Abstract
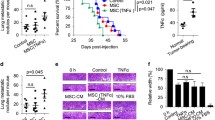
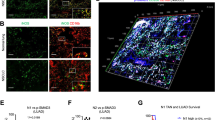
Myeloid-derived suppressor cells (MDSCs) suppress antitumor immune activities and facilitate cancer progression. Although the concept of immunosuppressive MDSCs is well established, the mechanism that MDSCs regulate non-small cell lung cancer (NSCLC) progression through the paracrine signals is still lacking. Here, we reported that the infiltration of MDSCs within NSCLC tissues was associated with the progression of cancer status, and was positively correlated with the Patient-derived xenograft model establishment, and poor patient prognosis. Intratumoral MDSCs directly promoted NSCLC metastasis and highly expressed chemokines that promote NSCLC cells invasion, including CCL11. CCL11 was capable of activating the AKT and ERK signaling pathways to promote NSCLC metastasis through the epithelial-mesenchymal transition (EMT) process. Moreover, high expression of CCL11 was associated with a poor prognosis in lung cancer as well as other types of cancer. Our findings underscore that MDSCs produce CCL11 to promote NSCLC metastasis via activation of ERK and AKT signaling and induction of EMT, suggesting that the MDSCs-CCL11-ERK/AKT-EMT axis contains potential targets for NSCLC metastasis treatment.






Similar content being viewed by others
Data availability
RNA-Seq of human NSCLC samples, MDSCs and A549 cells have been uploaded to public databases (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE136949). Other data generated or analyzed during this study are included either in this article or in the supplementary information files.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30.
Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69:363–85.
Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168:670–91.
Yang Z, Guo J, Weng L, Tang W, Jin S, Ma W. Myeloid-derived suppressor cells-new and exciting players in lung cancer. J Hematol Oncol. 2020;13:10.
Peranzoni E, Zilio S, Marigo I, Dolcetti L, Zanovello P, Mandruzzato S, et al. Myeloid-derived suppressor cell heterogeneity and subset definition. Curr Opin Immunol. 2010;22:238–44.
Marvel D, Gabrilovich DI. Myeloid-derived suppressor cells in the tumor microenvironment: expect the unexpected. J Clin Investig. 2015;125:3356–64.
Zhang G, Huang H, Zhu Y, Yu G, Gao X, Xu Y, et al. A novel subset of B7-H3(+)CD14(+)HLA-DR(-/low) myeloid-derived suppressor cells are associated with progression of human NSCLC. Oncoimmunology. 2015;4:e977164.
Meyer C, Cagnon L, Costa-Nunes CM, Baumgaertner P, Montandon N, Leyvraz L, et al. Frequencies of circulating MDSC correlate with clinical outcome of melanoma patients treated with ipilimumab. Cancer Immunol Immunother. 2014;63:247–57.
Huang A, Zhang B, Wang B, Zhang F, Fan KX, Guo YJ. Increased CD14(+)HLA-DR (-/low) myeloid-derived suppressor cells correlate with extrathoracic metastasis and poor response to chemotherapy in non-small cell lung cancer patients. Cancer Immunol Immunother. 2013;62:1439–51.
Diaz-Montero CM, Salem ML, Nishimura MI, Garrett-Mayer E, Cole DJ, Montero AJ. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin-cyclophosphamide chemotherapy. Cancer Immunol Immunother. 2009;58:49–59.
Sun HL, Zhou X, Xue YF, Wang K, Shen YF, Mao JJ, et al. Increased frequency and clinical significance of myeloid-derived suppressor cells in human colorectal carcinoma. World J Gastroenterol. 2012;18:3303–9.
Wang L, Chang EW, Wong SC, Ong SM, Chong DQ, Ling KL. Increased myeloid-derived suppressor cells in gastric cancer correlate with cancer stage and plasma S100A8/A9 proinflammatory proteins. J Immunol. 2013;190:794–804.
Lindau D, Gielen P, Kroesen M, Wesseling P, Adema GJ. The immunosuppressive tumour network: myeloid-derived suppressor cells, regulatory T cells and natural killer T cells. Immunology. 2013;138:105–15.
Mao Y, Poschke I, Wennerberg E, Pico de Coana Y, Egyhazi Brage S, Schultz I, et al. Melanoma-educated CD14+ cells acquire a myeloid-derived suppressor cell phenotype through COX-2-dependent mechanisms. Cancer Res. 2013;73:3877–87.
Rodriguez PC, Quiceno DG, Zabaleta J, Ortiz B, Zea AH, Piazuelo MB, et al. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 2004;64:5839–49.
Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–52.
