Abstract
BRAF inhibitors (BRAFi) have been approved for the clinical treatment of BRAF-mutant metastatic melanoma. Although initial responses to BRAFi are generally favorable, acquired BRAFi resistance emerges rapidly, resulting in treatment failure. Only some of the underlying mechanisms responsible for BRAFi resistance are currently understood. Here, we showed that the genetic inhibition of histone acetyltransferase 1 (HAT1) in BRAF-mutant melanoma cells resulted in BRAFi resistance. Using quantitative immunofluorescence analysis of patient sample pairs, consisting of pre-treatment along with matched progressed BRAFi + MEKi-treated melanoma samples, HAT1 downregulation was observed in 7/11 progressed samples (~63%) in comparison with pre-treated samples. Employing NanoString-based nCounter PanCancer Pathway Panel-based gene expression analysis, we identified increased MAPK, Ras, transforming growth factor (TGF)-β, and Wnt pathway activation in HAT1 expression inhibited cells. We further found that MAPK pathway activation following the loss of HAT1 expression was partially driven by increased insulin growth factor 1 receptor (IGF1R) signaling. We showed that both MAPK and IGF1R pathway inhibition, using the ERK inhibitor SCH772984 and the IGF1R inhibitor BMS-754807, respectively, restored BRAFi sensitivity in melanoma cells lacking HAT1. Collectively, we show that the loss of HAT1 expression confers acquired BRAFi resistance by activating the MAPK signaling pathway via IGF1R.
Similar content being viewed by others
Introduction
Melanoma is the deadliest form of skin cancer, accounting for ~80% of cancer-related deaths1. Both genetic and nongenetic factors contribute to melanoma initiation and progression, and exposure to UV radiation is the leading cause of nongenetic melanoma development2,3. Mutations in a number of genes have been shown to be involved in the initiation and progression of melanoma4,5. In particular, large-scale genomic DNA sequencing has identified activating BRAF mutations in ~50% of all melanoma cases6,7. In addition, mutations in NRAS, neurofibromin 1 (NF1), cyclin-dependent kinase inhibitor 2A (CDKN2A), microphthalmia-associated transcription factor (MITF), and phosphatase and tensin homolog (PTEN) have been shown to play important roles in melanomagenesis8,9.
BRAF is a member of the Raf kinase family, and oncogenic mutations in BRAF activate the mitogen-activating protein kinase (MAPK)/extracellular signal-related kinase (ERK) kinase (MEK)→ERK signaling pathway, which is required for melanoma growth and metastasis10,11,12. More than 30 different mutations have been reported in BRAF, associated with melanoma and other cancers7,13. The V600E mutation in BRAF has been identified in 90% of cases, followed by the V600K mutation, which has been found in 5% of cases, whereas other mutations, such as V600R, V600E2, and V600D, are found at even lower frequencies7,14.
Because over 50% of melanoma patients harbor oncogenic mutations in BRAF and BRAF-mutant melanoma cells depend upon BRAF mutations for the growth and survival, several BRAF inhibitors (BRAFi) have been approved by the US Food and Drug Administration (US FDA), including vemurafenib and dabrafenib, for the clinical treatment of metastatic melanoma15,16. Although, BRAFi produce impressive initial clinical responses against BRAF-mutant metastatic melanoma17,18, the durability of the BRAFi response is limited by the rapid emergence of acquired BRAFi resistance, often within a few months of treatment initiation19. Therefore, studies have been conducted to discern the mechanisms underlying acquired BRAFi resistance, resulting in the identification of several mechanisms associated with BRAFi resistance20,21,22. However, a subset of melanoma-resistance mechanisms that are responsible for acquired BRAFi resistance remain unknown23.
In order to determine the potential roles played by epigenetic regulators during the development of BRAFi resistance in melanoma, we had performed a large-scale, unbiased epigenome-wide short-hairpin RNA (shRNA) screen, targeting 363 epigenetic regulators24. This screen led to the identification of histone acetyltransferase 1 (HAT1) as one of candidate genes that mediates BRAFi resistance. In this study, we focused on determining the role of HAT1 in acquired BRAFi resistance and understanding the mechanism behind HAT1 loss-induced acquired BRAFi resistance. We also performed experiments to establish the clinical relevance of HAT1 during the development of BRAFi resistance in melanoma patients.
Our results showed that the loss of HAT1 expression resulted in the development of BRAFi resistance, in part due to the activation of the MAPK pathway by insulin growth factor 1 receptor (IGF1R). Using patient-derived melanoma samples, we found that a large majority of progressed samples (63%), from patients treated with BRAFi or BRAFi + MEK inhibitor (MEKi), showed reduced HAT1 expression levels compared with matched pre-treatment melanoma samples, indicating that HAT1 is clinically relevant. Thus our study indicates that loss of HAT1 is one of the crucial mechanism that drives BRAFi resistance in melanoma.
Results
Loss of histone acetyltransferase 1 (HAT1) expression confers acquired resistance to BRAFi
Epigenetic gene regulation mechanisms play important roles in key aspects of tumor growth and metastasis and are associated with the development of drug resistance25,26,27. Therefore, to determine the potential roles played by epigenetic regulators during the development of BRAFi resistance, we previously performed a large-scale, unbiased epigenome-wide, shRNA screen that targeted 395 known and predicted epigenetic regulators using 1875 shRNAs24. This shRNA screen resulted in the identification of HAT1, but the role of HAT1 during acquired BRAFi resistance was not characterized.
