Abstract
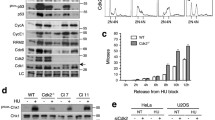
In addition to its classical role in apoptosis, accumulating evidence suggests that caspase-2 has non-apoptotic functions, including regulation of cell division. Loss of caspase-2 is known to increase proliferation rates but how caspase-2 is regulating this process is currently unclear. We show that caspase-2 is activated in dividing cells in G1-phase of the cell cycle. In the absence of caspase-2, cells exhibit numerous S-phase defects including delayed exit from S-phase, defects in repair of chromosomal aberrations during S-phase, and increased DNA damage following S-phase arrest. In addition, caspase-2-deficient cells have a higher frequency of stalled replication forks, decreased DNA fiber length, and impeded progression of DNA replication tracts. This indicates that caspase-2 protects from replication stress and promotes replication fork protection to maintain genomic stability. These functions are independent of the pro-apoptotic function of caspase-2 because blocking caspase-2-induced cell death had no effect on cell division, DNA damage-induced cell cycle arrest, or DNA damage. Thus, our data supports a model where caspase-2 regulates cell cycle and DNA repair events to protect from the accumulation of DNA damage independently of its pro-apoptotic function.








Similar content being viewed by others
Change history
15 June 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41388-022-02373-z
References
Boice A, Bouchier-Hayes L. Targeting apoptotic caspases in cancer. Biochimica Et Biophysica Acta Mol Cell Res. 2020;1867:118688.
Bouchier-Hayes L. The role of caspase-2 in stress-induced apoptosis. J Cell Mol Med. 2010;14:1212–24.
Bouchier-Hayes L, Green DR. Caspase-2: the orphan caspase. Cell Death Differ. 2012;19:51–7.
Ho LH, Taylor R, Dorstyn L, Cakouros D, Bouillet P, Kumar S. A tumor suppressor function for caspase-2. Proc Natl Acad Sci USA. 2009;106:5336–41.
Puccini J, Shalini S, Voss AK, Gatei M, Wilson CH, Hiwase DK, et al. Loss of caspase-2 augments lymphomagenesis and enhances genomic instability in Atm-deficient mice. Proc Natl Acad Sci USA. 2013;110:19920–5.
Terry MR, Arya R, Mukhopadhyay A, Berrett KC, Clair PM, Witt B, et al. Caspase-2 impacts lung tumorigenesis and chemotherapy response in vivo. Cell Death Differ. 2015;22:719–30.
Parsons MJ, McCormick L, Janke L, Howard A, Bouchier-Hayes L, Green DR. Genetic deletion of caspase-2 accelerates MMTV/c-neu-driven mammary carcinogenesis in mice. Cell Death Differ. 2013;20:1174–82.
Ren K, Lu J, Porollo A, Du C. Tumor-suppressing function of caspase-2 requires catalytic site Cys-320 and site Ser-139 in mice. J Biol Chem. 2012;287:14792–802.
Tinel A, Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–6.
Berube C, Boucher LM, Ma W, Wakeham A, Salmena L, Hakem R, et al. Apoptosis caused by p53-induced protein with death domain (PIDD) depends on the death adapter protein RAIDD. Proc Natl Acad Sci USA. 2005;102:14314–20.
Oliver TG, Meylan E, Chang GP, Xue W, Burke JR, Humpton TJ, et al. Caspase-2-mediated cleavage of Mdm2 creates a p53-induced positive feedback loop. Mol Cell. 2011;43:57–71.
Fava LL, Schuler F, Sladky V, Haschka MD, Soratroi C, Eiterer L, et al. The PIDDosome activates p53 in response to supernumerary centrosomes. Genes Dev. 2017;31:34–45.
Baptiste-Okoh N, Barsotti AM, Prives C. A role for caspase 2 and PIDD in the process of p53-mediated apoptosis. Proc Natl Acad Sci USA. 2008;105:1937–42.
Lassus P, Opitz-Araya X, Lazebnik Y. Requirement for caspase-2 in stress-induced apoptosis before mitochondrial permeabilization. Science. 2002;297:1352–4.
Robertson JD, Enoksson M, Suomela M, Zhivotovsky B, Orrenius S. Caspase-2 acts upstream of mitochondria to promote cytochrome c release during etoposide-induced apoptosis. J Biol Chem. 2002;277:29803–9.
