Abstract
Dental primary afferent (DPA) neurons and proprioceptive mesencephalic trigeminal nucleus (MTN) neurons, located in the trigeminal ganglion and the brainstem, respectively, are essential for controlling masticatory functions. Despite extensive transcriptomic studies on various somatosensory neurons, there is still a lack of knowledge about the molecular identities of these populations due to technical challenges in their circuit-validated isolation. Here, we employed high-depth single-cell RNA sequencing (scRNA-seq) in combination with retrograde tracing in mice to identify intrinsic transcriptional features of DPA and MTN neurons. Our transcriptome analysis revealed five major types of DPA neurons with cell type-specific gene enrichment, some of which exhibit unique mechano-nociceptive properties capable of transmitting nociception in response to innocuous mechanical stimuli in the teeth. Furthermore, we discovered cellular heterogeneity within MTN neurons that potentially contribute to their responsiveness to mechanical stretch in the masseter muscle spindles. Additionally, DPA and MTN neurons represented sensory compartments with distinct molecular profiles characterized by various ion channels, receptors, neuropeptides, and mechanoreceptors. Together, our study provides new biological insights regarding the highly specialized mechanosensory functions of DPA and MTN neurons in pain and proprioception.
Similar content being viewed by others
Introduction
Eating is an evolutionarily conserved behavior that requires a constant input of sensory information from the teeth and oral cavity, as well as proprioceptive feedback to the central nervous system (CNS) for coordinating chewing.1,2] Healthy teeth are equipped with specific sensory nerve fibers that respond to mechanical stimuli such as food texture, vibration, and pressure.1,2 However, in the event of tooth damage, these fibers can transmit pain in response to normally innocuous stimuli, including an air puff or a water spray, to alert the organism of the damage before it affects the tooth’s survival.3 Additionally, muscle spindles within the jaw muscles convey stretch and proprioceptive information from the jaw muscles by monitoring changes in muscle length during the rhythmic movements of chewing.4
Two distinct pseudo-unipolar sensory afferents of the trigeminal nerve relay sensory information from the teeth and jaw-closing muscle spindles.5 Dental primary afferent (DPA) neurons innervate the teeth and have cell bodies located in the trigeminal ganglion (TG), a part of the peripheral nervous system (PNS), along with dorsal root ganglion (DRG) neurons.5,6 By contrast, the cell bodies of proprioceptive sensory afferents from the jaw-closing muscle spindles reside in the mesencephalic trigeminal nucleus (MTN) located within the brainstem of the CNS.4,7 Nguyen and colleagues were pioneers in characterizing the transcriptional identity of TG neurons using single-cell RNA sequencing (scRNA-seq) where they found 13 unique clusters.8 These identified clusters notably differed from DRG neurons, presumably due to the absence of proprioceptive neurons in the TG, as well as a subset of TG neurons innervating unique environments, such as the oral cavity and facial organs.8 Although a recent study has classified DPA neurons using fluorescence in situ hybridization (FISH) with multiple probes in mice,9 comprehensive transcriptomic profiling has not yet been performed on specific subsets of trigeminal neurons, including DPA neurons and proprioceptive MTN neurons. Given that DPA neurons primarily serve nociceptive functions in response to diverse noxious and even innocuous stimuli,3,5 while MTN neurons exclusively handle mechanoreceptive functions for proprioception,4 transcriptome profiling of these two distinct somatosensory neurons (DPA neurons vs. MTN neurons), differing in function and location, will help us to understand the nature of somatosensation (e.g., pain and proprioception) and their unique properties at the molecular and cellular level. It will also establish a baseline for future studies that will infer molecular changes in the face of injury.
In this study, we employed scRNA-seq to generate transcriptomic profiles of both DPA and MTN neurons, which were selected based on retrograde dyes from the maxillary first molar teeth and the masseter muscle, respectively. We applied scRNA-seq with high-depth coverage to individually isolated neurons10 to detect more genes with very low expression levels in individual cells and to identify cell type classification, cell type-specific enrichment, and molecular signatures in DPA and MTN neurons. Recently, rapid advances in scRNA-seq have facilitated the classification of various somatosensory neurons, such as those in the TG8,11 and DRG.11,12,13 More recently, high-throughput methods for single-nucleus RNA sequencing (snRNA-seq) have been employed to easily sort cells and prepare libraries from TG somatosensory neurons displaying heterogeneity in size and shape.14,15 Though these studies have provided valuable insights, it has been noted that the number of transcripts in the nucleus is considerably lower than that in the soma, suggesting that nuclear transcripts might not comprehensively capture the entire transcriptome profile of an individual cell.16,17,18 Moreover, this approach potentially excludes localized subcellular RNA transcripts that play essential roles in cellular functions.16,17,18 Therefore, scRNA-seq of individually isolated neurons stands as an appropriate approach for detecting biologically meaningful transcriptional changes from samples with low RNA inputs, owing to its higher sensitivity, larger number of detectable genes, and elimination of multiplets.19 Furthermore, manual isolation allows additional characterization such as cell morphology. We thus constructed the DPA and MTN transcriptome datasets to identify the transcriptional landscapes of two distinct sensory distinct sensory afferent constituents that are highly specialized in the trigeminal nerves.
Results
Performing scRNA-seq in DPA neurons innervating the tooth pulp
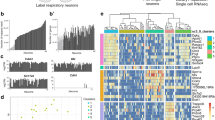
We performed scRNA-seq on DiI-labeled DPA neurons manually collected from the culture of the TG using SMART-Seq protocol, as illustrated in Fig. 1a. To ensure the DPA neuron identity, we selected cell bodies of DPA neurons with DiI labeling in the TG, specifically innervating the maxillary first molars (Fig. 1a and Fig. S1a). Additionally, we confirmed that all DiI-labeled cells expressed Advillin, a marker for somatic sensory neurons20 (Fig. S1b).
Single-cell RNA sequencing (scRNA-seq) identified dental primary afferent (DPA) neuron subtypes. a Schematic workflow of scRNA-seq for DPA neurons. b A t-distributed stochastic neighbor embedding (t-SNE) plot showing six distinct clusters, C4, C6, C7, C8, C10, and C13, identified in a total of 83 DPA neurons from 10 mice. The cluster labels are referenced from a previously published scRNA-seq dataset of the TG.8 Separated clusters are shown in different colors. c Violin plots showing expression of subtype-specific enriched genes [e.g., Trpm8, Cd34, S100b, Piezo2, Calca, Trpv1, Nppb, and Mrgprd, presented in TG8] in DPA neurons. d Two new sub-clusters exhibiting in C6, separated by C6A or C6B. e, f Most enriched genes in C6A [e.g., Chodl, Pvalb, Gfra3, and Tmem233] and C6B [e.g., Gpr83, Ntrk3, Rgs9, and Scn11a] (for more details, refer to Table S1)
We first assigned the uniquely mapped reads to the RefSeq annotated genes to generate gene counts (Fig. S1c), then normalized the dataset to mitigate differences in sequencing library depth.21,22 The transcriptomes of a total of 83 DPA neurons collected from 10 adult mice showed an average of 11 936 detected genes per neuron. We observed no significant cross-sample contamination (data not shown) and noted no noticeable correlation among the number of detected genes (Fig. S1d). The dataset also passed our quality control criteria (Fig. S1e, f) and underwent validation with reference genes, including housekeeping genes [e.g., Gapdh and Actb (β-actin)], sensory neuronal markers [e.g., Tubb3 (β-tubulin III), Uchl1 (PGP9.5), Avil (Advillin), and Rbfox3 (NeuN)],12,13 glial markers [Gfap (Glial fibrillary acidic protein), Kcnj10 (Kir4.1), and Gja1 (Connexin-43)],23 and motor neuronal markers [e.g., Chat (ChAT) and Neurog2 (Neurogenin-2)]24 (Fig. S1g).
Identification of DPA neuron subtypes
Since it is generally accepted that somatosensory neurons are a heterogeneous cell population that encompasses touch (or low-threshold mechanosensitive)- and pain-related neurons, we leveraged a previously published scRNA-seq dataset of the TG,8 of which DPA neurons are a subset. This TG dataset used droplet-based sequencing on 3 500 neurons to identify and characterize 13 distinct classes of cells within the TG. Using Seurat’s TransferAnchor function,25 we assigned cell subtype cluster labels to our DPA neuron datasets based on previously established cluster identities, predicted to have the following functional classifications: C1-C2 cool-responsive neurons, C3 C-low threshold mechanoreceptors (C-LTMRs), C4-C6 Aβ-LTMRs/Aδ-LTMRs/Aδ-nociceptors respectively, C7-C10 different subtypes of polymodal peptidergic (PEP) nociceptors, C12 itch-transmitting neurons, and C13 non-peptidergic (NP) nociceptors.8 The 2-dimensional visualizations of well-separated clusters with different colors are displayed in Fig. 1b.