In this study, we examined whether the loss of HAT1 expression confers BRAFi resistance in melanoma cells. We first knocked down HAT1 expression, using shRNAs in BRAF-mutant melanoma cell lines (A375 and SKMEL-28) (Fig. 1a, b). As a control, a nonspecific (NS) shRNA was used.
a Indicated melanoma cell lines, expressing either nonspecific (NS) shRNA or HAT1 shRNAs, were analyzed for HAT1 protein expression levels by immunoblotting. ACTINB was used as a loading control. b Indicated melanoma cell lines, expressing either a nonspecific (NS) shRNA or HAT1 shRNAs, were analyzed by qRT-PCR. Actin was used as an internal control. c Relative survival rates of A375 and SKMEL-28 cells, expressing either nonspecific (NS) shRNA or the indicated HAT1 shRNA, upon treatment with vemurafenib for 3 days, as measured by MTT assay. d Relative survival rates of A375 and SKMEL-28 cells, expressing either nonspecific (NS) shRNA or the indicated HAT1 shRNA, upon treatment with dabrafenib for 3 days, were measured by MTT assay. e IC50 values for the MTT data presented in panels c and d. f A375 or SKMEL-28 cells, expressing either a nonspecific (NS) shRNA or HAT1 shRNAs, were treated with DMSO or vemurafenib (1 μM) and analyzed by the soft-agar assay. Scale bar is 100 µm. g Relative colony sizes for the data presented in panel f. Data are presented as the mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.000, calculated using Student’s t test.
Melanoma cells expressing either NS or HAT1 shRNAs (HAT1 knockdown) were then tested for their sensitivity to the BRAFi vemurafenib, in a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT)-based short-term cell-survival assay. Our results showed that the knockdown of HAT1 conferred resistance to vemurafenib (Fig. 1c, e). Next, we determined whether these findings were specific to vemurafenib or could be extended to another BRAFi, dabrafenib. Dabrafenib is a more selective, reversible, ATP-competitive kinase inhibitor that inhibits BRAFV600E and is currently approved for the clinical treatment of melanoma patients18,28. We found that the knockdown of HAT1 also conferred resistance to dabrafenib (Fig. 1d, e).
To further fortify our results, we performed a soft-agar assay to determine the anchorage-independent growth abilities of HAT1-knockdown melanoma cells in the presence of vemurafenib. Our results showed that HAT1-knockdown BRAF-mutant melanoma cells were resistant to vemurafenib treatment compared with cells expressing a control NS shRNA (Fig. 1f, g). Taken together, these results demonstrated that the loss of HAT1 expression resulted in acquired BRAFi resistance in melanoma cells.
HAT1 knockout in melanoma cells confers resistance to BRAFi in a long-term survival assay
To emulate the clinical scenario, we performed a clonogenic survival assay to examine the long-term effects of reduced HAT1 expression on the development of BRAFi resistance. As shown in Supplementary Fig. S1, we found that HAT1-knockdown cells did not show BRAFi resistance in the long-term survival assay, suggesting that the partial reduction of HAT1 protein levels may not be sufficient to cause BRAFi resistance in the long-term. Because a partial reduction in HAT1 protein levels, following the shRNA-mediated HAT1 knockdown in melanoma cells, was unable to cause BRAFi resistance to vemurafenib in a clonogenic long-term survival assay, we generated a complete HAT1 knockout (HAT1-KO) in BRAF-mutant melanoma cells using a clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated (Cas)9-based approach29,30. We observed that HAT1-KO melanoma cells showed complete loss of HAT1 protein expression (Fig. 2a). Using HAT1-KO cells, we performed a long-term survival assay and found that these melanoma cells showed resistance to both vemurafenib and dabrafenib (Fig. 2b, c). These results confirmed that the complete loss of HAT1 expression resulted in BRAFi resistance in the long-term.
a Immunoblotting for the indicated proteins in A375 and SKMEL-28 HAT1-knockout (HAT1-KO) cells. b Clonogenic assay results for A375 and SKMEL-28 cells, expressing non-targeting (NT) or HAT1 sgRNAs, in the presence of vemurafenib (3 μM) (left). Colony number for the data are shown (right). c Clonogenic assay results for A375 and SKMEL-28 cells, expressing NT or HAT1 sgRNAs, in the presence of dabrafenib (100 nM) (left). Colony number for the data presented (right). Data are presented as the mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001, calculated using Student’s t test.
HAT1 protein is downregulated in patient-derived melanoma samples after BRAFi and BRAFi + MEKi treatment
The clonogenic long-term survival assay mimics the clinical circumstances, in which patients are treated for relatively long periods of time with a given drug. Because HAT1-KO melanoma cells became BRAFi resistant during a clonogenic long-term survival assay, we next tested whether HAT1 expression was downregulated in clinical samples taken from patients who experienced disease progression following treatment with BRAFi or BRAFi+MEKi. We analyzed 11 sets of matched, patient-derived, melanoma samples, consisting of a pre-treatment sample and a progressed sample, taken after treatment with either BRAFi or BRAFi+MEKi (Supplementary Table 1). To quantitatively monitor changes in HAT1 protein levels, we performed an automated quantitative analysis (AQUA)-based immunofluorescence analysis. Our results showed that 7 out of 11 cases of progressed melanoma samples, from patients previously treated with BRAFi or BRAFi+MEKi, showed lower expression levels of HAT1 protein compared with their respective, matched, pre-treatment samples (Fig. 3a, b; Supplementary Fig. S2 and Supplementary Table 1). These results further established that BRAFi resistance following the loss of HAT1 expression represents a clinically relevant mechanism among BRAFi- and BRAFi + MEKi-treated melanoma patients.
a Average AQUA scores for pre-treatment and progressed melanoma samples from patients treated with BRAFi or BRAFi + MEKi. b Representative AQUA immunofluorescence scores and merged images of the indicated matched pre-treatment and progressed melanoma samples, stained for DAPI, S100/HMB45, and HAT1. Data are presented as the mean ± SEM. ***p < 0.001, ****p < 0.0001, calculated using Student’s t test.