Lin S, Huang G, Xiao Y, Sun W, Jiang Y, Deng Q, et al. CD215+ Myeloid Cells Respond to Interleukin 15 Stimulation and Promote Tumor Progression. Front Immunol. 2017;8:1713.
Zhao F, Hoechst B, Duffy A, Gamrekelashvili J, Fioravanti S, Manns MP, et al. S100A9 a new marker for monocytic human myeloid-derived suppressor cells. Immunology. 2012;136:176–83.
Lin S, Huang G, Cheng L, Li Z, Xiao Y, Deng Q, et al. Establishment of peripheral blood mononuclear cell-derived humanized lung cancer mouse models for studying efficacy of PD-L1/PD-1 targeted immunotherapy. MAbs. 2018;10:1301–11.
Ye W, Jiang Z, Li GX, Xiao Y, Lin S, Lai Y, et al. Quantitative evaluation of the immunodeficiency of a mouse strain by tumor engraftments. J Hematol Oncol. 2015;8:59.
Oh K, Lee OY, Shon SY, Nam O, Ryu PM, Seo MW. et al. A mutual activation loop between breast cancer cells and myeloid-derived suppressor cells facilitates spontaneous metastasis through IL-6 trans-signaling in a murine model. Breast Cancer Res. 2013;15:R79. https://doi.org/10.1186/bcr3473.
Ichikawa M, Williams R, Wang L, Vogl T, Srikrishna G. S100A8/A9 activate key genes and pathways in colon tumor progression. Mol Cancer Res. 2011;9:133–48.
Levina V, Nolen BM, Marrangoni AM, Cheng P, Marks JR, Szczepanski MJ, et al. Role of eotaxin-1 signaling in ovarian cancer. Clin Cancer Res. 2009;15:2647–56.
Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4:540–50.
Wang D, Sun H, Wei J, Cen B, DuBois RN. CXCL1 Is Critical for Premetastatic Niche Formation and Metastasis in Colorectal Cancer. Cancer Res. 2017;77:3655–65.
Tian W, Liu J, Pei B, Wang X, Guo Y, Yuan L. Identification of miRNAs and differentially expressed genes in early phase non-small cell lung cancer. Oncol Rep. 2016;35:2171–6.
Liu H, Zhao H. Prognosis related miRNAs, DNA methylation, and epigenetic interactions in lung adenocarcinoma. Neoplasma. 2019;66:487–93.
Zajkowska M, Mroczko B. Eotaxins and their receptor in colorectal cancer-a literature review. Cancers. 2020;12:1383. https://doi.org/10.3390/cancers12061383.
Alzahrani AS. PI3K/Akt/mTOR inhibitors in cancer: at the bench and bedside. Semin Cancer Biol. 2019;59:125–32.
Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 2011;36:320–8.
Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer cell. 2012;22:725–36.
Xu W, Yang Z, Lu N. A new role for the PI3K/Akt signaling pathway in the epithelial-mesenchymal transition. Cell Adh Migr. 2015;9:317–24.
Watanabe-Takano H, Takano K, Hatano M, Tokuhisa T, Endo T. DA-Raf-mediated suppression of the Ras-ERK pathway is essential for TGF-beta 1-induced epithelial-mesenchymal transition in alveolar epithelial type 2 cells. PLoS ONE. 2015;10:e0127888. https://doi.org/10.1371/journal.pone.0127888.
Ouzounova M, Lee E, Piranlioglu R, El Andaloussi A, Kolhe R, Demirci MF, et al. Monocytic and granulocytic myeloid derived suppressor cells differentially regulate spatiotemporal tumour plasticity during metastatic cascade. Nat Commun. 2017;8:14979.
Xu W, Qian J, Zeng F, Li S, Guo W, Chen L, et al. Protein kinase Ds promote tumor angiogenesis through mast cell recruitment and expression of angiogenic factors in prostate cancer microenvironment. J Exp Clin Cancer Res. 2019;38:114.
Miyagaki T, Sugaya M, Fujita H, Ohmatsu H, Kakinuma T, Kadono T, et al. Eotaxins and CCR3 interaction regulates the Th2 environment of cutaneous T-cell lymphoma. J Invest Dermatol. 2010;130:2304–11.
Miyagaki T, Sugaya M, Murakami T, Asano Y, Tada Y, Kadono T, et al. CCL11-CCR3 interactions promote survival of anaplastic large cell lymphoma cells via ERK1/2 activation. Cancer Res. 2011;71:2056–65.
Zhu F, Liu P, Li J, Zhang Y. Eotaxin-1 promotes prostate cancer cell invasion via activation of the CCR3-ERK pathway and upregulation of MMP-3 expression. Oncol Rep. 2014;31:2049–54.