Our DPA neurons were annotated to six of the 13 TG clusters: namely C4, C6, C7, C8, C10, and C13, as per the cluster numbers of the TG dataset (Fig. 1b), according to the highest Seurat prediction score (Fig. S2a). Of note, since only a single DPA neuron out of 83 was annotated to C13 (Fig. 1b), we could not show its density plot in Fig. S2a. This single neuron assigned to C13 exhibited especially high expression of Mrgprd, which is a mas-related G protein-coupled receptor channel implicated in noxious mechanical pain transduction (Fig. 1c and Fig. S1h). This gene Mrgprd was also suggested as a marker gene for C13, predicted to be NP mechanosensitive nociceptors.8 The other 82 DPA neurons were annotated to five of the 13 previously defined TG clusters, with C6 (putative Aδ-nociceptors) being particularly abundant (42.2%, n = 35 of 83 neurons, as shown in Fig. 1b). In addition, these five DPA neuron clusters consisted of neurons with distinct cell body sizes (C7/C8/C10, predictive of polymodal PEP nociceptors appearing smaller than C6, predictive of Aδ-nociceptors on average) that supported further classification (Fig. S1i).
To compare our clusters with the clusters reported for the TG, we investigated the transcript levels of representative marker genes of the 13 TG clusters8,9 (Fig. 1c and Fig. S1h). These markers included a menthol receptor, Trpm8 (C1-C2, cool-responsive); a cell surface antigen, Cd34 (C3, C-LTMRs); a calcium-binding protein, S100b and a mechanosensitive ion channel, Piezo2 (C4–C6, respectively Aβ-LTMRs/Aδ-LTMRs/Aδ-nociceptors); calcitonin gene-related peptide, Calca, a heat and capsaicin receptor, Trpv1, and a wasabi receptor, Trpa1 (C7-C10, polymodal PEP nociceptors); natriuretic polypeptide B, Nppb (C11, itch-related neuropeptide); and Mrgprd (C13, NP nociceptors). While Trpm8 and Cd34 were highly expressed in C7 and C6 neurons respectively, the expression patterns of other representative genes, such as S100b, Piezo2, Calca, Trpv1, Nppb, and Mrgprd, were comparable with those of TG neurons in general (Fig. 1c and Fig. S1h). Mrgpra3 (C12, itch related), another mas-related G protein-coupled receptor channel mediating chloroquine-evoked itch, was filtered out as it was expressed in fewer than three cells. Therefore, our findings indicate that retrograde tracer-labeled DPA neurons fall within a subset of all TG neurons but seem to exclude some cluster markers that are possibly more associated with oral sensory afferents from the gingiva, palate, or tongue, facial skin afferents, or other non-dental afferents.
We also observed that the C6 neurons, presumed to be Aδ-nociceptors,8,9 exhibited notable expression of both Piezo2 and Calca (Fig. 1c), which could contribute to rapid nociception elicited by low-threshold mechanical stimulation. However, given the relatively lower expression of Trpv1 in C6 compared to that in C7/C8/C10 (Fig. 1c), we inferred that C6 is more associated with Aβ-fibers than with Aδ-fibers. More specifically, the DPA neurons belonging to C6 were sub-divided into two new sub-clusters, namely C6A and C6B, based on distinct expression profiles (Fig. 1d). The C6A sub-cluster displayed significantly elevated expression of Chodl (Chondrolectin), Pvalb (Parvalbumin), Gfra3 (GDNF family receptor alpha 3), and Tmem233, which encodes a transmembrane protein involved in the regulation of a voltage-gated sodium channel26 (Fig. 1e and Table S1), which suggested the C6A sub-cluster are Aβ-fiber slowly adapting (SA) LTMRs.27 The C6B sub-cluster showed enrichment of Gpr83 (G protein-coupled receptor 83), Ntrk3 (TRKC), Rgs9 (Regulator of G protein signaling 9), and Scn11a (Nav1.9) (Fig. 1f and Table S1), suggesting its classification as Aβ-nociceptors.27 Together, these findings demonstrate that DPA neurons consist of five major types that correspond to C4, C6, C7, C8, and C10.
Subtype-specific gene signatures of DPA neurons
To assess the robustness of the assigned clusters, we conducted a multiscale bootstrap hierarchical clustering using Euclidean distance on a subset of the 3 000 most variable genes. Bootstrap analysis revealed that the top node of the hierarchical tree distinctly separated C6 from the other clusters, with C4 also forming a separate subtree, distinct from two subtrees containing mixtures of C7/C8 and C7/C10 at a bootstrap replication p-value level of 0.05 (Fig. 2a). Thus, while the bootstrap tree did not show significant separation of some of the finer cluster relationships, the separation of C4/C6 from C8/C10 was robustly reproduced. We identified many differentially expressed (DE) genes enriched in C4/C6, such as Pvalb, Kcna1 (Kv1.1), Scn1a (Nav1.1), Scn8a (Nav1.6), Ldhb (Lactate dehydrogenase B), Nefh (Neurofilament heavy chain), S100b, and Piezo2, which are all known for their high expression in myelinated A-fibers13 (Fig. 2b and Table S2). Conversely, pain-related DE genes, such as Trpa1, Gal (Galanin), Calcb (Calcitonin gene-related peptide 2), Trpv1, Tac1 (Substance P), Npy1r (Neuropeptide Y receptor type 1), Tlr4 (Toll-like receptor 4), Calca, and Scn11a, were enriched in C8/C10 (ref. 28) (Fig. 2b and Table S2).
Transcriptome analysis of DPA neurons. a Distribution of five major clusters of DPA neurons (excluding C13) through multiscale bootstrap hierarchical clustering analysis. The top node of the hierarchical tree is denoted by as a blue-colored dot. Numbers in red indicate the approximately unbiased (AU) p-value for the corresponding cluster. b Heatmap displaying the top 45 differentially expressed (DE) genes distinguishing C4/C6 from C8/C10. Gene of interest is highlighted in red (refer to Table S2). c–h Heatmaps depicting genes in categories relevant to intrinsic properties of somatosensory neurons, including mechanoreceptors/myelination, neurotropic factor receptors, pain, voltage-gated sodium channels, voltage-gated calcium channels, and taste receptors. Data represent the log transform of the mean DESeq2-normalized counts of transcripts in each cluster. Gene names are shown with both gene symbols (italicized, first letter uppercase) and protein symbols (not italicized, all letters uppercase). i Transcript levels of Ntrk1 across DPA clusters. j Summary table presenting reference genes and functional annotations of each DPA cluster. LTMRs low-threshold mechanoreceptors, PEP peptidergic, MNs mechanosensitive nociceptors
To further investigate the subtype classification of DPA neurons, more detailed gene families in somatosensory neurons were examined using the log transform of the mean (DESeq2 normalized) transcript counts in each cluster (Fig. 2c-h). By profiling genes specifically expressed or enriched in each cluster, we assigned clusters of DPA neurons to putative functional subtypes. C4 was assigned to Aβ-fiber LTMRs, marked by high expression of Calb1 (Calbindin 1, 28 kDa),11,29 Sfrp1 (Secreted frizzled-related protein 1),27 and Ntrk3 (Ref. 12,27,29) (Fig. 2c, d), along with low expression of Scn10a and Scn11a, specifically expressed in nociceptive C-fibers among the voltage-gated sodium channel subtypes27,28,29 (Fig. 2f). This indicates that C4 is not involved in nociceptive functions. Interestingly, with the exception of C4, all other DPA neuron clusters showed high expression of Ntrk1 (TRKA), which is associated with nerve growth factor (NGF) (Fig. 2d, i). NGF signaling plays an important role in determining the fate of sensory neurons that become nociceptors and increases mechanically activated currents specifically in PEP nociceptive Aδ- and C-fibers.30,31 Hence, this result suggests that most DPA neurons possess the potential to function as mechanosensitive nociceptors (MNs) under elevated NGF conditions. Since C6 neurons have high levels of canonical markers for myelinated neurons [e.g., Nefh, S100b, Ldhb, and Thy1 (Fig. 2c)] and Calca (Fig. 2e), but low expression of Trpv1 (Fig. 2e), we classified them as heat-insensitive Aβ-fiber MNs. This category could further be divided into SA LTMRs and nociceptors, based on their sub-clusters (Fig. 1d-f). C7 neurons similarly expressed myelination markers for myelinated neurons (Fig. 2c), but featured higher expression of nociceptive markers [e.g., Runx1, Trpa1, Trpv1, and Tac1] than C6, consistent with the previously established C7 cell type in the TG8 (Fig. 2d, e). Consequently, we classified them as intermediate/polymodal Aδ-fiber nociceptors. Finally, C10 and C8 exhibited features associated with polymodal C-fiber nociceptors. Given that C8 and C10 featured high levels of Gal and Tac1 (Fig. 2e), they might represent subclasses of C-fibers transmitting different pain modalities (e.g., nerve injury pain or nociceptive sensitization).12 Interestingly, only C8 showed pronounced expression of Chrna3 (Cholinergic receptor nicotinic alpha 3 subunit), a known marker gene for silent mechanosensitive nociceptors27,31 (Fig. 2e), suggesting that this cluster could play a critical role in mechanical nociception under high NGF conditions. With respect to mechanoreceptors, the DPA neurons had low expression of Piezo1 and Tmc1 (Transmembrane channel like 1) & 2, which are associated with itch32 and hearing33 respectively, but high expression of Asic3 (Acid-sensing ion channel subunit 3)34 and Piezo2 (ref. 35), consistent with previous studies9,36,37 (Fig. 2c). Several voltage-gated calcium channels were moderately expressed across all clusters (Fig. 2g). Taste receptors genes were lowly expressed and similarly not specific to any cluster (Fig. 2h). In summary, most DPA subtypes, excluding C4, can be characterized as potent PEP MNs that are affected by NGF signaling (Fig. 2i, j).