Baptiste-Okoh N, Barsotti A, Prives C. A role for caspase 2 and PIDD in the process of p53-mediated apoptosis. Proc Natl Acad Sci USA. 2008;105:1937–42.
Sidi S, Sanda T, Kennedy RD, Hagen AT, Jette CA, Hoffmans R, et al. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell. 2008;133:864–77.
Pan Y, Ren KH, He HW, Shao RG. Knockdown of Chk1 sensitizes human colon carcinoma HCT116 cells in a p53-dependent manner to lidamycin through abrogation of a G2/M checkpoint and induction of apoptosis. Cancer Biol Ther. 2009;8:1559–66.
Ando K, Parsons MJ, Shah RB, Charendoff CI, Paris SL, Liu PH, et al. NPM1 directs PIDDosome-dependent caspase-2 activation in the nucleolus. J Cell Biol. 2017;216:1795–810.
Robeson AC, Lindblom KR, Wojton J, Kornbluth S, Matsuura K. Dimer-specific immunoprecipitation of active caspase-2 identifies TRAF proteins as novel activators. EMBO J. 2018;37:e97072.
Bouchier-Hayes L, Oberst A, McStay GP, Connell S, Tait SW, Dillon CP, et al. Characterization of cytoplasmic caspase-2 activation by induced proximity. Mol Cell. 2009;35:830–40.
Manzl C, Krumschnabel G, Bock F, Sohm B, Labi V, Baumgartner F, et al. Caspase-2 activation in the absence of PIDDosome formation. J Cell Biol. 2009;185:291–303.
Nitiss J, Wang JC. DNA topoisomerase-targeting antitumor drugs can be studied in yeast. Proc Natl Acad Sci. 1988;85:7501–5.
Goldwasser F, Shimizu T, Jackman J, Hoki Y, O’Connor PM, Kohn KW, et al. Correlations between S and G2 arrest and the cytotoxicity of camptothecin in human colon carcinoma cells. Cancer Res. 1996;56:4430–7.
Nelson WG, Kastan MB. DNA strand breaks: the DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol Cell Biol. 1994;14:1815–23.
Saintigny Y, Delacote F, Vares G, Petitot F, Lambert S, Averbeck D, et al. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 2001;20:3861–70.
Gundry MC, Brunetti L, Lin A, Mayle AE, Kitano A, Wagner D, et al. Highly efficient genome editing of murine and human hematopoietic progenitor cells by CRISPR/Cas9. Cell Rep. 2016;17:1453–61.
Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods. 2015;12:982–8.
Lapytsko A, Kollarovic G, Ivanova L, Studencka M, Schaber J. FoCo: a simple and robust quantification algorithm of nuclear foci. BMC Bioinformatics. 2015;16:392.
Krull A, Buchholz T-O, Jug F. Noise2Void - learning denoising from single noisy images. 2019 Ieee Cvf Conf Comput Vis Pattern Recognit Cvpr. 2019;00:2124–32.
von Chamier L, Laine RF, Jukkala J, Spahn C, Krentzel D, Nehme E, et al. Democratising deep learning for microscopy with ZeroCostDL4Mic. Nat Commun. 2021;12:2276.
Stringer C, Wang T, Michaelos M, Pachitariu M. Cellpose: a generalist algorithm for cellular segmentation. Nat Methods. 2021;18:100–6.
Singh M, Hunt CR, Pandita RK, Kumar R, Yang CR, Horikoshi N, et al. Lamin A/C depletion enhances DNA damage-induced stalled replication fork arrest. Mol Cell Biol. 2013;33:1210–22.
Sharma GG, Hwang KK, Pandita RK, Gupta A, Dhar S, Parenteau J, et al. Human heterochromatin protein 1 isoforms HP1(Hsalpha) and HP1(Hsbeta) interfere with hTERT-telomere interactions and correlate with changes in cell growth and response to ionizing radiation. Mol Cell Biol. 2003;23:8363–76.
Hunt CR, Dix DJ, Sharma GG, Pandita RK, Gupta A, Funk M, et al. Genomic instability and enhanced radiosensitivity in Hsp70.1- and Hsp70.3-deficient mice. Mol Cell Biol. 2004;24:899–911.