The loss of HAT1 expression leads to the activation of multiple oncogenic signaling pathways in BRAF-mutant melanoma cells
HAT1 is a histone acetyltransferase that regulates gene expression31,32. Therefore, we speculated that changes in gene expression may underlie the development of BRAFi resistance following reduced HAT1 expression. To measure changes in gene expression, we performed a NanoString-based gene expression analysis, using the nCounter PanCancer Pathway Panel for Gene Expression. The nCounter PanCancer Pathway Panel monitors the expression of over 700 genes, spread across 13 canonical cancer hallmark pathways (Supplementary Fig. S3). These includes genes in chromatin modification, Hedgehog, Wnt, Notch, apoptosis, cell cycle, RAS, PI3K, STAT, MAPK, TGF-β, DNA damage control, transcriptional regulation that drive different aspect of cancer growth, progression, and treatment response. Our results showed that common pathways that were upregulated in both HAT1-knockdown and HAT1-KO melanoma cells, includes TGF-β, Wnt, Ras, and MAPK pathways (Fig. 4a–c; Supplementary Table 2). We then analyzed the expression levels of genes associated with the TGF-β, Wnt, Ras, and MAPK signaling pathways and found the following changes in both HAT1-knockdown and HAT1-KO melanoma cells: in the Ras pathway, phospholipase D1 (PLD1), Ras and Rab interactor 1 (RIN1), insulin-like growth factor 1 receptor (IGF1R), and Src homology 2 domain-containing (SHC) transforming protein 1 (SHC1) were significantly upregulated; for both the TGF-β and MAPK signaling pathways, TGFβ1 was significantly upregulated; and in the Wnt signaling pathway, glycogen synthase kinase 3 β (GSK3B) was significantly upregulated, whereas cyclin D1 (CCND1) and Fos-related antigen 1 (FOSL1) were significantly downregulated, (Fig. 4d, e; Supplementary Table 2).
a Heatmap for A375 cells, expressing non-targeting (NT) or HAT1 sgRNAs. b Heatmap for A375 cells, expressing nonspecific (NS) or HAT1 shRNAs. c. Heatmap for SKMEL-28 cells, expressing NS or HAT1 shRNAs. d Venn diagram showing the genes that were significantly up- and downregulated in A375 HAT1 knockout (KO), A375 HAT1 knockdown (KD), and SKMEL-28 HAT1 KD cells in the Ras, MAPK, Wnt, and TGF-β signaling pathways. e Fold changes of the genes associated with the Ras, MAPK, TGF-β, and Wnt signaling pathways that were significantly altered in A375 cells expressing NT or HAT1 sgRNAs, A375 cells expressing NS or HAT1 shRNAs, and SKMEL-28 cells expressing NS or HAT1 shRNAs. Data are presented as the mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001, calculated using Student’s t test.
In HAT1-KO melanoma cells, genes associated with other signaling pathways were also upregulated, including Hedgehog, notch, cell cycle-apoptosis, chromatin modification, and driver genes (Fig. 4a). These upregulated pathways potentially contribute to the increased resistance phenotype during long-term survival assays observed for HAT1-KO cells compared with that observed for HAT1-knockdown cells. Overall, our results demonstrated that the loss of HAT1 expression resulted in the upregulation of both MAPK-independent and MAPK-dependent pathways, leading to the acquisition of BRAFi resistance in melanoma cells.
The activation of IGF1R following the loss of HAT1 expression results in the activation of MAPK pathways in BRAF-mutant melanoma cells
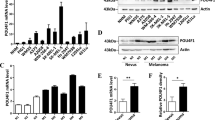
We next used immunoblotting analyses to measure the protein expression levels of the various signaling pathways identified during the NanoString analysis. We determined the levels of the following proteins: phospho- and total ERK1/2, to assess the MAPK signaling pathway; TGFβ1, to assess both the TGF-β and MAPK signaling pathways; and beta-catenin, to assess the Wnt signaling pathway. Our results showed that phospho-ERK1/2 protein levels were significantly upregulated in both HAT1-knockdown and HAT1-KO melanoma cells (Figs. 5a and 6a). However, we did not observe any significant changes in TGFβ1 or beta-catenin protein levels in either HAT1-knockdown or HAT1-KO melanoma cells (Supplementary Fig. S4).
a Immunoblotting for phospho- and the total ERK in A375 and SKMEL-28 cells expressing nonspecific NS or HAT1 shRNAs. b Relative IGF1R mRNA levels in A375 and SKMEL-28 cells expressing nonspecific NS or HAT1 shRNAs were analyzed by qRT-PCR. Actin was used as an internal control. c Immunoblotting for phospho- and the total IGF1R in A375 and SKMEL-28 cells expressing nonspecific NS or HAT1 shRNAs. d, f A375 or SKMEL-28 cells expressing a nonspecific (NS) shRNA or HAT1 shRNAs were treated with DMSO, BRAF inhibitor vemurafenib (1 μM), IGF1R inhibitor, BMS-754807 (0.1 μM) and ERK inhibitor, SCH772984 (0.2 μM), which was analyzed by soft-agar assay. e, g Relative colony size for data presented in panels d and f. Data are presented as the mean ± SEM, *p < 0.05, **p < 0.01, calculated using Student’s t test.
a Immunoblotting for phospho- and total ERK in A375 cells expressing non-targeting NT or HAT1 sgRNAs. b Relative IGF1R mRNA levels in A375 cells expressing non-targeting NT or HAT1 sgRNAs was analyzed by qRT-PCR. Actin was used as internal control. c, e A375 or SKMEL-28 cells expressing a non-targeting (NT) sgRNA or HAT1 sgRNAs were treated with DMSO, BRAF inhibitor vemurafenib (3 μM), IGF1R inhibitor, BMS-754807 (1 μM) and ERK inhibitor, SCH772984 (1 μM) was analyzed by clonogenic assay. d, f Relative colony number for data presented in panels c and e. Data are presented as the mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001, calculated using Student’s t test.
