59,60,61]. Interestingly, the ITPR3 gene has been associated to type 1 diabetes [62], which curiously shows a correlation with risk of ADHD in the offspring [63,64,65]. The other gene that was pointed by this analysis was dynamin 1 (DNM1), which expression significantly correlates not only with the CD-MA, but also with ADHD, ASD, MD, and SCZ, in all cases in the cortex and in the same direction. Rare variants in DNM1 have been identified in patients with a mendelian phenotype, epileptic encephalopathy [66], and in some cases of intellectual disability with seizures [67].
In the cross-disorder meta-analysis study [9], the eight psychiatric disorders used were classified into three groups using exploratory factor analysis (EFA): disorders characterized by compulsive/perfectionistic behaviors (AN, OCD, and TS), mood and psychotic disorders (MD, BIP, and SCZ), and early-onset neurodevelopmental disorders (ASD, ADHD, TS, as well as MD). Our results are in line with this classification, as we identified association of the serotonergic gene sets with BIP and MD, included in the same group, whereas the dopaminergic set was associated with ADHD and ASD, two disorders that conform the third factor.
Although SERT dysfunction has been proposed to be the common denominator in a wide range of neuropsychiatric illnesses [66], our gene-set results link this neurotransmitter system only with BIP (wide gene set) and with MD (core gene set). SERT signaling has been associated with concrete alterations in the intrinsic activity of the brain: an increase of the default-mode network and a decrease of the sensorimotor network. Interestingly, MD is characterized by an increase of internally focused thoughts and an inhibited psychomotor behavior and affectivity, an imbalance that has been also detected in BIP [66], but not in SCZ, the third of this genetically-correlated group of psychiatric disorders [9].
Gene-set analysis of the DA system showed association of the wide selection with ADHD and with ASD, two disorders that co-occur frequently [68] and with a high genetic correlation (rg = 0.37) [9]. Association of dopaminergic genes with these psychiatric conditions, especially the DA core genes, has been broadly studied using different strategies, including candidate-gene association studies or exome/genome sequencing [69,70,71,72]. Interestingly, the cross-disorder meta-analysis displayed association with the DA wide gene set, underscoring the importance that this neurotransmitter has in many psychiatric disorders [73, 74]. Finally, it is important to note that for SCZ, none of the studied gene sets were found significantly associated with the disorder, despite the large sample size of this study and the proven role of these neurotransmitter systems from pharmacological studies. However, we found many DA/SERT genes associated with SCZ at gene level (54 out of 376) and a nominal association with the DA wide gene set, highlighting the importance of this pathway in the pathophysiology of this disorder. Further studies are needed to confirm these results.
The primary drug target for antidepressants, antipsychotics, and stimulants are proteins that have a role in serotonin and dopamine neurotransmission and, accordingly, are encoded by genes belonging to these gene sets in our analysis. While serotonergic drugs have traditionally been used in affective disorders (e.g., selective serotonin reuptake inhibitors targeting the serotonin transporter) and dopaminergic drugs are used in schizophrenia (dopamine receptor antagonists) and ADHD (targeting the dopamine transporter), such distinction becomes more and more blurred. Compounds such as quetiapine or aripiprazole also affect monoaminergic transmitter systems, but are prescribed in depression, bipolar disorder, and schizophrenia. Therefore, the traditional grouping became obsolete and it was replaced by the Neuroscience-based Nomenclature [75]. Given that dopaminergic gene sets, in our analysis, were associated with most of the investigated traits (Fig. 3), such a trans-diagnostic efficacy of dopamine-modulating drugs might be explained on the genetic level. It must be mentioned however that association of the DA gene set with a phenotype does not indicate any direction so that both enhancers, as well as inhibitors of dopamine neurotransmission, are effective in distinct phenotypes. Further genetic studies might refine such findings in that phenotypes characterized by hypo-, hyper-, and mixed dopaminergic states can be discerned, and these phenotypes might well cut across traditional phenotypes. Given the broad, unspecific efficacy of drug targeting the monoaminergic system, genetic studies might also help to identify drug target that shares efficacy but add specificity.
