Abstract
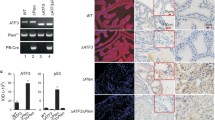
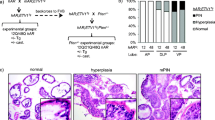
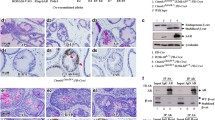
Recent genome analysis of human prostate cancers demonstrated that both AR gene amplification and TP53 mutation are among the most frequently observed alterations in advanced prostate cancer. However, the biological role of these dual genetic alterations in prostate tumorigenesis is largely unknown. In addition, there are no biologically relevant models that can be used to assess the molecular mechanisms for these genetic abnormalities. Here, we report a novel mouse model, in which elevated transgenic AR expression and Trp53 deletion occur simultaneously in mouse prostatic epithelium to mimic human prostate cancer cells. These compound mice developed an earlier onset of high-grade prostatic intraepithelial neoplasia and accelerated prostate tumors in comparison with mice harboring only the AR transgene. Histological analysis showed prostatic sarcomatoid and basaloid carcinomas with massive squamous differentiation in the above compound mice. RNA-sequencing analyses identified a robust enrichment of the signature genes for human prostatic basal cell carcinomas in the above prostate tumors. Master regulator analysis revealed SOX2 as a transcriptional regulator in prostatic basal cell tumors. Elevated expression of SOX2 and its downstream target genes were detected in prostatic tumors of the compound mice. Chromatin immunoprecipitation analyses implicate a coregulatory role of AR and SOX2 in the expression of prostatic basal cell signature genes. Our data demonstrate a critical role of SOX2 in prostate tumorigenesis and provide mechanistic insight into prostate tumor aggressiveness and progression mediated by aberrant AR and p53 signaling pathways.







Similar content being viewed by others
References
Kyprianou N, Isaacs JT. Activation of programmed cell death in the rat ventral prostate after castration. Endocrinology. 1988;122:552–62.
Huggins C, Hodges CV. Studies on prostatic cancer I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. 1941. J Urol. 2002;168:9–12.
Zhu C, Luong R, Zhuo M, Johnson DT, McKenney JK, Cunha GR, et al. Conditional expression of the androgen receptor induces oncogenic transformation of the mouse prostate. J Biol Chem. 2011;286:33478–88.
Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004;25:276–308.
Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28.
Quigley DA, Dang HX, Zhao SG, Lloyd P, Aggarwal R, Alumkal JJ, et al. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell. 2018;174:758–69.e759.
Koivisto P, Kononen J, Palmberg C, Tammela T, Hyytinen E, Isola J, et al. Androgen receptor gene amplification: a possible molecular mechanism for androgen deprivation therapy failure in prostate cancer. Cancer Res. 1997;57:314–319.
Aggarwal R, Huang J, Alumkal JJ, Zhang L, Feng FY, Thomas GV, et al. Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: a multi-institutional prospective study. J Clin Oncol. 2018;36:2492–503.
Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10:33–9.
Takeda DY, Spisak S, Seo JH, Bell C, O’Connor E, Korthauer K, et al. A somatically acquired enhancer of the androgen receptor is a noncoding driver in advanced prostate Cancer. Cell. 2018;174:422–32.e413.
Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–72.
Bookstein R, MacGrogan D, Hilsenbeck SG, Sharkey F, Allred DC. p53 is mutated in a subset of advanced-stage prostate cancers. Cancer Res. 1993;53:3369–73.
Kleihues P, Schauble B, zur Hausen A, Esteve J, Ohgaki H. Tumors associated with p53 germline mutations: a synopsis of 91 families. Am J Pathol. 1997;150:1–13.
Wang S, Gao J, Lei Q, Rozengurt N, Pritchard C, Jiao J, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4:209–21.
Zhou Z, Flesken-Nikitin A, Nikitin AY. Prostate cancer associated with p53 and Rb deficiency arises from the stem/progenitor cell–enriched proximal region of prostatic ducts. Cancer Res. 2007;67:5683–90.
Voeller HJ, Sugars LY, Pretlow T, Gelmann EP. p53 oncogene mutations in human prostate cancer specimens. J Urol. 1994;151:492–5.
Smith BA, Sokolov A, Uzunangelov V, Baertsch R, Newton Y, Graim K, et al. A basal stem cell signature identifies aggressive prostate cancer phenotypes. Proc Natl Acad Sci USA. 2015;112:E6544–52.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Grieshammer U, Agarwal P, Martin GR. A Cre transgene active in developing endodermal organs, heart, limb, and extra-ocular muscle. Genesis. 2008;46:69–73.
Ittmann M, Huang J, Radaelli E, Martin P, Signoretti S, Sullivan R, et al. Animal models of human prostate cancer: the consensus report of the New York meeting of the Mouse Models of Human Cancers Consortium Prostate Pathology Committee. Cancer Res. 2013;73:2718–36.
Zhou Z, Flesken-Nikitin A, Corney DC, Wang W, Goodrich DW, Roy-Burman P, et al. Synergy of p53 and Rb deficiency in a conditional mouse model for metastatic prostate cancer. Cancer Res. 2006;66:7889–98.
Frohwitter G, Buerger H, VAND PJ, Korsching E, Kleinheinz J, Fillies T. Cytokeratin and protein expression patterns in squamous cell carcinoma of the oral cavity provide evidence for two distinct pathogenetic pathways. Oncol Lett. 2016;12:107–13.