P2X3 expression in the PEP subtype of DPA neurons
Our FISH assay confirmed that several reference genes from the general TG population were also observed in DPA neurons retrogradely labeled with Fluoro-Gold (FG) (Fig. S3a–h and Fig. 3a–d). In line with nociceptive neurons, Tac1 and Trpv1 were predominantly expressed in small to medium-sized DPA neurons (Fig. S3a–d), whereas Calca and Piezo2 displayed high expression across DPA neurons of varying sizes (Fig. S3e–h). Moreover, we consistently observed a low proportion of Mrgprd-positive DPA neurons (Fig. 3a, b), in agreement with our sequencing results (Fig. 1b, c and Fig. S1h). Mrgprd-positive neurons, which bind to IB4, are required for response to noxious mechanical stimuli.38 Consistent with previous reports,9,36 we observed that the majority of collected DPA neurons did not bind FITC-conjugated IB4 (n = 3 positive of 96 neurons, Fig. 3e, f), supporting the notion that DPA neurons are primarily composed of PEP nociceptors (Fig. 2j). We also found that a relatively high proportion of DPA neurons exhibited expression of the extracellular ATP ionotropic receptor P2rx3 (P2X3), showing a bimodal distribution across medium to large sizes (Fig. 3c, d). Notably, since P2rx3 is generally present in NP nociceptors that are distinct from Trpv1-positive neurons,39 its relatively high expression in DPA neurons, which are Mrgprd- and IB4-nagative and Calca-positive PEP subtypes, is an atypical observation in DRG neurons. Our DPA neuron dataset also showed that P2rx3 expression predominated over other P2X (ligand-gated) and P2Y (G-protein coupled) purinergic receptors40 (Fig. 3g). Our immunohistochemical assays further demonstrated that the majority of P2X3-positive DPA neurons did not bind IB4-negative (Fig. 3h, i). These findings thus support the notion that ATP, acting through the P2X3 receptor, could be a crucial modulator of nociceptive tone in DPA neurons that could contribute to mechanical sensitivity in the context of injury or infection.41
P2X3 expression is frequently observed in IB4-negative DPA neurons. a–d Representative fluorescence images (20X magnification) and quantitative results showing DPA neurons labeled with Fluoro-Gold (FG, blue-dotted outline) and each marker gene, Mrgprd (C13) and P2rx3, shown in green after RNAscope assay. Arrowheads indicate DPA neurons expressing each marker gene. Scale bars: 25 μm. Bar graph represents the cell body size distribution (µm2) of DPA neurons expressing Mrgprd (n = 1 of 62 FG+ DPA neurons from n = 2 mice) or P2rx3 (n = 12 of 36 FG+ DPA neurons from n = 2 mice) as the average of two mice. e A representative image displaying an IB4-negative DiI-labeled DPA neuron (asterisk). A scale bar: 20 μm. f The number of IB4-positive (n = 3 of 96 neurons, green) or IB4-negative (n = 93 of 96 neurons, gray) DPA neurons during the collection process. g Transcript levels of P2Y (G-protein coupled) [e.g., P2ry1, P2ry2, and P2ry14] and P2X (ligand-gated) [e.g., P2rx3, P2rx4, P2rx5, P2rx6, and P2rx7] purinergic receptor family genes in DPA neuron transcriptomes. h Representative immunofluorescence images of FG-labeled DPA neurons (blue) showing P2X3 (red) and IB4-binding (green) in TG sections. Nissl stain (white) was used for cell body identification. Arrowheads indicate P2X3-expressing DPA neurons but do not bind to IB4. Scale bars: 25 μm. i Proportion of 4 categories [e.g., IB4−/P2X3+, IB4−/P2X3−, IB4+/P2X3+, IB4+/P2X3−] among all FG-labeled DPA neurons. Numbers represent mean ± SD. n = 100 neurons from n = 4 mice
Molecular diversity of MTN neurons innervating the masseter muscle spindles
As with DPA neurons, we performed scRNA-seq with the same protocol, but we specifically used juvenile mice to enhance the viability of MTN neurons cultured from brainstem slices (Fig. 4a). For sample preparation, we utilized the retrograde tracer DiI to label cell bodies originating from the masseter muscle spindles in a total of 8 juvenile mice (P21: 5 mice and P28: 3 mice) (Fig. 4a, b) and manually collected DiI-labeled MTN neurons expressing Advillin20 (Fig. 4c). Similar to DPA neurons, after quality control, we detected an average of 11 364 genes per MTN neurons from 108 MTN neurons (Fig. S4a–d). The MTN dataset was validated with the expression of specific marker genes associated with proprioceptive neurons [e.g., Pou4f1 (BRN3A), Ntrk3, Etv1(ER81), Pvalb, and Whrn (Whirlin)]42,43,44 in addition to the same set of reference genes that were previously employed for the validation of the DPA dataset12,13,23,24 (Fig. S4e). We conducted dimensionality reduction (t-distributed stochastic neighbor embedding; t-SNE) using expression data for the top 3 000 most variable genes, generated by Seurat, in pooled MTN samples of all ages. There was little batch effect between sequencing runs (Fig. S4f).
scRNA-seq analysis reveals heterogeneity of mesencephalic trigeminal nucleus (MTN) neurons. a Schematic diagram of MTN neuron preparation for scRNA-seq. b Representative fluorescence images for a couple of MTN neurons, which were retrogradely labeled with DiI (red) from the masseter muscle, in the brainstem. A scale bar: 100 µm. Insets represent the high-magnification images showing DiI (red) and Nissl staining (white). Scale bars: 20 µm. n = 1 mouse (P21). c Representative immunofluorescence images showing Advillin positivity (green) in cultured DiI-labeled MTN neurons. A scale bar: 20 µm. n = 1 mouse (P21). d Heatmap representing the distribution of two major subgroups of MTN neurons: those with high levels of both Ntrk3 and Ntrk2 (designated as Ntrk2high) and those with high levels of Ntrk3 but low levels of Ntrk2 (designated as Ntrk2low). e–h Heatmaps showing genes categorized according to intrinsic properties, including neurotropic factors and receptors, proprioceptor/mechanoreceptors, glutamate release and receptors, and acetylcholine receptors. Data represent the log transform of mean DESeq2-normalized counts of transcripts in each group (refer to Table S3). i–k Volcano plots showing the DE genes with a minimum log2 (fold change; FC) of 0.5 and an adjusted P-value < 0.05 that are enriched in the DPA neurons (left side) or MTN neurons (right side). For analysis, gene lists from Gene Ontology (GO) categories including sensory perception (GO:0007600), response to stimuli (GO:0050896), and a custom gene list of mechanosensitive ion channels were used. Genes of interest within the top 20 DE genes indicated with text labels), ranked by log2 FC in each GO term, are highlighted in blue (refer to Table S4)
Proprioceptive neurons innervating muscle spindles are typically classified into two major types: group Ia and group II.45 A third type of proprioceptive neuron, group Ib, innervates Golgi tendon organs.45 A recent scRNA-seq study focusing on proprioceptive neurons in limb skeletal muscle spindles identified three major marker genes, Lmcd1, Fxyd7, and Chad, representing group Ia, II, and Ib afferents, respectively.42 MTN neurons that innervate masticatory muscles are also known to comprise two main afferents, group Ia and II.7 Given these similarities, we investigated whether MTN neurons could be similarly classified based on these three marker genes. Our analysis confirmed the presence of comparable subtypes in MTN neurons, namely Lmcd1-enriched (similar to group Ia afferents, designated as Lmcd1high:Fxyd7low) and Fxyd7-enriched (similar to group II afferents, designated as Lmcd1low:Fxyd7high) subtypes (Fig. S5a). The cell body size of the Lmcd1low:Fxyd7high subtype tended to be smaller than that of the Lmcd1high:Fxyd7low subgroup, consistent with previous studies42 (Fig. S5b). We noted that Chad was not detected in our MTN samples, implying that our MTN samples did not contain neurons innervating Golgi tendon.42 Additionally, we observed two unidentified subtypes where both markers were enriched or both markers were rarely expressed (Fig. S5a). These four subgroups are visually depicted in a t-SNE plot of MTN neurons (Fig. S5c).
We notably observed the absence of Runx3 transcripts in MTN neurons (Fig. S4e), despite the predominant expression of other marker genes associated with the early development of MTN neurons [e.g., Ntrk3, Pou4f1, and Etv1].43,44 Runx3 suppresses the expression of Ntrk2, a receptor of brain-derived neurotrophic factor (BDNF) that is essential for the development and survival of proprioceptive neurons.46 Accordingly, we found that MTN neurons could be categorized into two subgroups with high or low Ntrk2 expression, termed Ntrk2high or Ntrk2low, respectively (Fig. 4d), and a t-SNE plot visualized the distribution of the two subgroups (Fig. S6a). While the expression levels of neurotrophic factor receptors, with the exception of Ntrk2 and Gfra1, were similar across the two subgroups (Fig. 4d and Table S3), we found higher expression of proprioceptive neuronal marker genes, including Pvalb and Etv1 (ref. 42), in Ntrk2low subgroup (Fig. 4f and Table S3). This suggests that the Ntrk2low subgroup is likely to be representative of typical proprioceptive neurons. Furthermore, we found evidence of glutamate release, which is required for static but not dynamic responses to stretch, and glutamate receptors in MTN neurons47 (Fig. 4g and Table S3). We also observed differential expression of nicotinic acetylcholine receptor subunits, which contribute to the sensitivity of a subset of MTN neurons to succinylcholine, an analog of acetylcholine, between the two subgroups (e.g., group Ia: succinylcholine-sensitive or group II: succinylcholine-insensitive)48 (Fig. 4h). These results suggest that the Ntrk2high subgroup is more likely to exhibit properties associated with glutamate-mediated static activity.47 Additionally, we found some differently expressed mechanosensitive ion channels, including Piezo1 (ref. 35) and Kcnk10 (ref. 29) (Fig. 4f and Table S3). Our further analysis revealed many features contributing to their differentiation (Fig. S7a-c).
