Abstract
Despite the high prevalence of osteoarthritis (OA) in older populations, disease-modifying OA drugs (DMOADs) are still lacking. This study was performed to investigate the effects and mechanisms of the small molecular drug salicin (SA) on OA progression. Primary rat chondrocytes were stimulated with TNF-α and treated with or without SA. Inflammatory factors, cartilage matrix degeneration markers, and cell proliferation and apoptosis markers were detected at the mRNA and protein levels. Cell proliferation and apoptosis were evaluated by EdU assays or flow cytometric analysis. RNA sequencing, molecular docking and drug affinity-responsive target stability analyses were used to clarify the mechanisms. The rat OA model was used to evaluate the effect of intra-articular injection of SA on OA progression. We found that SA rescued TNF-α-induced degeneration of the cartilage matrix, inhibition of chondrocyte proliferation, and promotion of chondrocyte apoptosis. Mechanistically, SA directly binds to IRE1α and occupies the IRE1α phosphorylation site, preventing IRE1α phosphorylation and regulating IRE1α-mediated endoplasmic reticulum (ER) stress by IRE1α-IκBα-p65 signaling. Finally, intra-articular injection of SA-loaded lactic-co-glycolic acid (PLGA) ameliorated OA progression by inhibiting IRE1α-mediated ER stress in the OA model. In conclusion, SA alleviates OA by directly binding to the ER stress regulator IRE1α and inhibits IRE1α-mediated ER stress via IRE1α-IκBα-p65 signaling. Topical use of the small molecular drug SA shows potential to modify OA progression.
Similar content being viewed by others
Introduction
Osteoarthritis (OA) is the most prevalent joint disease and affects an estimated 500 million people worldwide1. Identification of disease-modifying OA drugs (DMOADs) that modify the structural progression of OA is considered a potential strategy for the treatment of early- or middle-stage OA2,3.
Willow bark is regarded as a successful example of a modern drug developed from an herbal remedy, which was first described ~200 years ago. Salicin (SA) is the main chemically standardized willow bark extract; its chemical oxidation resulted in a new substance termed “salicylic acid”, and the acetylated derivative was finally turned into the famous drug called “aspirin”4,5. SA is metabolized into salicylic acid after oral ingestion and plays roles in the treatment of pain, headaches, and inflammatory conditions5,6. However, the formation of salicylic acid alone is unlikely to explain the analgesic or anti-rheumatic effects of willow bark7, which indicates potential mechanisms that need further clarification. In contrast, as a small molecule drug, SA is easy to manufacture and can be easily absorbed and directly cross cell membranes. Recent studies have shown that SA helps prevent inflammation and angiogenic effects8, prevents cellular senescence9, and exhibits anti-irritation and anti-aging effects in dermatological applications10. However, based on our research, although SA appears efficient and safe to use in the treatment of OA7,11,12,13, the mechanisms by which SA prevents OA cartilage degeneration have not been fully elucidated.
In this study, we investigated the effects of SA on cartilage degeneration by both in vitro and in vivo tests. We clarified that SA prevents cartilage degeneration by alleviating inositol-requiring enzyme 1α (IRE1α) signaling-mediated endoplasmic reticulum (ER) stress. Our findings indicated the potential therapeutic effects of SA on OA progression by intra-articular injection.
Materials and methods
Chondrocyte culture and chemicals
The experimental protocols were approved by the Institutional Review Board (IRB) of Chongqing Medical University (No. 2020-018). Male Sprague‒Dawley (SD) rats were fed in specific pathogen-free animal facilities. Primary rat knee chondrocytes were isolated from articular cartilage of 4-day-old neonatal rats, as described previously14. Chondrocytes at passage 0 or 1 were subjected to the following experiments.
The purity of SA (Selleck Chemicals, TX, USA) was 98.9523% according to the manufacturer’s examination report with the use of high-performance liquid chromatography (HPLC). SA was dissolved in 0.1% dimethylsulfoxide (DMSO), and 0.1% DMSO was used as the control. Tumor necrosis factor α (TNF-α) was purchased from Peprotech (NJ, USA), and APY29 was purchased from Selleck Chemicals (TX, USA) at a concentration of 10 μM, as previously described15,16. Unless indicated otherwise, all chemicals were purchased from Sigma-Aldrich or Corning.