Baliga BC, Read SH, Kumar S. The biochemical mechanism of caspase-2 activation. Cell Death Differ. 2004;11:1234–41.
Nishitani H, Lygerou Z, Nishimoto T. Proteolysis of DNA replication licensing factor Cdt1 in S-phase is performed independently of geminin through its N-terminal region. J Biol Chem. 2004;279:30807–16.
Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–98.
Nishitani H, Taraviras S, Lygerou Z, Nishimoto T. The human licensing factor for DNA replication Cdt1 accumulates in G1 and is destabilized after initiation of S-phase. J Biol Chem. 2001;276:44905–11.
Bianchi V, Pontis E, Reichard P. Changes of deoxyribonucleoside triphosphate pools induced by hydroxyurea and their relation to DNA synthesis. J Biol Chem. 1986;261:16037–42.
Singh DK, Pandita RK, Singh M, Chakraborty S, Hambarde S, Ramnarain D, et al. MOF Suppresses Replication Stress and Contributes to Resolution of Stalled Replication Forks. Mol Cell Biol. 2018;38:e00484–17.
Saintigny Y. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 2001;20:3861–70.
Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–68.
Mattoo AR, Pandita RK, Chakraborty S, Charaka V, Mujoo K, Hunt CR, et al. MCL-1 Depletion Impairs DNA Double-Strand Break Repair and Reinitiation of Stalled DNA Replication Forks. Mol Cell Biol. 2017;37:e00535–16.
Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–8.
Sanchez Y. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–501.
Mailand N. Rapid destruction of human Cdc25A in response to DNA damage. Science. 2000;288:1425–9.
Liu S, Shiotani B, Lahiri M, Marechal A, Tse A, Leung CC, et al. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol Cell. 2011;43:192–202.
Falck J, Petrini JHJ, Williams BR, Lukas J, Bartek J. The DNA damage-dependent intra–S phase checkpoint is regulated by parallel pathways. Nat Genet. 2002;30:290–4.
Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–7.
Ismail IH, Nyström S, Nygren J, Hammarsten O. Activation of ataxia telangiectasia mutated by DNA strand break-inducing agents correlates closely with the number of DNA double strand breaks. J Biol Chem. 2005;280:4649–55.
Guo Y, Srinivasula SM, Druilhe A, Fernandes-Alnemri T, Alnemri ES. Caspase-2 induces apoptosis by releasing proapoptotic proteins from mitochondria. J Biol Chem. 2002;277:13430–7.
Bonzon C, Bouchier-Hayes L, Pagliari LJ, Green DR, Newmeyer DD. Caspase-2-induced apoptosis requires bid cleavage: a physiological role for bid in heat shock-induced death. Mol Biol Cell. 2006;17:2150–7.
Howard A. Synthesis of deoxyribonucleic acid in normal and irradiated ceils and its relation to chromosome breakage. Heredity Suppl. 1953;6:261–73.
Kuzminov A. Collapse and repair of replication forks in Escherichia coli. Mol Microbiol. 1995;16:373–84.
Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9.
Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–46.
Beck H, Nahse-Kumpf V, Larsen MS, O’Hanlon KA, Patzke S, Holmberg C, et al. Cyclin-dependent kinase suppression by WEE1 kinase protects the genome through control of replication initiation and nucleotide consumption. Mol Cell Biol. 2012;32:4226–36.
Lopez-Garcia C, Sansregret L, Domingo E, McGranahan N, Hobor S, Birkbak NJ, et al. BCL9L dysfunction impairs caspase-2 expression permitting aneuploidy tolerance in colorectal cancer. Cancer cell. 2017;31:79–93.
Dawar S, Lim Y, Puccini J, White M, Thomas P, Bouchier-Hayes L, et al. Caspase-2-mediated cell death is required for deleting aneuploid cells. Oncogene. 2017;36:2704–14.
Paulovich AG, Hartwell LH. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–7.
Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–52.
Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: The trinity at the heart of the DNA damage response. Mol Cell. 2017;66:801–17.
Saldivar JC, Cortez D, Cimprich KA. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat Rev Mol Cell Biol. 2017;18:622–36.
Wang H, Wang H, Powell SN, Iliakis G, Wang Y. ATR affecting cell radiosensitivity is dependent on homologous recombination repair but independent of nonhomologous end joining. Cancer Res. 2004;64:7139–43.