Next, we conducted a comparative analysis between DPA and MTN neurons to better understand the distinctive characteristics of somatosensation associated with different functions, specifically pain perception and masticatory proprioception. In comparison to MTN neurons, we confirmed that DPA neurons differentially expressed pain-related genes, including Calca, P2rx3, Tlr4, Trpv1, Scn11a, Asic3, Tmem100 (ref. 49), Tac1, suggesting that DPA neurons are more involved in nociceptive functions (Fig. 4i and Table S4). Compared to DPA neurons, MTN neurons exhibited higher levels of Whrn,42 Scnn1a (Sodium channel epithelial 1 subunit alpha, regulated by Etv1/ER81),50 Onecut1 (One cut homeobox 1, required for normal development of MTN),51 Wnt7a (Wnt family member 7 A),12 Ntrk3 (ref. 44), as well as unexpectedly higher levels of neuropeptide-related genes, such as Gal and Nts (Neurotensin)52 (Fig. 4i, j and Table S4). We also observed differential expression of mechanosensitive ion channels, with higher expression in one or the other cell type (Fig. 4k and Table S4). Overall, these findings support that DPA and MTN neurons possess distinct intrinsic properties for somatosensation.
MTN neurons experience molecular changes during postnatal development
In rodents, the masseter muscle undergoes considerable changes in morphological properties and activity patterns between the pre-weaning (2 weeks) and post-weaning (4 weeks) stages, which coincide with alterations in feeding behavior.53,54 These changes could significantly impact MTN neurons, which coil around the masseter muscle spindles.55 This idea was reiterated in a recent study using bulk RNA-seq combined with proteomics in the masseter muscle of mice aged 3 to 25 days, revealing developmental shifts in the molecular composition of muscle spindle capsule cells.56 We also sought to investigate the possibility of changes occurring in MTN neurons as a result of the critical behavioral switch from suckling to chewing facilitated by the weaning process in mice. We initially attempted to work with mice younger than 1 week or older than 5 weeks; however, we encountered technical challenges in acquiring labeled neurons from these age groups. After several attempts, we developed an optimized protocol that enables DiI injection into the masticatory muscles of mice at P16 and P23, respectively, followed by cell collection at P21 and P28 (5 days post-injection). Previous research that video-monitored the behavioral patterns of rodent pups from P14 to P35 supports the choice of our P21 and P28 groups, representing an intermediate phase that involves a gradual decrease in suckling behavior followed by an increase in solid food intake within the overall weaning process.57 A t-SNE plot displayed the 2-dimensional location of MTN neurons at P21 and P28, respectively (Fig. S6b), and the two subgroups, Ntrk2high and Ntrk2low, showed distinct relative proportions at each postnatal age (Fig. S6c). Moreover, various DE genes were present in biological processes associated with neuron and muscle structure development, such as programmed cell death58 and synapse organization56 (Fig. 5a). Specifically, DE genes contributing to post-translational processes at synapses were found to be enriched in P28 MTN neurons (Fig. 5a). Furthermore, we observed a significant decrease in the cell body sizes of MTN neurons at P28 compared to P21, both in culture conditions (Fig. 5b) and within a specific subregion of the brainstem, the rostral part (Fig. 5c, d). This may suggest that the postnatal switch is coupled with changes in the cell size of muscle-afferent MTN neurons, which could potentially reflect adaptations to the environment (e.g., external or internal stimuli).
Postnatal changes in MTN neurons. a The number of DE genes enriched in P21 (left side) or P28 (right side) of MTN neurons (refer to Table S5). The gene list used for this analysis is within GO categories, including nervous system development (GO: 0007399), programmed cell death (GO: 0012501), muscle structure development (GO: 0061061), synapse organization (GO: 0050808), and translation at synapse (GO: 0140241). b Cell body size (μm2) of MTN neurons during the collection procedure. P21: n = 66 neurons and P28: n = 48 neurons. Bar graph presents as the mean ± SD. Unpaired nonparametric test (Kolmogorov-Smirnov test) was performed, *P= 0.015 6. c Representative fluorescence images showing DiI-labeled MTN neurons (red) and Nissl stain (green) located in the rostral or caudal part of the brainstem according to mouse age. Scale bars: 100 µm. d Violin plots quantitatively depicting neuronal cell body size (μm2) by age. n = 3 mice (P21), n = 2 mice (P28), and n = 2 mice (5-week). One-way analysis of variance with Bonferroni post hoc test was conducted, *P= 0.031 4
Discussion
To our knowledge, we are the first to perform scRNA-seq on retrograde-traced DPA and MTN neurons to generate a comprehensive transcriptional overview of oral sensory inputs. This study revealed transcriptional signatures specific to dental sensory transmission of external stimuli, which range from innocuous to noxious, and to proprioceptive transmission of stretching inputs from the masseter muscle spindles. Our single-cell transcriptomic analysis was used to identify cell types and their enriched genes and characterize the landscape of mechanosensory modulation that could be influenced by neurotropic factors such as NGF and BDNF. We could thus determine more specific features of mechanosensory neurons that underlie our perception of pain (nociception) and the regulation of jaw movements (proprioception).
Why tooth hypersensitivity is evoked by low-threshold mechanical stimulation
In healthy teeth, the tooth pulp innervating nerves are protected by hard tissues, enamel, which covers a layer of dentin around the tooth pulp. However, dental nociception, called dentin hypersensitivity, can be initiated in DPA neurons without inflammation through the mechanical stimulation of fluid movement in the dentinal tubules (known as hydrodynamic theory59) that have been exposed by tooth fracture or gingival recession. Dentin hypersensitivity produces a sharp, intense pain in response to not only noxious stimuli (e.g., hot, cold, acidic, or hyperosmolar), but also to innocuous mechanical stimuli to the affected tooth, such as air puffs, water spray, and sometimes even a breath.3,41 The hydrodynamic theory suggests that DPA neurons differ from other cutaneous sensory neurons where innocuous stimuli do not activate high-threshold nociceptors or evoke pain in a non-injured state.60 Several studies to date have demonstrated a specific type of myelinated A-fiber nociceptors (a subset of DPA neurons called algoneurons) that secrete CGRP (Calca) centrally, which enhances pain transmission.3,9,36 Through single-cell transcriptomics analysis, we found that C6/C7 are substantially consistent with algoneuron types, and make up the majority of all DPA neurons we sampled (Figs. 1b, 2j). This result supports the idea that most DPA neurons are myelinated PEP polymodal nociceptors, which can quickly transmit pain signals from all types of stimuli to prevent further damage. Furthermore, since Calca was found to be predominantly high throughout DPA neurons compared to other neurotransmitters and peptides involved in pain transmission (Fig. 2e and Fig. S3e, f), CGRP therapeutics for primary headache disorders may be beneficial for cases of intractable dental pain,61 although the impact of such therapeutics on concomitant infection and beneficial inflammation will need to be carefully examined.
We also identified a significant presence of the NGF receptor Ntrk1 that plays a critical role in determining the pathophysiological function of DPA neurons. In general, the expression of Ntrk1 is essential for cell growth, differentiation, survival, and the fate of sensory neurons to become nociceptors at the embryonic stage, but it has been reported that NGF signaling pathways potentiate electrophysiological properties of low-threshold mechanically activated currents induced by mechanosensitive ion channels in nociceptive PEP Aδ- and C-fibers, but not in NP C-fibers.30 In addition, it has been further demonstrated that this property of NGF mediates the acquisition of mechanical sensitivity in “silent” nociceptors (Chrna3+), normally unresponsive to mechanical stimuli, by upregulating Piezo2 (ref. 31). Thus, C8 with high expression of Chrna3 could be a strong match for this type (Fig. 2e, j). In addition, given that most DPA neurons have high expression of Ntrk1 (Fig. 2i, j), with the exception of C4, DPA neurons might have the potential to become sensitized to low-threshold mechanical stimuli by NGF signaling pathways in damaged teeth. NGF is also upregulated in odontoblast-like cells located close to DPA neurons for the repair of teeth and pulp cells in damaged teeth.62 This condition gives NGF release from odontoblasts a much greater potential to affect adjacent nerve cells. In addition to their mechanosensory functions, DPA neurons exhibit rapid responses to noxious stimuli, such as heat or cold.60 Our analysis demonstrated that the majority of nociceptive DPA neurons are polymodal, given that C7, C8, and C10 shared multiple nociceptors, such as Trpa1 and Trpv1 (Fig. 2e). Furthermore, the expression of myelination genes by these nociceptive neurons suggests rapid signaling.