Cell viability
Cell viability was determined by Cell Counting Kit-8 (CCK-8, Med Chem Express, Shanghai, China) assays following the manufacturer’s protocol. For identification of toxic dosages, chondrocytes were treated with SA or TNF-α in gradient concentrations (0–100 μM for SA and 0–200 ng/ml for TNF-α) for 48 h. For the sustained toxicity of SA or TNF-α, chondrocytes were cultured in medium containing SA at different concentrations for 5 days and TNF-α for 3 days. The optical density (OD) values were determined at a wavelength of 450 nm. Nonlinear regression analysis was used to calculate the half-maximal inhibitory concentration (IC50) values for SA (percent cell proliferation versus concentration). The concentration of TNF-α in the present study was 10 ng/ml for 48 h according to previous studies9,17,18,19,20,21. Cytotoxicity of TNF-α on primary articular chondrocytes was shown in Supplementary Fig. 1.
Reverse transcription-quantitative polymerase chain reaction and XBP1 mRNA splicing
Total RNA was extracted from the chondrocytes with TRIzol reagent (Thermo Fisher Scientific, MA, USA) according to the manufacturer’s instructions. Reverse transcription was carried out by using the EvoScript Universal cDNA Master Reagent Kit (Med Chem Express, Shanghai, China). SYBR Green qPCR Master Mix (Med Chem Express, Shanghai, China), cDNA and primers were mixed according to the manufacturer’s instructions. The qPCR protocols were as follows: 5 min at 95 °C, 40 cycles of 10 s at 95 °C, 20 s at the optimal temperature for each pair of primers, and 20 s at 72 °C. Melting curves were generated at every endpoint of amplification for 10 s at 95 °C before 30 increments of 0.5 °C from 65 to 95 °C. GAPDH was used as the reference gene. All sample values were normalized to GAPDH expression by using the 2−△△CT method. All qPCRs were performed with three independent experiments. The qPCR primer sequences are shown in Table 1.
XBP1 mRNA splicing was assessed with reverse transcription polymerase chain reaction (RT‒PCR). Total RNA was purified using an RNeasy Mini Kit (Qiagen) and then converted into cDNA by using an EvoScript Universal cDNA Master Reagent Kit (Med Chem Express, Shanghai, China) following the manufacturer’s protocols. cDNA samples were diluted (1:10) and amplified using the primers F: 5ʹ- TTACGAGAGAAAACTCATGGC-3ʹ and R: 5ʹ-GGGTCCAAGTTGTCCAGAATGC-3ʹ with RT‒PCR and assessed on 2.5% agarose gels as previously reported22.
Western blotting
Chondrocytes treated with or without SA were lysed in cold radioimmunoprecipitation (RIPA, Beyotime, Shanghai, China) buffer containing phosphatase and protease inhibitors and then centrifuged at 13,000 × g at 4 °C for 15 min. Equivalent quantities of protein (30 μg) in each group were separated by 10–12% SDS-polyacrylamide gel electrophoresis (SDS‒PAGE) and transferred to a polyvinylidene difluoride (PVDF) membrane (Roche, Basel, Switzerland). For the detection of aggrecan, samples were deglycosylated according to the manufacturer’s instructions. After the membranes were blocked with 5% skim milk, they were probed with primary antibodies against Col2α1 (Abcam), Aggrecan (Abcam), MMP13 (Zen Bio), B-cell lymphoma 2 (BCL2) (Zen Bio), Cyclin Dependent Kinase 1 (CDK1) (Abcam), total extracellular signal-regulated kinase 1/2 (ERK1/2) (CST), phospho-ERK1/2 (CST), total-IκBα (CST), phospho-IκBα (CST), phospho-IRE1α (Zen Bio), IRE1α (Bioss), GRP78 (Zen Bio), NF-κB p65 (CST), phospho-NF-κB p65 (CST), phospho-PERK (Thr982) (Zen Bio), PERK (Zen Bio), ATF6 (ABclonal), XBP-1s (CST), and GAPDH (Zen Bio) overnight at 4 °C. Horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG (Abcam) was used as the secondary antibody. An enhanced chemiluminescence (ECL, Thermo Fisher Scientific, MA, USA) detection system was used to detect the protein bands on the membrane. The intensity of the protein bands was analyzed by ImageJ software using GAPDH as a reference protein as previously described23,24.