Some strengths and limitations of our study should be discussed. (i) We performed a systematic study of the genes encoding key proteins for the dopaminergic and serotonergic neurotransmitter systems in eight psychiatric disorders and the meta-analysis of all of them. At gene level, we identified twelve genes significantly associated with more than one disorder. However, none of them was associated with more than two disorders, being BIP and/or SCZ one of these conditions in most cases (10 out to 12). This brings up the issue that these two disorders could drive some of the results obtained because of their large sample sizes. (ii) Also, some individual genes could be involved in other neurotransmitter systems and several neuronal functions. Therefore, we decided to perform a combined analysis to study the contribution of genes enriched in DA or SERT pathways to the pathophysiology of these disorders using a competitive gene-set analysis. Moreover, DA and SERT neurotransmission are functionally interconnected, with several genes participating in both pathways, and this could have some impact on the results obtained in the gene-set analysis. In addition, it is important to note that we might be missing information from some genes as these pathways are still not very well defined, and also because the X-chromosome genes are not included in most GWAS. (iii) As we know, assigning SNPs to genes based on their location with respect to those genes is not the best approach, and for that reason we also performed a more functional analysis on Metaxcan. However, as this software only provides information about those gene models where the predictive power is good enough, we could not retrieve information from most DA and SERT genes, especially in brain tissues. (iv) Finally, no significant results were found for ANO, OCD, and TS, neither in the gene-based nor in the gene-set analyses, maybe due to limited sample sizes. In addition, this study was performed on individuals with European ancestry, so, further studies should be performed in larger samples and in other populations to confirm these results.
To our knowledge, this is the first systematic genetic study of the dopaminergic and serotonergic neurotransmitter systems in eight psychiatric disorders (ADHD, ANO, ASD, BIP, MD, OCD, SCZ, and TS) and in the meta-analysis of all of them. At gene level, we identified association of 67 DA and/or SERT genes with at least one of the studied disorders, twelve of them associated with two conditions. Gene-set analysis revealed significant associations with the DA gene sets for ADHD, ASD, and the cross-disorder GWAS meta-analysis, and with SERT gene sets for MD and BIP. The results obtained support a cross-disorder contribution of these two neurotransmitters systems in several psychiatric conditions.
References
Steel Z, Marnane C, Iranpour C, Chey T, Jackson JW, Patel V, et al. The global prevalence of common mental disorders: a systematic review and meta-analysis 1980-2013. Int J Epidemiol. 2014;43:476–93.
Whiteford HA, Degenhardt L, Rehm J, Baxter AJ, Ferrari AJ, Erskine HE, et al. Global burden of disease attributable to mental and substance use disorders: Findings from the Global Burden of Disease Study 2010. Lancet 2013;382:1575–86.
Polderman TJ, Benyamin B, de Leeuw CA, Sullivan PF, van Bochoven A, Visscher PM, et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet. 2015;47:702–9.
Andrews G, Slade T, Issakidis C. Deconstructing current comorbidity: data from the Australian National Survey of Mental Health and Well-Being. Br J Psychiatry. 2002;181:306–14.
Plana-Ripoll O, Pedersen CB, Holtz Y, Benros ME, Dalsgaard S, de Jonge P, et al. Exploring comorbidity within mental disorders among a danish national population. JAMA Psychiatry. 2019;76:259–70.
Bijl RV, Ravelli A, van Zessen G. Prevalence of psychiatric disorder in the general population: results of The Netherlands Mental Health Survey and Incidence Study (NEMESIS). Soc Psychiatry Psychiatr Epidemiol. 1998;33:587–95.
Martin J, Taylor MJ, Lichtenstein P. Assessing the evidence for shared genetic risks across psychiatric disorders and traits. Psychol Med. 2018;48:1759–74.
Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, Duncan L, et al. Analysis of shared heritability in common disorders of the brain. Science 2018;360:eaap8757.
Lee. PH, Cross-Disorder Group of the Psychiatric Genomics Consortium V, Won H, Feng Y-CA, Rosenthal J, Zhu Z, Anttila V, Won H, et al. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell 2019;179:1469–1482.e11.
Ayano G. Dopamine: receptors, functions, synthesis, pathways, locations and mental disorders: review of literatures. J Ment Disord Treat. 2016;2:1–4.
Li P, Snyder GL, Vanover KE. Dopamine targeting drugs for the treatment of schizophrenia: past, present and future. Curr Top Med Chem. 2016;16:3385–403.
Tripp G, Wickens JR. Neurobiology of ADHD. Neuropharmacology 2009;57:579–89.
Sharma A, Couture J. A review of the pathophysiology, etiology, and treatment of attention-deficit hyperactivity disorder (ADHD). Ann Pharmacother. 2014;48:209–25.