We attempted to determine the mechanism through which phospho-ERK1/2 levels were elevated following the loss of HAT1 expression in BRAF-mutant melanoma cells. We first determined the levels of dual-specific phosphatases (DUSPs) in cells with reduced HAT1 expression because some DUSPs regulate MAPK signaling by inhibiting the activation of ERK1/233. Our results showed that DUSP levels were downregulated only in HAT1-KO melanoma cells, whereas in HAT1-knockdown melanoma cells, the DUSP expression levels did not change significantly (Supplementary Fig. S5).
In addition, because increased IGF1R expression levels were identified in the nCounter PanCancer Pathway Panel for Gene Expression, in both HAT1-knockdown and HAT1-KO BRAF-mutant melanoma cells (Fig. 4e), we analyzed the mRNA levels as well as protein levels of phospho- and total IGF1R in these samples. Strikingly, we found that IGF1R mRNA levels as well as phospho-IGF1R was significantly upregulated in both HAT1-knockdown and HAT1-KO cells (Figs. 5b, c and 6b). Previous studies have shown that the activation of IGF1R signaling can activate the MAPK/Ras pathway34,35. In our study, we found that in melanoma cells with reduced HAT1 expression levels, IGF1R levels were upregulated, leading to the activation of MAPK/Ras signaling pathway and conferring BRAFi resistance.
The pharmacological inhibition of IGF1R or ERK partially rescues the BRAFi-resistance phenotype in melanoma cells following the loss of HAT1 expression
Because our results showed that HAT1-knockdown and HAT1-KO BRAFi-resistant melanoma cells demonstrated increased IGF1R expression levels and increased MAPK pathway activation, we examined whether the ERK inhibitor SCH77298436 or the IGF1R inhibitor BMS-75480737 could resensitize HAT1-knockdown and HAT1-KO BRAFi-resistant melanoma cells to BRAF inhibitor vemurafenib. We found that treatment with either the ERK or IGF1R inhibitors was able to restore sensitivity to vermurafenib in both HAT1-knockdown and HAT1-KO BRAFi-resistant melanoma cells, in both the soft-agar assay (Fig. 5d–g) and in the clonogenic long-term survival assay (Fig. 6c–f). These results confirmed the loss of HAT1 expression resulted in the activation of the MAPK pathway by IGF1R, which conferred BRAFi resistance; therefore, IGF1R or ERK1/2 inhibitors represent pharmacologically tractable options for inhibiting the growth of BRAFi-resistant melanoma cells.
Discussion
Oncogenic mutations in BRAF have been identified in ~50% of melanoma cases. BRAFi, either alone or in combination with MEKi, represents therapeutic options for the treatment of BRAF-mutant metastatic melanomas. However, due to the rapid emergence of acquired BRAFi resistance, the clinical benefits of these therapies are often limited22,38,39.
Previous studies have identified several mechanisms associated with the development of BRAFi resistance. For example, the mutational activation of other oncogenes, such as NRAS, MEK1, and MEK2, the overexpression of CRAF, the dimerization of oncogenic BRAF, and the upregulation of the PTEN-PI3K-AKT signaling pathway have all been identified as potential mechanisms underlying the development of BRAFi/BRAFi + MEKi resistance39,40,41,42,43.
In this study, we investigated the role played by HAT1 in the regulation of BRAFi resistance in melanoma cells and showed that the loss of HAT1 expression contributed to the development of BRAFi resistance (Fig. 7). HAT1 is a type-B histone acetyltransferase that acetylates newly synthesized H3 and H4 histones and participates in chromatin assembly44,45. HAT1 has also been shown to induce apoptosis by upregulating Fas expression in lung cancer cells46. In addition, Ras-ERK1/2 signaling has been shown to promote the development of osteosarcoma through the regulation of H4K12Acetyl, via HAT147. We showed that the loss of HAT1 expression was associated with the development of resistance to BRAFi, such as vemurafenib and dabrafenib. We also confirmed that ~63% of progressed samples, from BRAF-mutant melanoma patients who experienced disease progression following BRAFi or BRAFi + MEKi therapy, showed significantly reduced HAT1 expression levels compared with matched pre-treatment samples. These results collectively demonstrated that the loss of HAT1 expression during the acquired resistance to BRAFi and BRAFi + MEKi represents a clinically relevant event.
We also performed gene expression NanoString analysis to further understand the underlying mechanisms that result in the development of BRAFi resistance following the loss of HAT1 expression. In this assay, we analyzed 700 genes, clustered into 13 hallmark cancer pathways. These studies identified the upregulation of the Ras, MAPK, Wnt, and TGF-β signaling pathways in melanoma cells lacking HAT1. We then confirmed that among these four upregulated signaling pathways, the MAPK pathway was significantly upregulated in BRAFi-resistant cells, following the loss of HAT1 expression. We further found that cells lacking HAT1 expression showed increased IGF1R signaling activity. IGF1R has been shown to activate the MAPK signaling pathway, promoting cell growth and proliferation, and can drive acquired resistance to various drugs48,49. Therefore, IGF1R signaling pathway inhibitors have been utilized as alternative strategies for the treatment of drug-resistant cancer cells and to counteract drug resistance41,50.