Cell immunofluorescence assays
Cells on coverslips were incubated with 6% normal goat serum and 4% bovine serum albumin in phosphate-buffered saline (PBS) for 1 h at 37 °C. Then, the cells were incubated with NF-κB p65 antibody (CST) and phospho-NF-κB p65 antibody (CST) overnight at 4 °C. The following day, fluorescein isothiocyanate (FITC/CY3)-labeled goat anti-rabbit IgG was used as a secondary antibody, and the cells were incubated for 1 h at room temperature. Finally, the cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and observed by fluorescence microscopy.
Cell proliferation assay
Chondrocyte proliferation was quantified by an ethynyl deoxyuridine (EdU) DNA in vitro proliferation detection kit (RiboBio, Guangzhou, China) according to the manufacturer’s instructions. The number of EdU-labeled cells was calculated from fields randomly selected in each well by two independent laboratory technicians in a blinded, random fashion.
Cell apoptosis assay
Chondrocytes in each treatment group were trypsinized, collected and washed three times with cold PBS. Approximately 4 × 105 cells/ml suspensions per group were mixed with Annexin V-FITC and PI binding buffer for 20 min according to the standard protocol of the Annexin V-FITC kit (Beyotime, Beijing, China). Finally, the mixtures were subjected to a flow cytometer (BD Biosciences, CA, USA) for apoptosis analysis.
ELISA
The levels of MMP13 in chondrocyte supernatants were measured using ELISA kits (E-EL-R0045c, Elabscience, Wuhan, China) according to the manufacturer’s instructions. The OD value was recorded at a wavelength of 450 nm.
RNA-sequencing and bioinformatics analysis
Total RNA extracted from chondrocytes treated with or without SA was subjected to RNA-sequencing (RNA-Seq). RNA samples were sent to OE Biomedical Technology Co., Ltd., (Shanghai, China) for RNA-Seq. RNA sequencing was performed on the HiSeq2500 system (Illumina, CA, USA). The R programming language program was used to analyze the raw data and identify differentially expressed genes (DEGs). Selection of differentially expressed transcripts with p values ≤ 0.05 and fold change ≥2 was followed by Gene Ontology (GO) biological function enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) signal pathway enrichment analysis.
Target prediction and molecular docking
The SA structure was downloaded from the PubChem database and converted to mol2 format. The three-dimensional structure of the IRE1α (PDB ID: 6HX1) protein was downloaded from the Research Collaboratory for Structural Bioinformatics (RCSB) protein database. AutoDock Vina 1.1.2 was used for semiflexible docking with SA and IRE1α. The parameter exhaustiveness was set as 20 to increase the calculation accuracy. The conformation with the best affinity (-8.567 kcal/mol) was selected as the final docking conformation.
Drug affinity responsive target stability
Drug affinity responsive target stability (DARTS) was performed as previously described25,26. Briefly, chondrocytes were treated with cold M-PER lysis buffer (Thermo Fisher, MA, USA) containing a protease inhibitor cocktail (1 mM Na3VO4 and 1 mM NaF). Protein lysates were first mixed with 10x TNC buffer (500 mM Tris–HCl pH=8.0, 500 mM NaCl and 100 mM CaCl2 at a ratio of 1:1) and then incubated with DMSO or SA for 1 h at room temperature. Next, the sample was proteolyzed in gradient concentrations of pronase (Roche, Basel, Switzerland) for 10 min, which was followed by the addition of 2 μl of cold 20x protease inhibitor cocktail. Coomassie blue staining (Beyotime, Shanghai, China) was carried out to estimate the relative abundance of proteins. An equal portion of each sample was loaded onto an SDS‒PAGE gel for Western blotting.