Albert PR, Benkelfat C. The neurobiology of depression-revisiting the serotonin hypothesis. II. Genetic, epigenetic and clinical studies. Philos Trans R Soc B Biol Sci 2013;368:20120535.
Coppen A. The biochemistry of affective disorders. Br J Psychiatry. 1967;113:1237–64.
Cipriani A, Furukawa TA, Salanti G, Chaimani A, Atkinson LZ, Ogawa Y, et al. Comparative efficacy and acceptability of 21 antidepressant drugs for the acute treatment of adults with major depressive disorder: a systematic review and network meta-analysis. Lancet 2018;391:1357–66.
Lin SH, Lee LT, Yang YK. Serotonin and mental disorders: a concise review on molecular neuroimaging evidence. Clin Psychopharmacol Neurosci. 2014;12:196–202.
Marazziti D. Understanding the role of serotonin in psychiatric diseases. F1000Research. 2017;6:180.
Gizer IR, Ficks C, Waldman ID. Candidate gene studies of ADHD: a meta-analytic review. Hum Genet. 2009;126:51–90.
Talkowski ME, Bamne M, Mansour H, Nimgaonkar VL. Dopamine genes and schizophrenia: case closed or evidence pending? Schizophr Bull. 2007;33:1071–81.
Sampaio AS, Lins RMP, Daltro-Oliveira R, Quarantini LC, Rosário MC, Miguel EC, et al. Genetic association studies in obsessive-compulsive disorder. Arch Clin Psychiatry. 2013;40:177–90.
Gatt JM, Burton KLO, Williams LM, Schofield PR. Specific and common genes implicated across major mental disorders: a review of meta-analysis studies. J Psychiatr Res. 2015;60:1–13.
Salatino-Oliveira A, Rohde LA, Hutz MH. The dopamine transporter role in psychiatric phenotypes. Am J Med Genet Part B Neuropsychiatr Genet. 2018;177:211–31.
Margoob MA, Mushtaq D. Serotonin transporter gene polymorphism and psychiatric disorders: is there a link? Indian J Psychiatry. 2011;53:289–99.
Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50:381–9.
Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014;511:421–7.
Howard DM, Adams MJ, Clarke TK, Hafferty JD, Gibson J, Shirali M, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22:343–52.
Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51:63–75.
Duncan L, Yilmaz Z, Gaspar H, Walters R, Goldstein J, Anttila V, et al. Significant locus and metabolic genetic correlations revealed in genome-wide association study of anorexia nervosa. Am J Psychiatry. 2017;174:850–8.
Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51:431–44.
Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51:793–803.
Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Genome- wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–81.
International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF-GC) and OCD Collaborative Genetics Association Studies (OCGAS). Revealing the complex genetic architecture of obsessive-compulsive disorder using meta-analysis. Mol Psychiatry. 2018;23:1181–8.
Yu D, Sul JH, Tsetsos F, Nawaz MS, Huang AY, Zelaya I, et al. Interrogating the genetic determinants of Tourette’s syndrome and other tiC disorders through genome-wide association studies. Am J Psychiatry. 2019;176:217–27.
Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–1.
Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature 2015;526:68–74.
Turner SD qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv. 2014;005165.
Barbeira AN, Dickinson SP, Bonazzola R, Zheng J, Wheeler HE, Torres JM, et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun. 2018;9:1825.
GTEx Consortium. et al. The Genotype- Tissue Expression (GTEx) project. Nat Publ Gr. 2013;45:580–5.
Yokokura M, Terada T, Bunai T, Nakaizumi K, Kato Y, Yoshikawa E, et al. Alterations in serotonin transporter and body image-related cognition in anorexia nervosa. NeuroImage Clin. 2019;23:101928.
Mohammadi A, Rashidi E, Amooeian VG. Brain, blood, cerebrospinal fluid, and serum biomarkers in schizophrenia. Psychiatry Res. 2018;265:25–38.
Banerjee E, Nandagopal K. Does serotonin deficit mediate susceptibility to ADHD? Neurochem Int. 2015;82:52–68.
Sinopoli VM, Burton CL, Kronenberg S, Arnold PD. A review of the role of serotonin system genes in obsessive-compulsive disorder. Neurosci Biobehav Rev. 2017;80:372–81.
Hou YW, Xiong P, Gu X, Huang X, Wang M, Wu J. Association of serotonin receptors with attention deficit hyperactivity disorder: a systematic review and meta-analysis. Curr Med Sci. 2018;38:538–51.