We also found that the pharmacological targeting of the MAPK signaling pathway via IGF1R and ERK1/2 inhibitors was able to partially restore the sensitivity of HAT1-knockdown and HAT1-KO melanoma cells to BRAFi. ERK1/2 is the most downstream kinase of the Ras–BRAF–MEK–ERK cascade, and targeting ERK1/2 can overcome resistance mechanisms caused by changes in upstream pathways51. In particular, ERK1/2 inhibitors have shown some activity in BRAFi- or BRAFi + MEKi-resistant melanomas52,53. Although not directly tested in this study, we predict that HAT1 expression levels in samples from patients with progressed disease states can be used as an indicator of IGF1R and ERK1/2 inhibitor sensitivity, due to the activation of the IGF1R and MAPK pathways in melanoma cells expressing low levels of HAT1. Although, we have not directly tested the BRAFi + MEKi combination in our cell culture experiments. However, based on the activation of IGF1R and our clinical findings that HAT1 is also downregulated in patients treated with the combination of BRAFi + MEKi, it is likely that HAT1 loss will also confer resistance to BRAFi + MEKi combination therapy. Collectively, our study showed that the loss of HAT1 expression results in acquired BRAFi resistance, in part by increasing MAPK pathway activation via IGF1R, and indicated that treatments with IGF1R and/or ERK1/2 inhibitors can enhance BRAFi efficacy and overcome the limitations associated with BRAFi and BRAFi + MEKi treatment.
Materials and methods
Cell culture and inhibitors
SKMEL-28, A375, and HEK293T cells were purchased from American Type Culture Collection (ATCC) and grown as recommended. Cell lines were used only after confirming the lack of Mycoplasma contamination using MycoAlert Mycoplasma detection kit (Lonza), and were also routinely tested for lack of mycoplasma contamination. All cell lines were passaged for 2–4 weeks between thawing and use in the described experiments. BRAFi (vemurafenib and dabrafenib), the ERK inhibitor SCH772984, and the IGF1R inhibitor BMS-754807 were purchased from Selleckem.
RNA preparation, cDNA preparation, quantitative PCR analysis
The total RNA was extracted with TRIzol Reagent (Invitrogen) and purified using the RNeasy Mini Kit (Qiagen). cDNA was generated using the M-MuLV First Strand cDNA Synthesis Kit (New England Biolabs), according to the manufacturer’s instructions. Quantitative reverse transcriptase (RT)-PCR was performed with gene-specific primers, using the Power SYBR-Green Master Mix (Applied Biosystems), according to the manufacturer’s instructions. Actin was used as an internal control. Primer sequences are provided in Supplementary Table 3.
shRNA and lentivirus preparation
pLKO.1 lentiviral vector-based shRNAs, targeting specific candidate genes, and NS shRNA controls were obtained from OpenBiosystems (Dharmacon). shRNA information is provided in Supplementary Table 3. Lentivirus particles were prepared by transfecting 293T cells with either gene-specific shRNA or NS shRNA plasmids, along with the lentiviral packaging plasmids, as described in detail at https://portals.broadinstitute.org/gpp/public/resources/protocols. All lentiviral transfections were performed using Effectene (Qiagen). Stable cell lines were generated by infecting melanoma cells with lentivirus particles, followed by selection with appropriate concentrations of puromycin (0.5–1.5 μg/mL), to enrich infected cells.
Preparation of the HAT1 single guide RNA (sgRNA) lentivirus and generation of stable cell lines
Gene-specific lentiviral HAT1 sgRNAs were cloned into the pLentiCRISPR v2 vector. The sgRNA sequences are provided in Supplementary Table 3. For lentivirus production, sgRNAs were transfected into 293T cells, along with the PDM2.G and psPAX2 packaging plasmids, using Effectene Transfection Reagent (Qiagen), according to the manufacturer’s instructions. After 48 h, the lentivirus-containing supernatants were harvested, filtered, and used for infections. Lentiviral sgRNA-infected melanoma cells were selected using 0.6 μg/ml puromycin.
Antibodies and immunoblot analysis
Whole-cell protein extracts were prepared using Pierce IP lysis buffer (Thermo Fisher Scientific), containing protease inhibitor mixture (Roche) and phosphatase inhibitor mixture (Sigma-Aldrich). Protein concentrations were estimated using the Bradford assay (Bio-Rad). Proteins were separated by 10 or 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride (PVDF) membranes by wet transfer. PVDF membranes were blocked with 5% nonfat dry milk or 5% bovine serum albumin (BSA), as recommended for each specific antibody, washed, and probed with primary antibodies. Membranes were washed again, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies (GE Healthcare). Immunoblots were developed using SuperSignal West Pico or Femto Substrates (Pierce), as necessary. All primary and secondary antibodies used in these studies are listed in Supplementary Table 3.
Soft-agar assay
Soft-agar assays were performed by seeding between 5 × 103 and 2 × 104 melanoma cells, which stably expressed the indicated shRNA or cDNA constructs, onto 0.4% low-melting-point agarose (Sigma-Aldrich), layered on top of 0.8% agarose. For drug-treatment experiments, cells were then treated with DMSO, vemurafenib (1 µM), or dabrafenib (50 nM), IGF1R inhibitor, BMS-754807 (0.1 μM) and ERK inhibitor, SCH772984 (0.2 μM), and cell culture media was changed every 3 days, adding fresh drug each time. After 3–4 weeks of incubation, colonies were stained with a 0.005% crystal violet solution and imaged using a microscope. Colony sizes were measured using ImageJ software (https://imagej.nih.gov/ij/) and plotted. This experiment was performed in triplicate.
