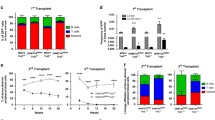
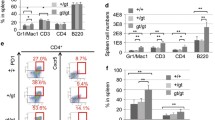
Abstract
DNA methylation-related genes, including TET2, IDH2, and DNMT3A are highly frequently mutated in angioimmunoblastic T-cell lymphoma (AITL), an aggressive malignancy of T follicular helper (Tfh) cells associated with aberrant immune features. It has been shown that TET2 loss cooperates with RHOAG17V to promote AITL in mice but the functional role of DNMT3A mutations in AITL remains unclear. Here, we report that DNMT3AR882H, the most common mutation of DNMT3A in AITL, accelerates the development of Tet2−/−; RHOAG17V AITL in mice, indicated by the expansion of malignant Tfh cells and aberrant B cells, skin rash, and significantly shortened disease-free survival. To understand the underlying cellular and molecular mechanisms, we performed single-cell transcriptome analyses of lymph nodes of mice transplanted with Tet2−/−, Tet2−/−; RHOAG17V or DNMT3AR882H; Tet2−/−; RHOAG17V hematopoietic stem and progenitor cells. These single-cell landscapes reveal that DNMT3A mutation further activates Tfh cells and leads to rapid and terminal differentiation of B cells, probably through enhancing the interacting PD1/PD-L1, ICOS/ICOSL, CD28/CD86, and ICAM1/ITGAL pairs. Our study establishes the functional roles of DNMT3A mutation in AITL and sheds light on the molecular mechanisms of this disease.






Similar content being viewed by others
Data availability
Single-cell RNA-seq data were deposited in the Gene Expression Omnibus database repository under accession number 142645. The private code of GSE142645 is mfuvgkuerburdqr.
Code availability
The analysis code can be found on GitHub (https://github.com/pangxueyu233/DNMT3AR882H-accelerates-angioimmunoblastic-T-cell-lymphoma).
References
Vose J, Armitage J, Weisenburger D. International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol: Off J Am Soc Clin Oncol. 2008;26:4124–30. https://doi.org/10.1200/jco.2008.16.4558.
Mourad N, Mounier N, Brière J, Raffoux E, Delmer A, Feller A, et al. Clinical, biologic, and pathologic features in 157 patients with angioimmunoblastic T-cell lymphoma treated within the Groupe d’Etude des Lymphomes de l’Adulte (GELA) trials. Blood. 2008;111:4463–70. https://doi.org/10.1182/blood-2007-08-105759.
Federico M, Rudiger T, Bellei M, Nathwani BN, Luminari S, Coiffier B, et al. Clinicopathologic characteristics of angioimmunoblastic T-cell lymphoma: analysis of the international peripheral T-cell lymphoma project. J Clin Oncol: Off J Am Soc Clin Oncol. 2013;31:240–6. https://doi.org/10.1200/jco.2011.37.3647.
Attygalle AD, Kyriakou C, Dupuis J, Grogg KL, Diss TC, Wotherspoon AC, et al. Histologic evolution of angioimmunoblastic T-cell lymphoma in consecutive biopsies: clinical correlation and insights into natural history and disease progression. Am J Surg Pathol. 2007;31:1077–88. https://doi.org/10.1097/PAS.0b013e31802d68e9.
de Leval L, Rickman DS, Thielen C, Reynies A, Huang YL, Delsol G, et al. The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood. 2007;109:4952–63. https://doi.org/10.1182/blood-2006-10-055145.
Piccaluga PP, Agostinelli C, Califano A, Carbone A, Fantoni L, Ferrari S, et al. Gene expression analysis of angioimmunoblastic lymphoma indicates derivation from T follicular helper cells and vascular endothelial growth factor deregulation. Cancer Res. 2007;67:10703–10. https://doi.org/10.1158/0008-5472.Can-07-1708.
Cortes JR, Ambesi-Impiombato A, Couronné L, Quinn SA, Kim CS, da Silva Almeida AC, et al. RHOA G17V induces T follicular helper cell specification and promotes lymphomagenesis. Cancer Cell. 2018;33:259–.e257. https://doi.org/10.1016/j.ccell.2018.01.001.
Chiba S, Sakata-Yanagimoto M. Advances in understanding of angioimmunoblastic T-cell lymphoma. Leukemia. 2020;34:2592–606. https://doi.org/10.1038/s41375-020-0990-y.
Mondragón L, Mhaidly R, De Donatis GM, Tosolini M, Dao P, Martin AR, et al. GAPDH overexpression in the T cell lineage promotes angioimmunoblastic T cell lymphoma through an NF-κB-dependent mechanism. Cancer Cell. 2019;36:268–.e210. https://doi.org/10.1016/j.ccell.2019.07.008.
Shi J, Hou S, Fang Q, Liu X, Liu X, Qi H. PD-1 controls follicular T helper cell positioning and function. Immunity. 2018;49:264–.e264. https://doi.org/10.1016/j.immuni.2018.06.012.