Intermediate trait of C7
In our bootstrap hierarchical clustering analysis of DPA neurons, C7 exhibited the most diverse distribution, mixed with either C8 or C10 (Fig. 2a). Seurat’s TransferAnchor function assigns a query sample to the cluster from the reference data with the highest prediction score, regardless of the numeric value of the score. Additionally, predictions are limited to known clusters in the reference data; no novel clusters can be identified in the query. When examining the prediction scores per sample, we observed that 30 cells (out of 83) had prediction scores below 0.80. While some of these samples represent cases of possible misassignment between two cluster types, many of the cells demonstrated low prediction scores to multiple clusters, which suggest some DPA neurons may have more ambiguous memberships. Notably, in our data set, every cell predicted to be in C7 had a low prediction score, which led us to hypothesize that C7 represents an intermediary/polymodal cluster (Fig. S2a, b). We note that since these cells were manually isolated, we avoided the possibility of multiplets. While these cells with low and ambiguous prediction scores may simply arise out of noise, they may also represent novel cell types, not in the reference data set or even transition states between cell type clusters. Transition states could arise within our data set either due to variation in the health of labeled mouse pulp (i.e., we cannot rule out prior dental injuries) or as a consequence of neuron dissociation and culture required to recover single neurons.
Comparison to previously established TG or DPA cell types
We aligned DPA neuron subtypes using the identified cluster information, defining functional subtypes based on published data from the TG as a whole.8 To validate our predicted labels, we compared them with the expression patterns of marker genes established in various studies (Fig. 1c, Fig. S1h, and Fig. 2). Emrick and colleagues recently determined that 76% of DPA neurons were C4, 20% were C6, and 4% fell within the other ten clusters defined by whole TG scRNA-seq data,8 using FISH assays with eight probes.9 Our results are generally in agreement. We found that approximately 50% of DPA neurons fall in C4/C6 while only one was a Mrgprd-positive NP C-nociceptors and no Nppb- and Mrgpra3-positive itch C-nociceptors were identified. However, unlike Emrick and colleagues who determined that C7 neurons are polymodal C-nociceptors,9 we believe that DPA neurons in C7 are more likely intermediate/polymodal Aδ-nociceptors based on moderate to high expression of myelinated A-fiber markers [e.g., Nefh, S100b, Ldhb, and Thy1] (Fig. 2c) and high expression of nociceptive genes [e.g., Runx1, Trpa1, Trpv1, Trpm8, Calca, and Tac1] (Fig. 2d, e). Therefore, adding to scRNA-seq of TG neurons8 and spatial RNA-seq of DRG neurons,27 our (increased depth) single-cell transcriptional profiling allowed for a more detailed classification of DPA neurons. However, as we determined the DPA neuron subtypes based on the predicted similarity to the TG cluster types8 (Fig. S2a, b) and exclusively examined neurons labeled with the retrograde tracer from molar teeth, we cannot completely exclude the presence of Mrgprd-positive DPA neurons based on our inability to find more than one, but they are likely to be rare. Indeed, a previous study has found that Mrgprd-green fluorescent protein (GFP) reporter mice exhibited positive signals in the molar pulp, but also only identified 7 cells out of 280 pulp-labeled TG neurons.63 Therefore, to confirm these putative classifications, additional experiments following functional assessments such as extracellular single-fiber recordings or in vivo calcium imaging, which can determine fiber-type-specific properties following various stimuli, are required.
Distinction between two proprioceptive neuronal types innervating the limb muscle spindles and the masseter muscle spindles
Almost all mammalian muscles contain muscle spindles, which are innervated by proprioceptive afferents. These afferents convey sensory information about changes in individual muscle lengths and stretching rates to the CNS, allowing the CNS to perceive the position and movement of our limbs and to regulate motor reflexes.45 MTN neurons, which innervate specifically to the masticatory muscle spindles, are comprised of two types of afferents, named group Ia and group II, according to their axonal conduction velocity.7 Group Ia afferents can be distinguished from group II afferents by their lower activation threshold, faster conduction velocity, and dynamic sensitivity.7,48 However, since MTN neurons exhibit a wide range of conduction velocities, it is challenging to practically differentiate between the two types of afferents.48 Another categorization of MTN neurons based on acetylcholine sensitivity has been proposed in previous studies, but this specific phenotype was observed in cats rather than rats.48,64 Therefore, to identify the molecular classification of MTN neurons, we first focused on the three major marker genes, such as Lmcd1, Fxyd7, and Chad,42 corresponding to group Ia, II, and Ib afferents, respectively, previously identified in limb skeletal muscle spindles. Our results showed that these genes could be applied to a subset of MTN neurons to help identify their subtypes (Fig. S5a). In addition, typical proprioceptive neuronal markers such as Pou4f1 and Ntrk3, were predominantly expressed in MTN neurons, whereas the transcript levels of other marker genes Etv1, Pvalb, and Whrn were highly heterogenous or rarely expressed (Fig. S4e). More surprisingly, Runx3, a representative marker for proprioceptive neurons, was also not found in MTN neurons (Fig. S4e). These findings are worth noting, given that proprioceptors have a mechanism to maintain proprioceptors by Runx3 suppression of Ntrk2 expression.46 However, the Ntrk2-negative traits are another important marker for classifying proprioceptive neurons in DRG neurons; therefore, careful consideration should be given to whether our results might be a temporary phenomenon that appears at a certain age of mice. These findings may also be an effect of their CNS localization, as Ntrk2 is the BDNF receptor. We were able to divide MTN neurons into two subgroups based on expression levels of Ntrk2 (Fig. 4d and Fig. S6a). We also found that Ret, a representative marker of Aβ SA LTMRs,29 was highly expressed throughout MTN neurons (Fig. 4e), suggesting that MTN neurons possess molecular features of both proprioceptors and Aβ SA LTMRs in response to stretch stimuli. It remains to be determined whether and how the Ntrk2high and Ntrk2low subgroups are related to group II and group Ia afferent, respectively. It should be noted that Ntrk2 levels are on a continuum, so subtle differences in its expression can affect classification. Furthermore, there is a possibility of differential expression of unidentified mechanosensitive molecules or mechanosensitive ion channels (namely transmembrane (TMEM) protein family), such as Tmem150c (Tentonin-3),65 between the two subgroups. Further research is needed to elucidate and understand these potential factors. Taken together, our deeply sequenced datasets not only increase our understanding of proprioceptive MTN neurons but also provide an important resource for exploring additional candidate genes responsible for aspects of mechanosensation.
Limitations of this study
This study takes into consideration the influence of in vitro experimental conditions, which could induce damage or stress-derived transcriptional factors and persistent morphological changes by neurite growth. It is also possible that the absence of neurotrophic factors that are crucial for the maintenance and survival of somatosensory neurons in vivo may affect our results. In addition, the presence of gene expression generally used for glial markers,23 such as Gfap, Kcnj10, and Gja1, in certain DPA and MTN neurons raises the possibility of glial cell contamination during the cell collection procedure. However, these samples included for the downstream analysis as glial marker gene expression did not significantly influence the classification of neuron subtypes. Interestingly, these markers were found to be more frequently expressed in P21 MTN neurons compared to DPA neurons or P28 MTN neurons. Recently, snRNA-seq and spatial transcriptomics have emerged as a powerful tool for characterizing somatosensory neurons under various conditions, including injury or pain,15,27 so further studies with this technology will be valuable to understand these neuronal population.
In conclusion, our transcriptome analysis revealed a new classification in which DPA neurons have a unique capacity to transmit mechano-nociceptive signals in response to innocuous mechanical stimuli in teeth, and MTN neurons exhibit distinct responses to mechanical stretching in the masseter muscle. These characteristics were based on the expression patterns of the neurotrophic factor receptors Ntrk1/TRKA, Ntrk2/TRKB, and Ntrk3/TRKC. The precise regulation of these receptors plays a critical role in conferring unique properties to the pain- and mechano-transducers of the dental and masseter nerves, setting it apart from the bodily somatosensory system.
Materials and methods
Animals
A total of 30 adults (5- to 7-week old) and 15 juvenile (P16 to P28) male C57BL/6J mice were obtained from the Doo Yeol Biotech (Republic of Korea) and Jackson Laboratories (Bar Harbor, Maine, USA). All surgical and experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at Seoul National University and University of Pennsylvania and conformed to the ARRIVE (Animal Research: Reporting In Vivo Experiments) guidelines for animal studies.