Zai G, Barta C, Cath D, Eapen V, Geller D, Grünblatt E. New insights and perspectives on the genetics of obsessive-compulsive disorder. Psychiatr Genet. 2019;29:142–51.
Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: from polygenic to omnigenic. Cell 2017;169:1177–86.
Paschou P. The genetic basis of Gilles de la Tourette Syndrome. Neurosci Biobehav Rev. 2013;37:1026–39.
Li MD, Ma JZ, Beuten J. Progress in searching for susceptibility loci and genes for smoking- related behaviour. Clin Genet. 2004;66:382–92.
Rady A, Elsheshai A, Mokhtar M, Abou el Wafa H, Elkholy O. The A1 allele of the DRD2 TaqA1/A2 polymorphism as risk factor for PTSD. Eur J Psychiatry. 2011;25:144–9.
Nisoli E, Brunani A, Borgomainerio E, Tonello C, Dioni L, Briscini L, et al. D2 dopamine receptor (DRD2) gene Taq1A polymorphism and the eatingrelated psychological traits in eating disorders (anorexia nervosa and bulimia) and obesity. Eat Weight Disord. 2007;12:91–6.
Colizzi M, Iyegbe C, Powell J, Ursini G, Porcelli A, Bonvino A, et al. Interaction between functional genetic variation of DRD2 and cannabis use on risk of psychosis. Schizophr Bull. 2015;41:1171–82.
He H, Wu H, Yang L, Gao F, Fan Y, Feng J, et al. Associations between dopamine D2 receptor gene polymorphisms and schizophrenia risk: a PRISMA compliant meta-analysis. Neuropsychiatr Dis Treat. 2016;12:3129–44.
Baselmans BML, Jansen R, Ip HF, van Dongen J, Abdellaoui A, van de Weijer MP, et al. Multivariate genome-wide analyses of the well-being spectrum. Nat Genet. 2019;51:445–51.
Nagel M, Jansen PR, Stringer S, Watanabe K, de Leeuw CA, Bryois J, et al. Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat Genet. 2018;50:920–7.
Heyes S, Pratt WS, Rees E, Dahimene S, Ferron L, Owen MJ, et al. Genetic disruption of voltage-gated calcium channels in psychiatric and neurological disorders. Prog Neurobiol. 2015;134:36–54.
Moon AL, Haan N, Wilkinson LS, Thomas KL, Hall J. CACNA1C: association with psychiatric disorders, behavior, and neurogenesis. Schizophr Bull. 2018;44:958–65.
Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, et al. CaV1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell 2004;119:19–31.
Lanciego JL, Luquin N, Obeso JA. Functional neuroanatomy of the basal ganglia. Cold Spring Harb Perspect Med. 2012;2:a009621.
Duan M, Chen X, He H, Jiang Y, Jiang S, Xie Q, et al. Altered basal ganglia network integration in schizophrenia. Front Hum Neurosci. 2015;9:561.
Macpherson T, Hikida T. Role of basal ganglia neurocircuitry in the pathology of psychiatric disorders. Psychiatry Clin Neurosci. 2019;73:289–301.
Ring HA, Serra-Mestres J. Neuropsychiatry of the basal ganglia. J Neurol Neurosurg Psychiatry. 2002;72:12–21.
Roach JC, Deutsch K, Li S, Siegel AF, Bekris LM, Einhaus DC, et al. Genetic mapping at 3- kilobase resolution reveals inositol 1,4,5-triphosphate receptor 3 as a risk factor for type 1 diabetes in Sweden. Am J Hum Genet. 2006;79:614–27.
Xiang AH, Wang X, Martinez MP, Getahun D, Page KA, Buchanan TA, et al. Maternal gestational diabetes mellitus, type 1 diabetes, and type 2 diabetes during pregnancy and risk of ADHD in offspring. Diabetes Care. 2018;41:2502–8.
Ji J, Chen T, Sundquist J, Sundquist K. Type 1diabetes in parents and risk of attention deficit/hyperactivity disorder in offspring: a population-based study in Sweden. Diabetes Care. 2018;41:770–4.
Nielsen PR, Benros ME, Dalsgaard S. Associations between autoimmune diseases and attention-deficit/hyperactivity disorder: a nationwide study. J Am Acad Child Adolesc Psychiatry. 2017;56:234–240.e1.