MTT assay
For this assay, 5 × 103 cells were plated in a 100-µl volume, in 96-well plates in triplicate. After 48 h, inhibitors (i.e., vemurafenib and dabrafenib), used at a range of concentrations, were mixed in 100 µl of medium and added to the cells. After 48 h of inhibitor treatment, the cell viability was evaluated by adding 20 µl of 5 mg/ml MTT solution dissolved in 1× PBS to each well and incubating for 1 h at 37 °C. The MTT solution was removed gently, and 100 µl of DMSO was added to each well. After mixing each well by pipetting, the absorbance was measured at 590 and 630 nm. An average was calculated for both readings, and then the measurement at 630 nm was subtracted from that at 590 nm. The relative growth rate was plotted with respect to vehicle-treated control cells.
Clonogenic assay
The clonogenic abilities of cells stably expressing control or gene-specific shRNAs were measured in untreated and vemurafenib- and dabrafenib-treated conditions. For clonogenic assay, 1 × 105 cells were seeded in a six-well plate in triplicate and after 24 h they were either vehicle treated or treated with vemurafenib (3 μM), dabrafenib (100 nM), IGF1R inhibitor, BMS-754807 (1 μM) and ERK inhibitor, SCH772984 (1 μM). After 3–4 weeks of treatment, colonies were fixed with a fixing solution, containing 50% methanol and 10% acetic acid, and then stained with 0.05% Coomassie blue (Sigma-Aldrich). The relative number of colonies was calculated first by counting the number of colonies for each samples and then by plotting the average numbers of colonies counted for triplicate in the indicated shRNAs versus NS shRNA.
Patient sample acquisition
Melanoma samples were obtained through biopsies and surgical resections, performed during the standard clinical care of melanoma patients. Excess samples not required for surgical pathology assessments were stored in the Vanderbilt University melanoma tumor repository, as formalin-fixed paraffin-embedded (FFPE) samples. All patients provided consent through an Institutional Review Board-approved protocol before tissue acquisition (Vanderbilt IRB# 030220), and samples were de-identified for the analysis (Supplementary Table 1).
Quantitative immunofluorescence analysis
Whole-tissue sections of paired pre-treatment and progressed samples from patients treated with targeted BRAFi, as described in Supplementary Table 1, were deparaffinized at 60 °C for 30 min, incubated in xylene (twice, for 20 min each), and rehydrated with ethanol (twice, in 100% ethanol for 1 min, and then in 70% ethanol for 1 min). Antigen retrieval was performed by boiling the samples for 20 min at 97 °C in citrate buffer, pH 6.0 (PT module, Lab Vision; Thermo Scientific). Slides were blocked with 30% hydrogen peroxide in methanol and then incubated with a blocking solution, containing 0.3% BSA in Tris-buffered saline and 0.05% Tween solution (TBST), for 30 min at room temperature. Slides were then incubated overnight, with a mixture of HAT1 rabbit antibodies, and S100 and HMB45 mouse antibodies (Supplementary Table 3). The next day, slides were washed and treated with Alexa 546-conjugated goat anti-mouse secondary antibody (Invitrogen), diluted 1:100 in rabbit Envision reagent (K4003, Dako), and incubated for 60 min at room temperature. For target detection, slides were treated with cyanine 5, directly conjugated to tyramide (FP1117; Perkin-Elmer), at a 1:50 dilution, for 10 min. ProLong gold mounting medium (Invitrogen), containing 4′,6-diamidino-2-phenylindole (DAPI), was used to stain nuclei. Control slides were processed for reproducibility alongside each experimental slide-staining run. Quantitative measurements of HAT1 and immunofluorescence analysis were performed using the AQUA method54. A tumor mask was created by binarizing the HAT1, S100, and HMB45 signals. Quantitative immunofluorescence scores were calculated by dividing the target pixel intensity by the area of the S100 and HMB45 compartments. All patient samples were scored using AQUA software, and in each sample 40–70 different spots were analyzed; the average tumor mask-normalized scores are shown in the figures.
NanoString analysis
RNA was analyzed using the NanoString nCounter platform (Seattle, WA), through the UAB NanoString Laboratory (http://www.uab.edu/medicine/radonc/en/nanostring). All RNA samples had A260/A280 and A260/A230 ratios between 1.8 and 2.3, as recommended by the manufacturer and determined using a DeNovix DS-11 spectrophotometer (Wilmington, DE). Briefly, 100 ng of each sample was hybridized for 18 h, using Reporter and Capture Probes specific to the human PanCancer Pathways panel, and processed on the NanoString nCounter Flex system, according to the manufacturer’s instructions. This premade panel contains 730 genes involved in 13 hallmark cancer pathways (apoptosis, cell cycle, chromatin modification, DNA damage control, Hedgehog, MAPK, Notch, P13K, Ras, STAT, TGF-β, transcriptional regulation, and Wnt) as well as housekeeping genes and negative and positive controls. The samples were read at the standard 280 FOV count, the resultant RCC data files were imported into NanoString nSolver 4.0, and the raw data was used to run through the advanced analysis module. This module selects the best housekeeping genes to use during the analysis, through the GNORM program, and those selected genes were used to normalize the data.
Statistical analysis
All experiments were conducted using at least three biological replicates. The sample size was determined based on previous experience for each experiments to detect specific effects and it was not predetermined with any statistical methods. Results for individual experiments are expressed as the mean ± standard error of the mean (SEM). For measurements of MTT assays, statistical analyses were performed by analyzing the area under the curve, using GraphPad Prism software, version 7.0, for Macintosh (GraphPad Software; https://www.graphpad.com). For the remaining experiments, P-values were calculated using a two-tailed, unpaired, Student’s t test, in GraphPad Prism software, version 7.0, for Macintosh.
References
Aladowicz, E. et al. Molecular networks in melanoma invasion and metastasis. Future Oncol. 9, 713–726 (2013).
Watson, M., Holman, D. M. & Maguire-Eisen, M. Ultraviolet radiation exposure and its impact on skin cancer risk. Semin Oncol. Nurs. 32, 241–254 (2016).