Tian M, Chen LN, Ma L, Wang DD, Shao B, Wu JY, et al. Expression and prognostic significance of CCL11/CCR3 in glioblastoma. Oncotarget. 2016;7:32617–27.
Blank S, Nienhuser H, Dreikhausen L, Sisic L, Heger U, Ott K, et al. Inflammatory cytokines are associated with response and prognosis in patients with esophageal cancer. Oncotarget. 2017;8:47518–32.
Zohny SF, Fayed ST. Clinical utility of circulating matrix metalloproteinase-7 (MMP-7), CC chemokine ligand 18 (CCL18) and CC chemokine ligand 11 (CCL11) as markers for diagnosis of epithelial ovarian cancer. Med Oncol. 2010;27:1246–53.
Agarwal M, He C, Siddiqui J, Wei JT, Macoska JA. CCL11 (eotaxin-1): a new diagnostic serum marker for prostate cancer. Prostate. 2013;73:573–81.
Lai Y, Wei X, Lin S, Qin L, Cheng L, Li P. Current status and perspectives of patient-derived xenograft models in cancer research. J Hematol Oncol. 2017;10:106.
DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert MT, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17:1514–20.
John T, Kohler D, Pintilie M, Yanagawa N, Pham NA, Li M, et al. The ability to form primary tumor xenografts is predictive of increased risk of disease recurrence in early-stage non-small cell lung cancer. Clin Cancer Res. 2011;17:134–41.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60.
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 2011;12:323.
Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PloS One. 2013;8:e82241.
Nagy A, Lanczky A, Menyhart O, Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227.
Acknowledgements
We thank the cancer patients who donated their tissues. This work was supported by Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19030205); and National Key Research and Development Plan (2017YFE0131600, 2019YFA0111500); and National Natural Science Foundation of China (81961128003; 81972672; 31872800; 81773301; 82003054); and China Postdoctoral Science Foundation (2018M640771); and Guangdong Provincial Significant New Drugs Development (2019B020202003); and Guangdong Basic and Applied Basic Research Foundation (2019A1515110084, 2019A1515010062, 2020A1515011516); and Guangdong Special Support Program (2017TX04R102); and Science and Technology Planning Project of Guangdong Province (2017B030314056); and Natural Science Foundation of Guangdong Province (2020A0505100062); and Guangdong Provincial Key Lab of Translational Medicine in Lung Cancer (2017B030314120); and Guangzhou City Science and Technology Key Topics Project (201904020025); and Guangzhou Science and Technology Plan Project (201907010042, 201904010473) and Foundation of Guangzhou Science and Information Technology of Guangzhou Key Project (201803040009); and Guangzhou Regenerative Medicine and Health Guangdong Laboratory Frontier Research Program (2018GZR110105003); and Clinical Innovation Research Program of Guangzhou Regenerative Medicine and Health Guangdong Laboratory (2018GZR0201002); and Research Program of the Hefei Institute of Stem Cell and Regenerative Medicine (2019YF001); and Science and Technology Program of Guangzhou (202002020083).
Author information
Authors and Affiliations
Contributions
SL and LC, contributed to the conception and design; the collection and/or assembly of data; data analysis and interpretation; and manuscript writing. XZ, GH, JL, and DZ contributed to the provision of study material or patient samples and the collection and/or assembly of data. BL, SL, SW, YL, and QW provided animal care and administrative support. WW, PL, DP, YY, SZC, ZW, and SC contributed to the conception and design of the study. XS, YY, YL, and PL contributed to the conception and design of the study; data analysis and interpretation; manuscript writing; and the final approval of the manuscript and provided financial support. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lin, S., Zhang, X., Huang, G. et al. Myeloid-derived suppressor cells promote lung cancer metastasis by CCL11 to activate ERK and AKT signaling and induce epithelial-mesenchymal transition in tumor cells. Oncogene 40, 1476–1489 (2021). https://doi.org/10.1038/s41388-020-01605-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-01605-4
- Springer Nature Limited
This article is cited by
-
The role of stromal cells in epithelial–mesenchymal plasticity and its therapeutic potential
Discover Oncology (2024)
-
Exploring CCL11 in breast cancer: unraveling its anticancer potential and immune modulatory effects involving the Akt-S6 signaling
Journal of Cancer Research and Clinical Oncology (2024)
-
SFRP1 decreases WNT-Mediated M2 macrophage marker expression in breast tissue
Cancer Immunology, Immunotherapy (2024)
-
Cancer-associated fibroblast-derived PAI-1 promotes lymphatic metastasis via the induction of EndoMT in lymphatic endothelial cells
Journal of Experimental & Clinical Cancer Research (2023)
-
MacroH2A restricts inflammatory gene expression in melanoma cancer-associated fibroblasts by coordinating chromatin looping
Nature Cell Biology (2023)