You S, Knudsen BS, Erho N, Alshalalfa M, Takhar M, Al-Deen Ashab H, et al. Integrated classification of prostate cancer reveals a novel luminal subtype with poor outcome. Cancer Res. 2016;76:4948–58.
Zhao SG, Chang SL, Erho N, Yu M, Lehrer J, Alshalalfa M, et al. Associations of luminal and basal subtyping of prostate cancer with prognosis and response to androgen deprivation therapy. JAMA Oncol. 2017;3:1663–72.
Mollaoglu G, Jones A, Wait SJ, Mukhopadhyay A, Jeong S, Arya R, et al. The lineage-defining transcription factors SOX2 and NKX2-1 determine lung cancer cell fate and shape the tumor immune microenvironment. Immunity. 2018;49:764–79.e769.
Chen Y, Chi P, Rockowitz S, Iaquinta PJ, Shamu T, Shukla S, et al. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat Med. 2013;19:1023–9.
Wyce A, Bai Y, Nagpal S, Thompson CC. Research Resource: The androgen receptor modulates expression of genes with critical roles in muscle development and function. Mol Endocrinol. 2010;24:1665–74.
Matusik RJ, Jin RJ, Sun Q, Wang Y, Yu X, Gupta A, et al. Prostate epithelial cell fate. Differentiation. 2008;76:682–98.
Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen CC, et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017;355:84–88.
Stoyanova T, Cooper AR, Drake JM, Liu X, Armstrong AJ, Pienta KJ, et al. Prostate cancer originating in basal cells progresses to adenocarcinoma propagated by luminal-like cells. Proc Natl Acad Sci USA. 2013;110:20111–6.
Sarkar A, Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12:15–30.
Esposito S, Russo MV, Airoldi I, Tupone MG, Sorrentino C, Barbarito G, et al. SNAI2/Slug gene is silenced in prostate cancer and regulates neuroendocrine differentiation, metastasis-suppressor and pluripotency gene expression. Oncotarget. 2015;6:17121–34.
Johnson DT, Luong R, Lee SH, Peng Y, Shaltouki A, Lee JT, et al. Deletion of leucine zipper tumor suppressor 2 (lzts2) increases susceptibility to tumor development. J Biol Chem. 2013;288:3727–38.
Kwak MK, Johnson DT, Zhu C, Lee SH, Ye DW, Luong R, et al. Conditional deletion of the Pten gene in the mouse prostate induces prostatic intraepithelial neoplasms at early ages but a slow progression to prostate tumors. PLoS ONE. 2013;8:e53476.
Lee SH, Luong R, Johnson DT, Cunha GR, Rivina L, Gonzalgo ML, et al. Androgen signaling is a confounding factor for beta-catenin-mediated prostate tumorigenesis. Oncogene. 2016;35:702–14.
Lee J, Beliakoff J, Sun Z. The novel PIAS-like protein hZimp10 is a transcriptional co-activator of the p53 tumor suppressor. Nucleic Acids Res. 2007;35:4523–34.
Ewels P, Magnusson M, Lundin S, Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–8.
Chamarthy MR, Williams SC, Moadel RM. Radioimmunotherapy of non-Hodgkin’s lymphoma: from the ‘magic bullets’ to ‘radioactive magic bullets’. Yale J Biol Med. 2011;84:391–407.
Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25.
You S, Yoo SA, Choi S, Kim JY, Park SJ, Ji JD, et al. Identification of key regulators for the migration and invasion of rheumatoid synoviocytes through a systems approach. Proc Natl Acad Sci USA. 2014;111:550–5.
Storey JD. A direct approach to false discovery rates. J R Stat Soc. 2002;64:478–9.
Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–25.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Yang JH, Li JH, Jiang S, Zhou H, Qu LH. ChIPBase: a database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic Acids Res. 2013;41:D177–87.
Linhart C, Halperin Y, Shamir R. Transcription factor and microRNA motif discovery: the Amadeus platform and a compendium of metazoan target sets. Genome Res. 2008;18:1180–9.
Chen L, Wu G, Ji H. hmChIP: a database and web server for exploring publicly available human and mouse ChIP-seq and ChIP-chip data. Bioinformatics. 2011;27:1447–8.
Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma’ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–44.
Cahan P, Li H, Morris SA, Lummertz da Rocha E, Daley GQ, Collins JJ. CellNet: network biology applied to stem cell engineering. Cell. 2014;158:903–15.
Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–40.
Acknowledgements
This work was supported by Public Health Service grants, R01CA070297, R01CA166894, R21CA190021, and R01DK104941.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
He, Y., Johnson, D.T., Yang, J.S. et al. Loss of the tumor suppressor, Tp53, enhances the androgen receptor-mediated oncogenic transformation and tumor development in the mouse prostate. Oncogene 38, 6507–6520 (2019). https://doi.org/10.1038/s41388-019-0901-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-0901-8
- Springer Nature Limited
This article is cited by
-
Aberrant androgen action in prostatic progenitor cells induces oncogenesis and tumor development through IGF1 and Wnt axes
Nature Communications (2022)
-
A comparative study of PCS and PAM50 prostate cancer classification schemes
Prostate Cancer and Prostatic Diseases (2021)