Couch FB, Bansbach CE, Driscoll R, Luzwick JW, Glick GG, Betous R, et al. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 2013;27:1610–23.
Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 2016;17:337–49.
Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–96.
Khanna KK, Keating KE, Kozlov S, Scott S, Gatei M, Hobson K, et al. ATM associates with and phosphorylates p53: mapping the region of interaction. Nat Genet. 1998;20:398–400.
Gatei M, Scott SP, Filippovitch I, Soronika N, Lavin MF, Weber B, et al. Role for ATM in DNA damage-induced phosphorylation of BRCA1. Cancer Res. 2000;60:3299–304.
Lim D-S, Kim S-T, Xu B, Maser RS, Lin J, Petrini JHJ, et al. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–7.
Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci. 2000;97:10389–94.
Chaturvedi P, Eng WK, Zhu Y, Mattern MR, Mishra R, Hurle MR, et al. Mammalian Chk2 is a downstream effector of the ATM-dependent DNA damage checkpoint pathway. Oncogene. 1999;18:4047–54.
Brown AL, Lee CH, Schwarz JK, Mitiku N, Piwnica-Worms H, Chung JH. A human Cds1-related kinase that functions downstream of ATM protein in the cellular response to DNA damage. Proc Natl Acad Sci. 1999;96:3745–50.
Zhong Y, Nellimoottil T, Peace JM, Knott SR, Villwock SK, Yee JM, et al. The level of origin firing inversely affects the rate of replication fork progression. J Cell Biol. 2013;201:373–83.
Andersen JL, Johnson CE, Freel CD, Parrish AB, Day JL, Buchakjian MR, et al. Restraint of apoptosis during mitosis through interdomain phosphorylation of caspase-2. Embo J. 2009;20:3216–27.
Lim Y, De Bellis D, Sandow JJ, Capalbo L, D'Avino PP, Murphy JM, et al. Phosphorylation by Aurora B kinase regulates caspase-2 activity and function. Cell Death Differ. 2020;28:349–66.
Nurse P. Universal control mechanism regulating onset of M-phase. Nature. 1990;344:503–8.
Glover DM, Leibowitz MH, McLean DA, Parry H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;81:95–105.
Gao Z, Shao Y, Jiang X. Essential roles of the Bcl-2 family of proteins in caspase-2-induced apoptosis. J Biol Chem. 2005;280:38271–5.
Lin Y-F, Shih H-Y, Shang Z-F, Kuo C-T, Guo J, Du C, et al. PIDD mediates the association of DNA-PKcs and ATR at stalled replication forks to facilitate the ATR signaling pathway. Nucleic Acids Res. 2018;46:1847–59.
Henry CM, Martin SJ. Caspase-8 acts in a non-enzymatic role as a scaffold for assembly of a pro-inflammatory “FADDosome” Complex upon TRAIL Stimulation. Mol Cell. 2017;65:715–29 e715.
Acknowledgements
We would like to thank Jennifer Martinez (NIEHS) for careful reading of this paper. Funding for this project includes NIH/NIGMS R01GM121389 (LBH), NIH/NCI R21CA256606 (LBH) and NIH/NIGMS T32GM008231 (KEL). This project was supported by the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the NIH (P30 AI036211, P30 CA125123, and S10 RR024574) and the expert assistance of J. M. Sederstrom. We would like to acknowledge the Texas Children’s Hospital William T. Shearer Center for Human Immunobiology for their generous support for this research and the expert assistance of Rebecca Kairis.
Author information
Authors and Affiliations
Contributions
AGB, KEL, MJP, TKP, and LB-H conceived and designed experiments. AGB, KEL, RKP, MJP, CIC, and VC performed the experiments. AFC developed the imaging analysis. AGB and LB-H wrote the paper.
Corresponding author
Ethics declarations
Competing interests
MJP is currently employed at BD Biosciences. The remaining authors have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Boice, A.G., Lopez, K.E., Pandita, R.K. et al. Caspase-2 regulates S-phase cell cycle events to protect from DNA damage accumulation independent of apoptosis. Oncogene 41, 204–219 (2022). https://doi.org/10.1038/s41388-021-02085-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-02085-w
- Springer Nature Limited