Retrograde labeling of neurons and single-cell isolation
DPA neurons were retrogradely labeled with a few 1,1′-Dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI, Invitrogen, USA) crystals from the maxillary first molars on both sides in each mouse (5-week-old), then prepared as described previously.66 Two weeks post-labeling, DPA neurons were isolated in acute culture from the TG tissues as previously described.66 DPA neurons were incubated with FITC-conjugated isolectin B4 (10 μg·mL−1 in culture media, Sigma, USA) for 15 min at 37 °C to detect NP C-fibers. Similarly, MTN neurons were retrogradely labeled with a total volume of 5 to 10 µL DiI (3% in Dimethyl sulfoxide; DMSO) from the masseter muscles on both sides in each mouse (either P16 or P23). Five days post-labeling, mice were sacrificed, and the brainstem tissue from the midbrain to the medulla oblongata was dissected out and embedded in agarose. The tissue was cut into 300 to 500 μm thickness coronal slices with a vibrating blade microtome (VT1000P, Leica, USA) while immersed in an artificial cerebrospinal fluid solution. Unlike DPA neurons, MTN neurons should be dissociated from brainstem slices, so we used young mice in order to increase cell viability.67 MTN neurons were acutely cultured by modifying a previous method used for rats67 to be more appropriate for mice. Briefly, we placed chunks of MTN tissue in papain solution (20 U·mL−1, Worthington Biochemical, USA) in Ca2+- and Mg2+-free Hanks’ Balanced Salt Solution (HBSS, Gibco, USA), added a few crystals of L-cysteine (Sigma, Germany), adjusted to pH 7.4, and incubated the tissue chunks for 15 to 25 min at 37 °C. After aspirating fluid, we added Ham’s F12 culture media (Gibco, USA) containing 10% fetal bovine serum (FBS), 1% Penicillin/Streptomycin (P/S), and NGF 2.5S (50 ng·mL−1, Invitrogen, USA) and triturated the tissues extremely gently until chunks were diminished. Afterward, cells were placed on poly-D-lysine/laminin-coated glass coverslips, and culture media were changed to Ham’s F12 feeding media containing 10% FBS and 1% P/S but no neurotrophic factors. Cells were incubated for at least four hours at 37 °C (5% CO2) in a humid incubator until the start of the cell collection.
scRNA-seq
Sample collection
To minimize cell-to-cell variation under in vitro conditions, cells were collected only during the 6-hour incubation period from the start of cell collection. During the collection procedure, we continuously flowed a freshly prepared diethyl pyrocarbonate (DEPC, Sigma, Switzerland)-treated 2 mM Ca2+/Na+ solution (bath solution). DPA and MTN neurons were manually collected using a glass micropipette filled with a pipette solution containing RNase inhibitor (2 U·μL−1, Takara, China) in bath solution while minimizing non-neuronal contaminations. Each neuron was photographed prior to collection, and the images were used for cell body area (μm2) measurements using ImageJ software (National Institutes of Health, USA). We also collected the bath solution without cells as the negative control for each batch.
cDNA library preparation and sequencing
Once the DPA neurons were separated into individual tubes, they were digested and reverse-transcribed to non-stranded cDNA using poly-T primers. The resulting cDNA was amplified using SMART-Seq v4 Ultra Low Input RNA kit (Clontech, China) into cDNA libraries according to the manufacturer’s instructions, but with the following modifications: 1) ERCC RNA Spike-In controls (1/4 000 000 dilution, Invitrogen, Lithuania) were added at the cell lysis step; 2) 18 PCR cycles were performed to amplify cDNA libraries. The libraries were diluted accordingly and used to make DNA sequencing libraries by following the Nextera XT DNA Library Preparation Kit protocol (Illumina, USA). Before sequencing, the libraries were analyzed using a Bioanalyzer High Sensitivity DNA Kit (Agilent Technologies, USA) to determine their quality. Libraries that passed quality control filters (for size distributions) were sequenced using a NextSeq 500 (Illumina, USA) with single-end 75 base pair reads. Reads were trimmed for adapters and poly-A sequences using in-house software and then mapped to the mouse genome (mm10) using STAR.68 In the resulting dataset, the average sample has a depth of 5.8 million reads, with 4.1 million uniquely aligned to exons. Uniquely mapped reads were used for feature quantification using VERSE.69
Sample quality control
We collected a total of 96 DPA neurons from 10 mice (7-week-old) in two independent batches. A total of 75 MTN neurons (P21, 5 mice) and 47 MTN neurons (P28, 3 mice) were collected in single batches separated by age. Based on summary statistics from in-house software, 10 samples with “Percent Spike-In” > 5%, indicating relatively low RNA concentration compared to the spike-in standard, were removed from downstream analyses. Also, three additional samples with a concentration of less than 0.5 ng·μL−1 after reverse transcription and amplification were removed. Consequently, a total of 83 DPA samples were used in the downstream analysis. Similarly, from the MTN neurons, two samples that had “Percent Spike-In” > 5%, two samples that had “Percent mitochondrial transcript” > 10%, and two samples that had a concentration of less than 0.5 ng·μL−1 after reverse transcription and amplification were removed. Also removed were four additional samples without expression of Avil and another four samples with no detectable expression of Ntrk3, Etv1, or Pvalb all previously known markers of proprioceptive neurons.42 Consequently, a total of 108 MTN samples were used in the downstream analyses.
scRNA-seq data analysis
DPA cluster identification
This paper made extensive use of the Seurat package developed by the Satija lab70; in essence, the recommended (TransferData) methods were used to project reference data onto a query object.25 Briefly, single-cell TG transcriptome data, to be used as the reference,8 were log-normalized and center scaled; variable features were identified and used for linear dimensional reduction (principal components analysis; PCA). As previously described, informative principal components were used for clustering, and multidimensional data were displayed in a t-SNE 2-dimensional representation. Similarly, DPA transcriptome data (query) were log-normalized and center scaled; variable features were identified. Next, anchors were identified in the reference data set, and a weights matrix was constructed that defined the association between each query cell and each anchor. Afterward, a binary classification matrix was created, where the rows correspond to possible cluster identities, and the columns correspond to the anchors, and each matrix cell is 1 if the anchor pair is a member of a certain cluster. The classification matrix was then multiplied by the weights matrix as mentioned above to calculate prediction scores to transfer cluster labels onto the DPA data. Each query sample was assigned to the cluster with the highest prediction score. When transferred cluster labels were visualized in a 2-dimensional non-linear dimensionality reduction (t-SNE) of the top 3 000 most variably expressed genes (normalized by DESeq2), then log transformed. The neurons were well separated into their respective assigned clusters.
We wanted to verify the validity of the transferred labels with an independent clustering method, using the same subset of log-transformed, DESeq2-normalized counts of the top 3 000 variable features. Given the limited number of samples compared to that of droplet-based capture and sequencing and the uncertainty of hierarchical clustering, we used R package pvclust71 to perform multiscale bootstrap hierarchical clustering. Briefly, pvclust generates bootstrap samples by randomly sampling varying proportions of the data, then computes a hierarchical clustering on each bootstrap copy. Then, for each cluster observed in the hierarchical clustering of the original data, it calculates a P-value between 0 and 1, corresponding to how strongly the data supports the cluster, based on how often the cluster was observed across hierarchical clustering of the bootstrap copies. Based on the approximately unbiased (AU) p-values in the bootstrap hierarchical clustering, there we found strong evidence of clusters composed of samples assigned to a single cluster, except for samples predicted to be C7, as well as separation of samples across C4/C6 and C8/C10, with a single exception. To summarize, this multiscale bootstrap hierarchical clustering analysis independently reproduces the transferred cluster label from Seurat.
Differentially expressed (DE) gene analysis
For differential expression analysis, only genes that contained at least three reads across five samples were considered. Read counts were normalized for library size using DESeq2 and DE genes were identified with DESeq2. A minimum fold change of 1.2 and an adjusted P-value < 0.05 after multiple hypothesis correction (Benjamini–Hochberg) was used to identify significant DE genes. Gene set enrichment analysis (GSEA) was performed using the R package clusterProfiler (gseGO) 72.
Fluorescence in situ hybridization (FISH)
Retrograde labeling of DPA neurons
Since the lipophilic dye DiI was inadequate for the FISH method, we used another retrograde neuronal tracer Fluoro-Gold (FG, Cayman Chemical, USA) for labeling DPA neurons. All procedures of tooth dentin and pulp exposure surgery, including anesthesia, anti-inflammatory analgesic treatment, and surgery in mice, were followed as previously described.73 A total volume of 0.5–2 µL FG (4% in distilled water) was directly applied to an exposed dental pulp of the maxillary molar (either a left side or both sides of the jaw) in mice, and then the dental cavity was sealed with the GC Fuji II dental cement (Cat. #21, GC Corporation, Japan) according to the manufacturer’s instructions. Upon anesthetic recovery, animals were returned to their home cages.
Sample preparation and RNAscope assay
Five days post-labeling, mice were deeply anesthetized and were perfused with phosphate-buffered saline (PBS) solution, followed by ice-cold 4% paraformaldehyde in PBS (PFA solution). TG tissues were post-fixed in 4% PFA solution for 4 h at 4 °C and then were transferred to 30% sucrose solution in PBS at 4 ℃. Serial frozen transverse sections (thickness: 14 µm) of TG were mounted onto superfrost plus slides and were stored at −80 °C until later use. FG-positive cells were verified microscopically prior to FISH. The sections were used for RNAscope Multiplex Fluorescent v2 assay (Advanced Cell Diagnostics, USA), as previously described,73 using the following probes: Mm-Tac1 (Cat. #410351-C2), Mm-Trpv1 (Cat. #313331-C2), Mm-Calca (Cat. #420361-C2), Mm-Piezo2 (Cat. #400191-C3), Mm-Mrgprd (Cat. #417921-C2), and Mm-P2rx3 (Cat. #521611-C3). All z-stack images were acquired using a fluorescence microscope (Leica) controlled by LasX software.