Animal model
All animal experiments were approved by the Institutional Review Board (IRB) of Chongqing Medical University (No. 2020-018). After anesthesia and standard aseptic surgical procedures, ACLT was performed in the right knee of rats (200 ± 20 g) to establish an OA model (duration: four weeks)20,21, and rats with only open knee joints were regarded as a sham group (n = 5). Poly(lactic-co-glycolic acid) (PLGA) (Sigma-Aldrich, MO, USA) was dissolved in CH2Cl2 and mixed with ultrasonically dissolved SA, and then, PLGA-loaded SA was obtained by the double-emulsion method as reported previously27,28. The animals that received ACLT were randomly divided into three groups: ACLT (PBS only, n = 5), vehicle (PLGA vehicle, n = 5), and SA (PLGA vehicle with SA, n = 5). Four weeks after surgery, intra-articular injection of 100 μL of PLGA + SA suspension (the equivalent of 1 mM SA, PLGA loaded), PLGA only and PBS only was performed. Four weeks post-intra-articular injection, the rats were sacrificed, and knee joints were harvested.
Histological analysis
Samples were fixed in 4% paraformaldehyde (Beyotime, Beijing, China), decalcified in 0.5 M ethylenediaminetetraacetic acid (EDTA) and embedded in paraffin. Serial 5-μm-thick sections were obtained and subjected to histological and other specialty staining evaluations, including hematoxylin-eosin (H&E) and safranin O/green staining, as previously described22,24,29. The articular surface of the femur and tibia in coronal sections of the knee joint was qualitatively and semiquantitatively analyzed as previously described20,21. Each specimen was scored by three orthopedic surgeons following the double-blind principle according to the Osteoarthritis Research Society International (OARSI) criteria for rats30.
For tissue IF staining, paraffin sections were dehydrated in ethanol solutions of different concentrations, permeabilized with 0.1% Triton X-100 and blocked with 5% BSA. Then, the sections were incubated overnight at 4 °C with primary phospho-NF-κB p65 (CST), Ki-67 (D3B5) (CST) and p-IRE1α (Zen Bio) antibodies. Sections were treated with appropriate fluorescein-conjugated secondary antibodies. DAPI was utilized for nuclear staining. The TUNEL assay was carried out using a Click-iTTM Plus TUNEL Apoptosis Assay Kit (Invitrogen) according to the manufacturer’s instructions. The images of positive cells were captured by a fluorescence microscope.
Sample size determinations
For in vitro experiments with fold change or quantitative analysis, we calculated a minimum sample size (n ≥ 3) with three independent experiments. For in vivo experiments, based on previous experiments, we selected a sample size of n ≥ 4 at an alpha level of 0.05 and a power of 80%31,32.
Statistical analysis
All datasets were compared using GraphPad Prism (GraphPad Software, La Jolla, CA) version 9.0. Data are shown as the mean ± standard deviation (SD). For in vivo and vitro studies, unpaired Student’s t test (for two groups) and one-way ANOVA (for multiple groups) were used followed by the Tukey‒Kramer test. p < 0.05 was considered statistically significant.
Results
Phenotypic characterization of primary articular chondrocytes and the cytotoxicity of SA on chondrocytes
Primary articular chondrocytes were characterized by toluidine blue staining and IF staining with Col2a1 (red) and ACAN (green) (Fig. 1a). We first identified the effects of SA on the cell cycle and found that SA promoted chondrocyte proliferation (Fig. 1bi). Specifically, quantitative analysis (Fig. 1bii) showed that SA significantly increased S-phase chondrocytes. Next, the cytotoxicity of SA to chondrocytes was assessed. As shown in Fig. 1c, 5 μM, 10 μM and 20 μM SA resulted in significant effects, and 10 μM had the strongest effect. However, 50 μM and 100 μM SA dramatically reduced cell viability. Furthermore, we found that 10 μM SA treatment promoted chondrocyte proliferation from Day 1 to Day 4 compared with the control treatment (Fig. 1d). For these experiments, the IC50 of SA was 42.30 μM (Fig. 1e). Thus, 10 μM SA was considered not toxic to chondrocytes and selected as the optimal dose for the next investigations.