Euroepinomics-Res C, Epilepsy Phenome/Genome P, Consortium. De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am J Hum Genet. 2014;95:360–70.
Deciphering Developmental Disorders S. Large- scale discovery of novel genetic causes of developmental disorders. Nature 2015;519:223–8.
Antshel KM, Zhang-James Y, Wagner KE, Ledesma A, Faraone SV. An update on the comorbidity of ADHD and ASD: A focus on clinical management. Expert Rev Neurother. 2016;16:279–93.
Faraone SV, Larsson H. Genetics of attention deficit hyperactivity disorder. Mol Psychiatry. 2019;24:562–75.
Hamilton PJ, Campbell NG, Sharma S, Erreger K, Herborg Hansen F, Saunders C, et al. De novo mutation in the dopamine transporter gene associates dopamine dysfunction with autism spectrum disorder. Mol Psychiatry. 2013;18:1315–23.
Hettinger JA, Liu X, Hudson ML, Lee A, Cohen IL, Michaelis RC, et al. DRD2 and PPP1R1B (DARPP-32) polymorphisms independently confer increased risk for autism spectrum disorders and additively predict affected status in male-only affected sib-pair families. Behav Brain Funct. 2012;8:19.
Gadow KD, Pinsonneault JK, Perlman G, Sadee W. Association of dopamine gene variants, emotion dysregulation and ADHD in autism spectrum disorder. Res Dev Disabil. 2014;35:1658–65.
Schmidt WJ, Reith MEA (eds.). Dopamine and glutamate in psychiatric disorders. (Totowa, NJ: Humana Press, 2005).
Ptácek R, Kuzelová H, Stefano GB. Dopamine D4 receptor gene DRD4 and its association with psychiatric disorders. Med Sci Monit. 2011;17:RA215–20.
Zohar J, Stahl S, Moller HJ, Blier P, Kupfer D, Yamawaki S, et al. A review of the current nomenclature for psychotropic agents and an introduction to the Neuroscience-based Nomenclature. Eur Neuropsychopharmacol. 2015;25:2318–25.
Acknowledgements
The authors thank the PGC ADHD, ANO, ASD, BIP, MD, OCD, SCZ, TS and cross-disorder working groups for making accessible the data from their studies.
Funding
Major financial support for this research was received by BC from the Spanish ‘Ministerio de Ciencia, Innovación y Universidades’ (RTI2018-100968-B-100), ‘Ministerio de Ciencia e Innovación’ (PID2021-127776OB-I00), ‘Ministerio de Economía y Competitividad’ (SAF2015-68341-R), ‘Ministerio de Sanidad, Servicios Sociales e Igualdad/Plan Nacional Sobre Drogas’ (PNSD 2017I050) and ‘AGAUR/Generalitat de Catalunya’ (2017-SGR-738). The research leading to these results has also received support from the ECNP network “ADHD across the lifespan”, the European Union H2020 Program [H2020/2014-2020] under grant agreements n° 667302 (CoCA), 643051 (MiND) and 728018 (Eat2beNICE) to BC, ‘Ministerio de Sanidad, Servicios Sociales e Igualdad/Plan Nacional Sobre Drogas’ (PNSD 2020I042) to NF-C and Federal Ministry of Education and Research (BMBF, grant number: 01EE1404A) to AR. BT and JC-D were supported by the H2020 CoCA project, and NF-C by ‘Centro de Investigación Biomédica en Red de Enfermedades Raras’ (CIBERER) and by an EMBO short-term fellowship (ASTF 573-2016).
Author information
Authors and Affiliations
Contributions
AR, NF-C and BC conceived and designed the study. JC-D and BT obtained the data, performed the bioinformatic analyses and wrote the manuscript. JC-D, BT and NF-C prepared Figures and Tables. BC and NF-C coordinated the study and supervised the manuscript preparation. All authors contributed to and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interests. This publication reflects only the author’s view and the European Commission is not responsible for any use that may be made of the information it contains.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cabana-Domínguez, J., Torrico, B., Reif, A. et al. Comprehensive exploration of the genetic contribution of the dopaminergic and serotonergic pathways to psychiatric disorders. Transl Psychiatry 12, 11 (2022). https://doi.org/10.1038/s41398-021-01771-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41398-021-01771-3
- Springer Nature Limited
This article is cited by
-
The influence of the COMT Val158Met polymorphism on prefrontal TDCS effects on aggression
Scientific Reports (2024)