RNAscope assay image analysis
Cell body area (µm2) of labeled neurons was all measured with ImageJ. In particular, since the FG signal of DPA neurons was significantly weakened by wash steps in the RNAscope assay, we took images of FG-positive cells using the DAPI filter cube before the assay, then imaged again after the assay with different probe images using the Cy3 or Cy5 filter cube, using memory function in the microscope to ensure the same field of view was captured both times. We used Fiji Plugins of ImageJ to align and merge FG images (in the DAPI range) with the corresponding probe image (in the Cy3 or Cy5 range) obtained after the assay. A total of three to six DPA sections for each probe were randomly selected and converted to 8-bit grayscale images, and the brightness/contrast and threshold adjustments were applied to each fluorophore channel separately. Positive neurons were determined if there were at least 10 puncta dots inside the FG-positive somata or if their average intensity values were above the threshold.
Immunocytochemistry
Cells on poly-D-lysine-coated glass coverslips were washed three times with ice-cold PBS, followed by fixation with 4% PFA solution for 15 min. After washing three times with PBS for 5 min each, the cells were incubated with a blocking solution containing 2% bovine serum albumin (BSA) and 0.2% Tween 20 at room temperature (RT) for 1 h, and then anti-advillin antibodies (1:1 000, Cat. #ab72210, Abcam, USA) diluted in blocking solution were incubated overnight at 4 °C. The next day, cells were washed with PBS containing 0.1% Tween 20 for 10 min, washed twice with PBS for 5 min each, then were co-incubated with donkey anti-rabbit Alexa Fluor 488 (1:200, Cat. #A-21206, Invitrogen, USA) and DAPI (1:1 000, Cat. #D9542, Sigma, Israel) diluted in PBS at RT for 2 h. After washing in the same way, the coverslips were mounted with a mounting medium (Vector Laboratories, USA). All images were examined using a confocal microscope (LSM700, Carl Zeiss Microscopy, Germany).
Immunohistochemistry
Mice were deeply anesthetized and were perfused with PBS, followed by ice-cold 4% PFA solution. TG and brainstem tissues were dissected out, post-fixed in 4% PFA solution at 4 °C overnight, and then transferred to 30% sucrose solution in PBS at 4 °C. We prepared TG and brainstem sections, which were transversely sectioned (TG thickness: 14 µm and brainstem thickness: 30 to 40 µm). For experiments in Fig. S1a, Fig. 4b, and Fig. 5c, the sections were washed three times with PBS for 10 min each. For permeabilization, the sections were washed with PBS containing 0.1% Tween 20 for 10 min, washed twice with PBS for 5 min each, and then incubated with NeuroTrace 500/525 Green Fluorescent Nissl Stain (1:100, Invitrogen, USA) diluted in PBS for 30 min at RT. After washing in the same way, the slides were mounted with the mounting medium. All images were examined using a confocal microscope. For subsequent immunohistochemical analyses in Fig. 5c, d, a total of two to three sections corresponding to the rostral part (Bregma −4.9 mm to −5.1 mm) and caudal part (Bregma −5.4 mm to −5.5 mm) of the brainstem of each mouse were randomly selected. The images were converted to 8-bit grayscale and analyzed with ImageJ for their cross-sectional area (µm2) measurements.
For experiments in Fig. 3h, i, TG sections were washed three times with PBS for 10 min each and were incubated with a blocking solution containing 10% normal donkey serum (NDS) and 0.3% Triton X-100 in PBS for 1 h at RT and followed by rabbit anti-P2X3 receptor (1:1 000, Cat. #APR-016, Alomone labs, Israel) and Isolectin B4, biotin conjugate (20 μg·mL−1, Cat. #L2140, Sigma, Israel) in PBS containing 1% NDS and 0.3% Triton X-100 for overnight at 4 °C. On the next day, the slides were washed with PBS containing 0.1% Triton X-100 for 10 min and washed twice with PBS for 5 min each, followed by donkey anti-rabbit Alexa Fluor 594 (1:200, Invitrogen, USA), Alexa Fluor 488 Streptavidin (1:500, Jackson ImmunoResearch, USA), and NeuroTrace 640/660 Deep-Red Fluorescent Nissl Stain (1:100, Invitrogen, USA) incubated in PBS for 2 h at RT. After washing in the same way, the slides were mounted with the mounting medium. Images were obtained using a confocal microscope, and all images were converted to 8-bit grayscale with ImageJ, brightness/contrast and color thresholds were equalized, and positive neurons were counted in three to six sections obtained from each mouse.
Statistics and data presentation
Bar graphs/statistical data are presented as the mean or mean ± SD. Comparisons between the two groups in Fig. 5b were analyzed using an unpaired nonparametric test, and comparisons between the three groups in Fig. 5d were analyzed using a one-way analysis of variance. Differences were considered statistically significant at P-value < 0.05. Bar graphs and plots were generated using the Prism software (v.7.00, GraphPad), RStudio (v.1, RStudio, Inc.), or R (v.4, R Project for Statistical Computing).
Data availability
The raw read count data of genes, which are provided in the supplementary tables, are supplied in the supplementary information. The datasets supporting the conclusions of this article are available in the GEO and SRA repositories under accessions (GSE216835).
References
Fried, K. & Gibbs, L. J. In The Dental Pulp Biology, Pathology, and Regenerative Therapies (ed Goldberg, M.) Ch. 6 (Springer, 2014).
Haggard, P. & de Boer, L. Oral somatosensory awareness. Neurosci. Biobehav. Rev. 47, 469–484 (2014).
Fried, K., Sessle, B. J. & Devor, M. The paradox of pain from tooth pulp: low-threshold “algoneurons”? Pain 152, 2685–2689 (2011).
Lazarov, N. E. Neurobiology of orofacial proprioception. Brain Res. Rev. 56, 362–383 (2007).
Oh, S. B., Lee, P. R. & Ettlin, D. A. In Cell-to-cell Communication: Cell-atlas-visual Biology In Oral Medicine 1st edn, Vol. 7 (eds Gruber, R., Stadlinger, B. & Terheyden, H.) Nerve Cells (Quintessence Publishing Deutschland, 2022).
Furstman, L., Saporta, S. & Kruger, L. Retrograde axonal transport of horseradish peroxidase in sensory nerves and ganglion cells of the rat. Brain Res. 84, 320–324 (1975). (75)90987-7.
Shigenaga, Y., Mitsuhiro, Y., Yoshida, A., Cao, C. Q. & Tsuru, H. Morphology of single mesencephalic trigeminal neurons innervating masseter muscle of the cat. Brain Res. 445, 392–399 (1988). (88)91206-1.
Nguyen, M. Q., Wu, Y., Bonilla, L. S., von Buchholtz, L. J. & Ryba, N. J. P. Diversity amongst trigeminal neurons revealed by high throughput single cell sequencing. PLoS ONE 12, e0185543 (2017).
Emrick, J. J., von Buchholtz, L. J. & Ryba, N. J. P. Transcriptomic classification of neurons innervating teeth. J Dent. Res. https://doi.org/10.1177/0022034520941837 (2020).
Tang, F. et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382 (2009).
Sharma, N. et al. The emergence of transcriptional identity in somatosensory neurons. Nature 577, 392–398 (2020).
Li, C. L. et al. Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res. 26, 83–102 (2016).
Usoskin, D. et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 18, 145–153 (2015).
Yang, L. et al. Human and mouse trigeminal ganglia cell atlas implicates multiple cell types in migraine. Neuron https://doi.org/10.1016/j.neuron.2022.03.003 (2022)
Nguyen, M. Q., Le Pichon, C. E. & Ryba, N. Stereotyped transcriptomic transformation of somatosensory neurons in response to injury. Elife 8, https://doi.org/10.7554/eLife.49679 (2019).
Middleton, S. A., Eberwine, J. & Kim, J. Comprehensive catalog of dendritically localized mRNA isoforms from sub-cellular sequencing of single mouse neurons. BMC Biol. 17, 5 (2019).
Zaghlool, A. et al. Characterization of the nuclear and cytosolic transcriptomes in human brain tissue reveals new insights into the subcellular distribution of RNA transcripts. Sci. Rep. 11, 4076 (2021).
Yu, H. et al. Single-soma deep RNA sequencing of human DRG neurons reveals novel molecular and cellular mechanisms underlying somatosensation. bioRxiv https://doi.org/10.1101/2023.03.17.533207 (2023)
Ding, J. et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat. Biotechnol. 38, 737–746 (2020).
Hunter, D. V. et al. Advillin is expressed in all adult neural crest-derived neurons. eNeuro 5, https://doi.org/10.1523/ENEURO.0077-18.2018 (2018)
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Dueck, H. et al. Deep sequencing reveals cell-type-specific patterns of single-cell transcriptome variation. Genome Biol. 16, 122 (2015).
Avraham, O. et al. Profiling the molecular signature of satellite glial cells at the single cell level reveals high similarities between rodents and humans. Pain https://doi.org/10.1097/j.pain.0000000000002628 (2022)
Solomon, E. et al. Global transcriptome profile of the developmental principles of in vitro iPSC-to-motor neuron differentiation. BMC Mol. Cell Biol. 22, 13 (2021).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 e1821 (2019).
Jami, S. et al. Pain-causing stinging nettle toxins target TMEM233 to modulate Na(V)1.7 function. Nat. Commun. 14, 2442 (2023).
Tavares-Ferreira, D. et al. Spatial transcriptomics of dorsal root ganglia identifies molecular signatures of human nociceptors. Sci. Transl. Med. 14, eabj8186 (2022).