a Staining of articular chondrocytes in primary cultures with toluidine blue. Immunostaining of primary articular chondrocytes with Col2a1 (red), ACAN (green) and DAPI (blue). b Flow cytometry for cell cycle progression of chondrocytes with or without SA (i). Quantitative analysis (ii) found that SA significantly promoted S-phase cells (n = 3, Student’s t test). c Chondrocytes were treated with SA at gradient concentrations (0-100 μM) for 48 h and subjected to CCK-8 analysis. From 5 μM to 20 μM SA treatment, the OD values were significantly higher than those of the 0 μM (DMSO) group, and for 50 μM and 100 μM SA treatment, the OD values were significantly lower than those of the 0 μM (DMSO) group (n = 5, one-way ANOVA). d Chondrocyte proliferation curve with SA treatment from Day 1 to Day 5. Compared with DMSO treatment, 10 μM SA treatment significantly promoted cell proliferation from Day 1 to Day 5 (n = 5, Student’s t test). e IC50 of SA on chondrocytes; concentrations were transferred to Log(c). The data are expressed as the mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, *compared with the DMSO group, #p < 0.05, ##p < 0.01, ###p < 0.001, # compared with the indicated group.
SA inhibits TNF-α-induced chondrocyte inflammatory factor expression and extracellular matrix degeneration
We first explored the effects of SA on TNF-α-induced chondrocyte inflammatory factors and matrix gene expression. As shown in Fig. 2a, TNF-α dramatically increased interleukin-1β (IL-1β), interleukin-6 (IL-6), and matrix metalloproteinase 13 (MMP13) gene expression and inhibited Col2a1 and ACAN gene expression in chondrocytes compared with that of the control group. However, these effects could be diminished by 48 h of treatment with 10 μM SA in vitro. Moreover, we found that TNF-α induced chondrocyte matrix degeneration by elevating the expression and secretion of MMP13, which was reversed by SA at the protein level (Fig. 2b-c). In addition, we found that SA could rescue TNF-α-induced cartilage matrix degradation by detecting the expression of Col2a1 and ACAN at the protein level (Fig. 2ci). Quantitative analysis showed the same trend (Fig. 2cii-iv). These results indicated that SA inhibits TNF-α-induced chondrocyte degeneration in vitro.
Chondrocytes were stimulated with TNF-α and then treated with or without 10 μM SA for 48 h. a Relative mRNA expression levels of IL1β, IL-6, MMP13, COL2A1 and ACAN were detected by reverse transcription-quantitative polymerase chain reaction (n = 3, one-way ANOVA). b Expression of active MMP13 protein was reduced by the chondrocytes when stimulated with SA (n = 3, one-way ANOVA). c WB analysis for detecting the cartilage matrix markers MMP13, ACAN and COL2A1 in different groups (i). Quantitative analysis of MMP13 (ii), ACAN (iii), and COL2A1 (iv) at the protein level. GAPDH was used as a reference protein (n = 3, one-way ANOVA). The data are expressed as the mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, and ns not significant.
SA ameliorates cartilage degeneration by enhancing chondrocyte proliferation and inhibiting chondrocyte apoptosis
Next, we investigated the effects of SA on TNF-α-mediated chondrocyte apoptosis and proliferation. We found that TNF-α significantly inhibited the expression of the antiapoptotic gene BCL-2 and the cell cycle genes CDK1 and MKi67 in chondrocytes. However, when the chondrocytes were treated with 10 μM SA, these inhibitory effects were eliminated (Fig. 3a). Regarding the phosphorylation of ERK, we found that TNF-α did not influence total ERK (t-ERK) expression, but it significantly inhibited p-ERK expression. SA blocked the downregulation of p-ERK and elevated the ratio of p-ERK and t-ERK in comparison to the TNF-α treatment (Fig. 3b). In addition, we determined the effects of SA on TNF-α-mediated inhibition of BCL2 and CDK1 expression at the protein level. We found that SA eliminated TNF-α-induced inhibition of BCL2 (Fig. 3ci), but TNF-α did not inhibit the expression of CDK1 at the protein level, and SA promoted the expression of CDK1 in the presence of TNF-α (Fig. 3ci). Quantitative analysis showed the same trend (Fig. 3cii–iii). These results suggested that SA eliminated TNF-α-mediated inhibition of cell cycle-related marker expression.