Anna, B. et al. Mechanism of UV-related carcinogenesis and its contribution to nevi/melanoma. Expert Rev. Dermatol 2, 451–469 (2007).
Potrony, M. et al. Update in genetic susceptibility in melanoma. Ann. Transl. Med. 3, 210 (2015).
Sini, M. C. et al. Genetic alterations in main candidate genes during melanoma progression. Oncotarget 9, 8531–8541. (2018).
Ascierto, P. A. et al. The role of BRAF V600 mutation in melanoma. J. Transl. Med. 10, 85 (2012).
Cheng, L., Lopez-Beltran, A., Massari, F., MacLennan, G. T. & Montironi, R. Molecular testing for BRAF mutations to inform melanoma treatment decisions: a move toward precision medicine. Mod. Pathol. 31, 24–38 (2018).
Hodis, E. et al. A landscape of driver mutations in melanoma. Cell 150, 251–263 (2012).
Garman, B. et al. Genetic and genomic characterization of 462 melanoma patient-derived xenografts, tumor biopsies, and cell lines. Cell Rep. 21, 1936–1952 (2017).
Cantwell-Dorris, E. R., O’Leary, J. J. & Sheils, O. M. BRAFV600E: implications for carcinogenesis and molecular therapy. Mol. Cancer Ther. 10, 385–394 (2011).
Yaeger, R. & Corcoran, R. B. Targeting alterations in the RAF-MEK pathway. Cancer Discov. 9, 329–341 (2019).
Sullivan, R. J. & Flaherty, K. MAP kinase signaling and inhibition in melanoma. Oncogene 32, 2373–2379 (2013).
Hussain, M. R. et al. BRAF gene: from human cancers to developmental syndromes. Saudi J. Biol. Sci. 22, 359–373 (2015).
Heinzerling, L. et al. Rare BRAF mutations in melanoma patients: implications for molecular testing in clinical practice. Br. J. Cancer 108, 2164–2171 (2013).
Ryu, S. et al. Therapeutic inhibitors against mutated BRAF and MEK for the treatment of metastatic melanoma. Chonnam. Med. J. 53, 173–177 (2017).
Khunger, A., Khunger, M. & Velcheti, V. Dabrafenib in combination with trametinib in the treatment of patients with BRAF V600-positive advanced or metastatic non-small cell lung cancer: clinical evidence and experience. Ther. Adv. Respir. Dis. 12, 1753466618767611 (2018).
Karoulia, Z., Gavathiotis, E. & Poulikakos, P. I. New perspectives for targeting RAF kinase in human cancer. Nat. Rev. Cancer 17, 676–691 (2017).
Holderfield, M., Deuker, M. M., McCormick, F. & McMahon, M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat. Rev. Cancer 14, 455–467 (2014).
Sun, C. et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 508, 118–122 (2014).
Arozarena, I. & Wellbrock, C. Overcoming resistance to BRAF inhibitors. Ann. Transl. Med. 5, 387 (2017).
Yaeger, R. et al. Mechanisms of acquired resistance to BRAF V600E inhibition in colon cancers converge on RAF dimerization and are sensitive to its inhibition. Cancer Res. 77, 6513–6523 (2017).
Villanueva, J., Vultur, A. & Herlyn, M. Resistance to BRAF inhibitors: unraveling mechanisms and future treatment options. Cancer Res. 71, 7137–7140 (2011).
Johnson, D. B. et al. Acquired BRAF inhibitor resistance: a multicenter meta-analysis of the spectrum and frequencies, clinical behaviour, and phenotypic associations of resistance mechanisms. Eur. J. Cancer 51, 2792–2799 (2015).
Gupta, R. et al. Loss of BOP1 confers resistance to BRAF kinase inhibitors in melanoma by activating MAP kinase pathway. Proc. Natl Acad. Sci. USA 116, 4583–4591 (2019).
Akar, R. O., Selvi, S., Ulukaya, E. & Aztopal, N. Key actors in cancer therapy: epigenetic modifiers. Turk. J. Biol. 43, 155–170 (2019).
Wilting, R. H. & Dannenberg, J. H. Epigenetic mechanisms in tumorigenesis, tumor cell heterogeneity and drug resistance. Drug Resist Updat 15, 21–38 (2012).
Moosavi, A. & Motevalizadeh Ardekani, A. Role of epigenetics in biology and human diseases. Iran. Biomed. J. 20, 246–258 (2016).
Menzies, A. M. & Long, G. V. Dabrafenib and trametinib, alone and in combination for BRAF-mutant metastatic melanoma. Clin. Cancer Res. 20, 2035–2043 (2014).
Campenhout, C. V. et al. Guidelines for optimized gene knockout using CRISPR/Cas9. Biotechniques 66, 295–302 (2019).
Spiegel, A., Bachmann, M., Jurado Jimenez, G. & Sarov, M. CRISPR/Cas9-based knockout pipeline for reverse genetics in mammalian cell culture. Methods 164-165, 49–58 (2019).
Verdone, L., Caserta, M. & Di Mauro, E. Role of histone acetylation in the control of gene expression. Biochem Cell Biol. 83, 344–353 (2005).
Gruber, J. J. et al. HAT1 coordinates histone production and acetylation via H4 promoter binding. Mol. Cell 75, 711–724 e715 (2019).
Lake, D., Correa, S. A. & Muller, J. Negative feedback regulation of the ERK1/2 MAPK pathway. Cell Mol. Life Sci. 73, 4397–4413 (2016).
Iams, W. T. & Lovly, C. M. Molecular pathways: clinical applications and future direction of insulin-like growth factor-1 receptor pathway blockade. Clin. Cancer Res. 21, 4270–4277 (2015).