Chiu, I. M. et al. Transcriptional profiling at whole population and single cell levels reveals somatosensory neuron molecular diversity. Elife 3, https://doi.org/10.7554/eLife.04660 (2014)
Zheng, Y. et al. Deep sequencing of somatosensory neurons reveals molecular determinants of intrinsic physiological properties. Neuron 103, 598–616 e597 (2019).
Di Castro, A., Drew, L. J., Wood, J. N. & Cesare, P. Modulation of sensory neuron mechanotransduction by PKC- and nerve growth factor-dependent pathways. Proc. Natl Acad. Sci. USA 103, 4699–4704 (2006).
Prato, V. et al. Functional and molecular characterization of mechanoinsensitive “silent” nociceptors. Cell Rep. 21, 3102–3115 (2017).
Hill, R. Z., Loud, M. C., Dubin, A. E., Peet, B. & Patapoutian, A. PIEZO1 transduces mechanical itch in mice. Nature https://doi.org/10.1038/s41586-022-04860-5 (2022)
Pan, B. et al. TMC1 and TMC2 are components of the mechanotransduction channel in hair cells of the mammalian inner ear. Neuron 79, 504–515 (2013).
Chen, C. C. & Wong, C. W. Neurosensory mechanotransduction through acid-sensing ion channels. J. Cell Mol. Med. 17, 337–349 (2013).
Coste, B. et al. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 330, 55–60 (2010).
Won, J. et al. Piezo2 expression in mechanosensitive dental primary afferent neurons. J. Dent. Res. 96, 931–937 (2017).
Hermanstyne, T. O., Markowitz, K., Fan, L. & Gold, M. S. Mechanotransducers in rat pulpal afferents. J. Dent. Res. 87, 834–838 (2008).
Cavanaugh, D. J. et al. Distinct subsets of unmyelinated primary sensory fibers mediate behavioral responses to noxious thermal and mechanical stimuli. Proc. Natl Acad. Sci. USA 106, 9075–9080 (2009).
Zylka, M. J., Rice, F. L. & Anderson, D. J. Topographically distinct epidermal nociceptive circuits revealed by axonal tracers targeted to Mrgprd. Neuron 45, 17–25 (2005).
Puchalowicz, K., Tarnowski, M., Baranowska-Bosiacka, I., Chlubek, D. & Dziedziejko, V. P2X and P2Y receptors-role in the pathophysiology of the nervous system. Int J. Mol. Sci. 15, 23672–23704 (2014).
Cook, S. P., Vulchanova, L., Hargreaves, K. M., Elde, R. & McCleskey, E. W. Distinct ATP receptors on pain-sensing and stretch-sensing neurons. Nature 387, 505–508 (1997).
Wu, H. et al. Distinct subtypes of proprioceptive dorsal root ganglion neurons regulate adaptive proprioception in mice. Nat. Commun. 12, 1026 (2021).
Dykes, I. M., Lanier, J., Eng, S. R. & Turner, E. E. Brn3a regulates neuronal subtype specification in the trigeminal ganglion by promoting Runx expression during sensory differentiation. Neural Dev. 5, 3 (2010).
Hasegawa, H. & Wang, F. Visualizing mechanosensory endings of TrkC-expressing neurons in HS3ST-2-hPLAP mice. J. Comp. Neurol. 511, 543–556 (2008).
Matthews, P. B. Muscle spindles and their motor control. Physiol. Rev. 44, 219–288 (1964).
Inoue, K. et al. The transcription factor Runx3 represses the neurotrophin receptor TrkB during lineage commitment of dorsal root ganglion neurons. J. Biol. Chem. 282, 24175–24184 (2007).
Than, K. et al. Vesicle-released glutamate is necessary to maintain muscle spindle afferent excitability but not dynamic sensitivity in adult mice. J. Physiol. 599, 2953–2967 (2021).
Inoue, H., Morimoto, T. & Kawamura, Y. Response characteristics an classification of muscle spindles of the masseter muscle in the cat. Exp. Neurol. 74, 548–560 (1981).
Weng, H. J. et al. Tmem100 is a regulator of TRPA1-TRPV1 complex and contributes to persistent pain. Neuron 85, 833–846 (2015).
Ohmoto, M., Jyotaki, M., Yee, K. K. & Matsumoto, I. A transcription factor Etv1/Er81 is involved in the differentiation of sweet, umami, and sodium taste cells. eNeuro 10, https://doi.org/10.1523/ENEURO.0236-22.2023 (2023).
Espana, A. & Clotman, F. Onecut factors control development of the Locus Coeruleus and of the mesencephalic trigeminal nucleus. Mol. Cell Neurosci. 50, 93–102 (2012).
Lazarov, N. E. Comparative analysis of the chemical neuroanatomy of the mammalian trigeminal ganglion and mesencephalic trigeminal nucleus. Prog. Neurobiol. 66, 19–59 (2002).
Miyata, H., Sugiura, T., Wada, N., Kawai, Y. & Shigenaga, Y. Morphological changes in the masseter muscle and its motoneurons during postnatal development. Anat. Rec. 244, 520–528 (1996).
Maeda, N., et al. in Mechanoreceptors (eds Hník, P., Soukup, T., Vejsada, R. & Zelená, J.) Postnatal Development and Aging of Muscle Spindles in the Mouse Masseter Muscle and Effects of a Fine-Grained Diet on Them (Springer, 1988).
Paik, S. K. et al. Gamma-aminobutyric acid-, glycine-, and glutamate-immunopositive boutons on mesencephalic trigeminal neurons that innervate jaw-closing muscle spindles in the rat: ultrastructure and development. J. Comp. Neurol. 520, 3414–3427 (2012).
Bornstein, B. et al. Molecular characterization of the intact mouse muscle spindle using a multi-omics approach. Elife 12, https://doi.org/10.7554/eLife.81843 (2023)
Thiels, E., Alberts, J. R. & Cramer, C. P. Weaning in rats: II. Pup behavior patterns. Dev. Psychobiol. 23, 495–510 (1990).
Kristiansen, M. & Ham, J. Programmed cell death during neuronal development: the sympathetic neuron model. Cell Death Differ. 21, 1025–1035 (2014).
Brännström, M. & Astroem, A. A study on the mechanism of pain elicited from the dentin. J. Dent. Res. 43, 619–625 (1964).
Chung, G., Jung, S. J. & Oh, S. B. Cellular and molecular mechanisms of dental nociception. J. Dent. Res, 92, 948–955 (2013).
Russo, A. F. Calcitonin gene-related peptide (CGRP): a new target for migraine. Annu, Rev. Pharm. Toxicol. 55, 533–552 (2015).
Mitsiadis, T. A. & Pagella, P. The versatile roles of nerve growth factor in neuronal attraction, odontoblast differentiation, and mineral deposition in human teeth. Adv. Exp. Med. Biol. 1331, 65–75 (2021).
Chung, M. K., Jue, S. S. & Dong, X. Projection of non-peptidergic afferents to mouse tooth pulp. J. Dent. Res. 91, 777–782 (2012).
Masri, R., Ro, J. Y., Dessem, D. & Capra, N. Classification of muscle spindle afferents innervating the masseter muscle in rats. Arch. Oral. Biol. 51, 740–747 (2006).
Hong, G. S. et al. Tentonin 3/TMEM150c confers distinct mechanosensitive currents in dorsal-root ganglion neurons with proprioceptive function. Neuron 91, 107–118 (2016).
Lee, P. R., Lee, J. Y., Kim, H. B., Lee, J. H. & Oh, S. B. TRPM8 mediates hyperosmotic stimuli-induced nociception in dental afferents. J. Dent. Res. 99, 107–114 (2020).
Eckert, S. P., Taddese, A. & McCleskey, E. W. Isolation and culture of rat sensory neurons having distinct sensory modalities. J. Neurosci. Methods 77, 183–190 (1997).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Zhu, Q., Fisher, S. A., Shallcross, J. & Kim, J. VERSE: a versatile and efficient RNA-Seq read counting tool. bioRxiv https://doi.org/10.1101/053306 (2016)
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Suzuki, R. & Shimodaira, H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542 (2006).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb.) 2, 100141 (2021).
Rossi, H. L. et al. Evoked and spontaneous pain assessment during tooth pulp injury. Sci. Rep. 10, 2759 (2020).
Acknowledgment
The authors thank Dr. Youngnam Kang (Osaka University) for critical advice throughout the manuscript. This research was supported by the National Research Foundation (NRF) of Korea (grant number: 2018R1A5A2024418, 2022M3E5E8081190 and RS-2023-00302281 to S.B.O., and 2022R1C1C2003317 to P.R.L.) funded by the Korean government (Ministry of Education, Science and Technology). All authors gave their final approval and agreed to be accountable for all aspects of the work.
Author information
Authors and Affiliations
Contributions
P.R.L. and J.K. designed and performed experiments, analyzed the data, generated figures, interpreted the results, and wrote the manuscript; H.L.R., S.C., and S.Y.H. contributed to sample preparation; H.L.R., Dr. J.K., and Dr. S.B.O. critically revised the manuscript; Dr. J.K. and Dr. S.B.O. supervised all parts of the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lee, P.R., Kim, J., Rossi, H.L. et al. Transcriptional profiling of dental sensory and proprioceptive trigeminal neurons using single-cell RNA sequencing. Int J Oral Sci 15, 45 (2023). https://doi.org/10.1038/s41368-023-00246-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41368-023-00246-z
- Springer Nature Limited