a SA rescued TNF-α-induced inhibition of cell cycle marker expression at the mRNA level. Chondrocytes were stimulated with TNF-α and then treated with or without 10 μM SA for 48 h, and the relative mRNA expression levels of BCL2, CKD1, and MKi67 were detected (n = 3, one-way ANOVA). b Western blot analysis for detecting t-ERK and p-ERK in different groups treated with SA for 48 h (a). Quantitative analysis of the ratio of p-ERK/t-ERK. GAPDH was used as a reference protein (b) (n = 3, one-way ANOVA). c WB for detecting the proliferation markers BCL2 and CDK1 in different groups 48 hours after treatment at the protein level. Quantitative analysis of BCL2 (ii) and CDK1 (iii) at the protein level. GAPDH was used as a reference protein (n = 3, one-way ANOVA). d EdU assays for detecting DNA synthesis indicating cell proliferation with 24 h and 48 h treatment (i), scale bar 400 μm. Percentage of EdU (red)-positive staining at 24 h (ii) and 48 h (c) (n = 5, random fields, one-way ANOVA). e Flow cytometry for cell apoptosis analysis of chondrocytes in each treatment group at 24 h and 48 h (i). Percentage of apoptotic cells in each treatment group at 24 h (ii) and 48 h (c) (n = 3, one-way ANOVA). The data are expressed as the mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, and ns not significant.
Furthermore, cell proliferation by EdU staining showed that SA could rescue TNF-α-induced inhibition of DNA duplication in a dose-dependent manner at both 24 h and 48 h (Fig. 3di). Quantitative analysis showed the same trend (Fig. 3dii–iii). Cell apoptosis and quantitative analysis also confirmed that SA eliminated the TNF-α-induced promotion of cell apoptosis at both 24 h and 48 h (Fig. 3e).
SA inhibits IRE1α-IκBα-p65 signaling-mediated ER stress by directly binding to IRE1α
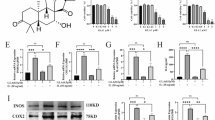
RNA-seq was performed in chondrocytes cultured with or without SA under TNF-α stimulation. As shown in Fig. 4a, b, for differentially expressed genes (DEGs), we found that 94 DEGs were upregulated, and 96 DEGs were downregulated. Then, DEGs were subjected to GO and KEGG signaling pathway enrichment analysis (Fig. 4c, d). Through GO analysis, we found that DEGs were highly enriched in biological processes (BPs) such as cellular responses to ER stress and oxygen levels. In addition, KEGG signaling pathway enrichment analysis showed that protein processing in the ER (Fig. 4c, d marked with blue) was one of the most highly enriched signaling pathways.
a Heatmap for global gene expression with group clusters (n = 3). b Volcano map of DEGs in the SA group vs. the control group (upregulation: 75 genes and downregulation: 32 genes). DEGs with FC (fold change) ≥ 2 were accepted as positive DEGs. c Pathway enrichment bubble map based on the KEGG enrichment analysis. A rich factor indicates a higher degree of enrichment, a larger p value (−log10) indicates a higher statistical significance, and a larger bubble indicates a higher degree of enrichment. d GO enrichment of those DEGs. A rich factor indicates a higher degree of enrichment, a larger p value (−log10) indicates a higher statistical significance, and a larger bubble indicates a higher degree of enrichment, BP: biological process, CC: cellular component. e Molecular docking for estimating the binding site of IRE1α and SA. The three-dimensional (3D) structures of IRE1α and SA were subjected to molecular docking, and the binding site was obtained according to the software scoring system. Upper left panel: SA was located in the cavity formed by the IRE1α 3D structure, upper right panel: magnification of the cavity; lower left panel: binding site of SA and IRE1α, lower left panel: magnification showing the hydrogen bonds (yellow dot line) formed between SA and IRE1α. f DARTS assay for confirmation of IRE1α and SA binding. Protease was used to digest IRE1α protein. With the addition of SA, the digestion of IRE1α was significantly blocked at each concentration of protease compared with the DMSO group.