Yamauchi, K. & Pessin, J. E. Insulin receptor substrate-1 (IRS1) and Shc compete for a limited pool of Grb2 in mediating insulin downstream signaling. J. Biol. Chem. 269, 31107–31114 (1994).
Morris, E. J. et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 3, 742–750 (2013).
Awasthi, N., Zhang, C., Ruan, W., Schwarz, M. A. & Schwarz, R. E. BMS-754807, a small-molecule inhibitor of insulin-like growth factor-1 receptor/insulin receptor, enhances gemcitabine response in pancreatic cancer. Mol. Cancer Ther. 11, 2644–2653 (2012).
Sullivan, R. J. & Flaherty, K. T. Resistance to BRAF-targeted therapy in melanoma. Eur. J. Cancer 49, 1297–1304 (2013).
Villanueva, J. et al. Concurrent MEK2 mutation and BRAF amplification confer resistance to BRAF and MEK inhibitors in melanoma. Cell Rep. 4, 1090–1099 (2013).
Smalley, K. S. et al. Increased cyclin D1 expression can mediate BRAF inhibitor resistance in BRAF V600E-mutated melanomas. Mol. Cancer Ther. 7, 2876–2883 (2008).
Villanueva, J. et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell 18, 683–695 (2010).
Chen, S. H. et al. Oncogenic BRAF deletions that function as homodimers and are sensitive to inhibition by RAF dimer inhibitor LY3009120. Cancer Disco. 6, 300–315 (2016).
Perna, D. et al. BRAF inhibitor resistance mediated by the AKT pathway in an oncogenic BRAF mouse melanoma model. Proc. Natl Acad. Sci. USA 112, E536–E545 (2015).
Parthun, M. R. Histone acetyltransferase 1: more than just an enzyme? Biochim. Biophys. Acta 1819, 256–263 (2012).
Qin, S. & Parthun, M. R. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol. Cell Biol. 22, 8353–8365 (2002).
Han, N. et al. HAT1 induces lung cancer cell apoptosis via up regulating Fas. Oncotarget 8, 89970–89977 (2017).
Zhang, J., Liu, M., Liu, W. & Wang, W. Ras-ERK1/2 signalling promotes the development of osteosarcoma through regulation of H4K12ac through HAT1. Artif. Cells Nanomed. Biotechnol. 47, 1207–1215 (2019).
Yuan, J., Yin, Z., Tao, K., Wang, G. & Gao, J. Function of insulin-like growth factor 1 receptor in cancer resistance to chemotherapy. Oncol. Lett. 15, 41–47 (2018).
Worrall, C. et al. Novel mechanisms of regulation of IGF-1R action: functional and therapeutic implications. Pediatr. Endocrinol. Rev. 10, 473–484 (2013).
Alcala, A. M. & Flaherty, K. T. BRAF inhibitors for the treatment of metastatic melanoma: clinical trials and mechanisms of resistance. Clin. Cancer Res. 18, 33–39 (2012).
Liu, F., Yang, X., Geng, M. & Huang, M. Targeting ERK, an Achilles’ Heel of the MAPK pathway, in cancer therapy. Acta Pharm. Sin. B 8, 552–562 (2018).
Hatzivassiliou, G. et al. ERK inhibition overcomes acquired resistance to MEK inhibitors. Mol. Cancer Ther. 11, 1143–1154 (2012).
Li, Y., Dong, Q. & Cui, Y. Synergistic inhibition of MEK and reciprocal feedback networks for targeted intervention in malignancy. Cancer Biol. Med. 16, 415–434. (2019).
Camp, R. L., Dolled-Filhart, M., King, B. L. & Rimm, D. L. Quantitative analysis of breast cancer tissue microarrays shows that both high and normal levels of HER2 expression are associated with poor outcome. Cancer Res. 63, 1445–1448 (2003).
Acknowledgements
We gratefully acknowledge grants from the National Institutes of Health [R03CA221926, R03CA230815, R21CA229927, R01CA196566, and R01CA200919].
Author information
Authors and Affiliations
Contributions
R.G. designed the experiments. S.B., K.B., S.C., R.P., D.L.D.M., D.S., and G.C. performed the experiments. E.S.Y. and D.B.J. are collaborators. R.G. interpreted the data and wrote the paper. All authors have read and approved the final version of the paper.
Corresponding author
Ethics declarations
Conflict of interest
D.B.J. serves on advisory boards for Array Biopharma, BMS, Jansen, Merck, and Novartis, and receives research funding from BMS and Incyte. E.S.Y. serves on advisory boards for Strata Oncology, AstraZeneca and Bayer, serves as consultant for Eli Lilly and AstraZeneca, and receives research funding from Eli Lilly, Novartis, and PUMA.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bugide, S., Parajuli, K.R., Chava, S. et al. Loss of HAT1 expression confers BRAFV600E inhibitor resistance to melanoma cells by activating MAPK signaling via IGF1R. Oncogenesis 9, 44 (2020). https://doi.org/10.1038/s41389-020-0228-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41389-020-0228-x
- Springer Nature Limited
This article is cited by
-
TGF-β Signaling Activation Confers Anlotinib Resistance in Gastric Cancer
Pharmaceutical Research (2023)
-
Histone acetyltransferase 1 promotes gemcitabine resistance by regulating the PVT1/EZH2 complex in pancreatic cancer
Cell Death & Disease (2021)
-
Disruptor of telomeric silencing 1-like promotes ovarian cancer tumor growth by stimulating pro-tumorigenic metabolic pathways and blocking apoptosis
Oncogenesis (2021)
-
The association of immunosurveillance and distant metastases in colorectal cancer
Journal of Cancer Research and Clinical Oncology (2021)
-
Blockade of EIF5A hypusination limits colorectal cancer growth by inhibiting MYC elongation
Cell Death & Disease (2020)