Odejide O, Weigert O, Lane AA, Toscano D, Lunning MA, Kopp N, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014;123:1293–6. https://doi.org/10.1182/blood-2013-10-531509.
Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46:171–5. https://doi.org/10.1038/ng.2872.
Palomero T, Couronné L, Khiabanian H, Kim MY, Ambesi-Impiombato A, Perez-Garcia A, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. 2014;46:166–70. https://doi.org/10.1038/ng.2873.
FlowJo™ Software. Version 10.3. Ashland, OR: Becton, Dickinson and Company. 2019.
Ng SY, Brown L, Stevenson K, deSouza T, Aster JC, Louissaint A Jr., et al. RhoA G17V is sufficient to induce autoimmunity and promotes T-cell lymphomagenesis in mice. Blood. 2018;132:935–47. https://doi.org/10.1182/blood-2017-11-818617.
Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N. Engl J Med. 2010;363:2424–33. https://doi.org/10.1056/NEJMoa1005143.
Shah MY, Licht JD. DNMT3A mutations in acute myeloid leukemia. Nat Genet. 2011;43:289–90. https://doi.org/10.1038/ng0411-289.
Wang C, McKeithan TW, Gong Q, Zhang W, Bouska A, Rosenwald A, et al. IDH2R172 mutations define a unique subgroup of patients with angioimmunoblastic T-cell lymphoma. Blood. 2015;126:1741–52. https://doi.org/10.1182/blood-2015-05-644591.
Yao WQ, Wu F, Zhang W, Chuang SS, Thompson JS, Chen Z, et al. Angioimmunoblastic T-cell lymphoma contains multiple clonal T-cell populations derived from a common TET2 mutant progenitor cell. J Pathol. 2020;250:346–57. https://doi.org/10.1002/path.5376.
Nguyen TB, Sakata-Yanagimoto M, Asabe Y, Matsubara D, Kano J, Yoshida K, et al. Identification of cell-type-specific mutations in nodal T-cell lymphomas. Blood Cancer J. 2017;7:e516 https://doi.org/10.1038/bcj.2016.122.
Scourzic L, Couronné L, Pedersen MT, Della Valle V, Diop M, Mylonas E, et al. DNMT3A(R882H) mutant and Tet2 inactivation cooperate in the deregulation of DNA methylation control to induce lymphoid malignancies in mice. Leukemia. 2016;30:1388–98. https://doi.org/10.1038/leu.2016.29.
Crescenzo R, Abate F, Lasorsa E, Tabbo F, Gaudiano M, Chiesa N, et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell. 2015;27:516–32. https://doi.org/10.1016/j.ccell.2015.03.006.
Lu KT, Kanno Y, Cannons JL, Handon R, Bible P, Elkahloun AG, et al. Functional and epigenetic studies reveal multistep differentiation and plasticity of in vitro-generated and in vivo-derived follicular T helper cells. Immunity. 2011;35:622–32. https://doi.org/10.1016/j.immuni.2011.07.015.
Attygalle A, Al-Jehani R, Diss TC, Munson P, Liu H, Du M-Q, et al. Neoplastic T cells in angioimmunoblastic T-cell lymphoma express CD10. Blood. 2002;99:627–33. https://doi.org/10.1182/blood.V99.2.627.
Hippen AA, Falco MM, Weber LM, Erkan EP, Zhang K, Doherty JA, et al. miQC: an adaptive probabilistic framework for quality control of single-cell RNA-sequencing data. PLOS Comput Biol. 2021;17:e1009290 https://doi.org/10.1371/journal.pcbi.1009290.
Na F, Pan X, Chen J, Chen X, Wang M, Chi P, et al. KMT2C deficiency promotes small cell lung cancer metastasis through DNMT3A-mediated epigenetic reprogramming. Nat Cancer. 2022;3:753–67. https://doi.org/10.1038/s43018-022-00361-6.
Wang M, Chen X, Tan P, Wang Y, Pan X, Lin T, et al. Acquired semi-squamatization during chemotherapy suggests differentiation as a therapeutic strategy for bladder cancer. Cancer Cell. 2022;40:1044–.e1048. https://doi.org/10.1016/j.ccell.2022.08.010.
Pritchett JC, Yang ZZ, Kim HJ, Villasboas JC, Tang X, Jalali S, et al. High-dimensional and single-cell transcriptome analysis of the tumor microenvironment in angioimmunoblastic T cell lymphoma (AITL). Leukemia. 2022;36:165–76. https://doi.org/10.1038/s41375-021-01321-2.
Yoo HY, Sung MK, Lee SH, Kim S, Lee H, Park S, et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46:371–5. https://doi.org/10.1038/ng.2916.
Cortes JR, Palomero T. The curious origins of angioimmunoblastic T-cell lymphoma. Curr Opin Hematol. 2016;23:434–43. https://doi.org/10.1097/MOH.0000000000000261.
Lemonnier F, Couronné L, Parrens M, Jaïs JP, Travert M, Lamant L, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood. 2012;120:1466–9. https://doi.org/10.1182/blood-2012-02-408542.
Couronné L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. N. Engl J Med. 2012;366:95–96. https://doi.org/10.1056/NEJMc1111708.
Fukumoto K, Nguyen TB, Chiba S, Sakata-Yanagimoto M. Review of the biologic and clinical significance of genetic mutations in angioimmunoblastic T-cell lymphoma. Cancer Sci. 2018;109:490–6. https://doi.org/10.1111/cas.13393.
Li Z, Cai X, Cai CL, Wang J, Zhang W, Petersen BE, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–18. https://doi.org/10.1182/blood-2010-12-325241.
Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. https://doi.org/10.1016/j.ccr.2011.06.001.
Yang L, Rodriguez B, Mayle A, Park HJ, Lin X, Luo M, et al. DNMT3A loss drives enhancer hypomethylation in FLT3-ITD-associated leukemias. Cancer Cell. 2016;29:922–34. https://doi.org/10.1016/j.ccell.2016.05.003.
Chen C, Liu Y, Lu C, Cross JR, Morris JPT, Shroff AS, et al. Cancer-associated IDH2 mutants drive an acute myeloid leukemia that is susceptible to Brd4 inhibition. Genes Dev. 2013;27:1974–85. https://doi.org/10.1101/gad.226613.113.
Leca J, Lemonnier F, Meydan C, Foox J, El Ghamrasni S, Mboumba DL et al. IDH2 and TET2 mutations synergize to modulate T Follicular Helper cell functional interaction with the AITL microenvironment. Cancer Cell. 2023; https://doi.org/10.1016/j.ccell.2023.01.003.
Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207:339–44. https://doi.org/10.1084/jem.20092506.
Janin M, Mylonas E, Saada V, Micol JB, Renneville A, Quivoron C, et al. Serum 2-hydroxyglutarate production in IDH1- and IDH2-mutated de novo acute myeloid leukemia: a study by the acute leukemia French association group. J Clin Oncol. 2014;32:297–305. https://doi.org/10.1200/jco.2013.50.2047.
Chotirat S, Thongnoppakhun W, Promsuwicha O, Boonthimat C, Auewarakul CU. Molecular alterations of isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) metabolic genes and additional genetic mutations in newly diagnosed acute myeloid leukemia patients. J Hematol Oncol. 2012;5:5 https://doi.org/10.1186/1756-8722-5-5.
Schwartz FH, Cai Q, Fellmann E, Hartmann S, Mäyränpää MI, Karjalainen-Lindsberg ML, et al. TET2 mutations in B cells of patients affected by angioimmunoblastic T-cell lymphoma. J Pathol. 2017;242:129–33. https://doi.org/10.1002/path.4898.
Fujisawa M, Nguyen TB, Abe Y, Suehara Y, Fukumoto K, Suma S, et al. Clonal germinal center B cells function as a niche for T-cell lymphoma. Blood. 2022;140:1937–50. https://doi.org/10.1182/blood.2022015451.
Kelley J, de Bono B, Trowsdale J. IRIS: a database surveying known human immune system genes. Genomics. 2005;85:503–11. https://doi.org/10.1016/j.ygeno.2005.01.009.
Pan X, Wang J, Guo L, Na F, Du J, Chen X, et al. Identifying a confused cell identity for esophageal squamous cell carcinoma. Signal Transduct Target Ther. 2022;7:122 https://doi.org/10.1038/s41392-022-00946-8.
Liu P, Pan X, Chen C, Niu T, Shuai X, Wang J, et al. Nivolumab treatment of relapsed/refractory Epstein-Barr virus-associated hemophagocytic lymphohistiocytosis in adults. Blood. 2020;135:826–33. https://doi.org/10.1182/blood.2019003886.
Acknowledgements
We thank all the members of the Chen and Liu laboratory for their invaluable suggestions and technical support. We thank Dr. Yuquan Wei for his generous support. We thank Dr. Renzhan Tong for his technical support. We thank the Core Facilities of West China Hospital for technical support.
Funding
This work was supported by the National Natural Science Foundation of China (82130007), the Sichuan Science and Technology Program (2018RZ0140, 2018JZ0077, 2022YFS0205), the Incubation Program for Clinical Trials (19HXFH030), the Achievement Transformation Project (CGZH21001), the 1.3.5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (ZYJC21009, ZYGD22012, ZYJC21007), and the Translational Research Grant of NCRCH (2021WWB03).
Author information
Authors and Affiliations
Contributions
CC and YL conceived the project and designed experiments. ZW, JZ, ZZ, HL, PL, QZ, XD and FN performed experiments. XP performed bioinformatic analyses. ZW, JZ, XP, CC, TN and YL analyzed data. ZW, JZ, XP, TN and YL prepared and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zheng, J., Wang, Z., Pan, X. et al. DNMT3AR882H accelerates angioimmunoblastic T-cell lymphoma in mice. Oncogene 42, 1940–1950 (2023). https://doi.org/10.1038/s41388-023-02699-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-023-02699-2
- Springer Nature Limited