Three main pathways mediated the transduction of ER stress. As shown in Supplementary Fig. 2, SA did not influence TNF-α induced PERK phosphorylation and the expression of ATF6. However, SA dramatically rescued TNF-α induced IRE1-α phosphorylation. Therefore, we analyzed the potential combination of SA and IRE1α.
Three-dimensional (3D) structures of SA and IRE1α were subjected to molecular docking. The score of SA docking with IRE1α was −8.567 kcal/mol, and the theoretical binding mode is shown in Fig. 4e. The compound and the amino acid residue TYR628 formed a hydrogen bond with a bond length of 3.3 Å; the amino acid residue CYS645 formed a hydrogen bond with a bond length of 3.2 Å; and the amino acid residue SER724 (phosphorylation site) formed a hydrogen bond with a bond length of 3.6 Å. These interactions guaranteed the stability of the combination of IRE1α and SA. Furthermore, through the DARTS assay, we found that SA protected IRE1α from dosage-dependent protease digestion compared with the control (Fig. 4f), which indicates the direct combination of SA and IRE1α protein.
Next, we investigated the regulatory effects of SA on IRE1α-mediated ER stress. First, we detected the expression level of the ER stress marker GRP78 and found that SA inhibited TNF-α-initiated GRP78 expression by western blotting (WB; Fig. 5ai) and quantitative analysis (Fig. 5aii). Total IRE1α and p-IRE1α levels were also detected by WB. Compared to the control, TNF-α did not influence total IRE1α expression but downregulated phosphorylated IRE1α expression, which could be rescued by SA treatment (Fig. 5aiii). Then, we detected IRE1α downstream gene NF-kappa-B inhibitor alpha (IκBα) and p65 expression at the protein level. The results showed that TNF-α induced high expression of p-IκBα and p65, which could be diminished by SA treatment (Fig. 5aiv–v).
a WB analysis for detecting IRE1α and downstream gene expression at the protein level. The ER stress-associated proteins GRP78, pIRE1α, IRE1α, p-IκBα, t-IκBα, p-p65 and p65 were detected in each group (i). Quantitative analysis of GRP78 (ii), the ratio of p-IRE1α/t-IRE1α (iii), the ratio of p-IκBα/t-IκBα (iv), and the ratio of p-p65/t-p-p65 (v) at the protein level. GAPDH was used as a reference protein (n = 3, one-way ANOVA). b P65 nuclear translocation in each treatment group. IF was used to detect p-65 in the nucleus, and DAPI was used to stain the cell nucleus; scale bar: 50 μm. c The IRE1α inhibitor APY29 blocked TNF-α-mediated ER stress. Chondrocytes were stimulated with TNF-α and then treated with 10 μM SA or 10 μM APY29 for 48 h. XPB1u and XPB1s were detected by RT-QPCR (i), and XPB1s, MMP13, GRP78, IRE1α, p-IRE1α, p-p65 and p65 were detected by WB analysis (ii). Quantitative analysis of XPB1s (iii), MMP13 (iv), GRP78 (v), the ratio of p-IRE1α/t-IRE1α (vi), and the ratio of p-p65/t-p65 (vii) at the protein level. GAPDH was used as a reference protein (n = 3, one-way ANOVA). The data are expressed as the mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, and ns, not significant.
Subsequently, we explored whether SA influenced the TNF-α-induced distribution of p65 protein in chondrocytes by IF. As expected, TNF-α-induced p65 translocation from the cytoplasm into the nucleus was eliminated by SA treatment (Fig. 5b). These findings indicated that SA inhibited IRE1α-IκBα-p65 signaling-mediated ER stress by occupying the phosphorylation site of IRE1α.
Finally, the IRE1α inhibitor APY29 was utilized to confirm the function of SA. We found that the IRE1-regulated transcription factors XBP1u and XBP1s were upregulated with the stimulation of TNF-α, and SA and APY29 did not influence XBP1u or XBP1s expression at the mRNA level (Fig. 5ci) or XPB1s (Fig. 5cii) at the protein level, which indicated that SA does not regulate IRE1α-mediated ER stress through XPB1s signaling. However, APY29 dramatically inhibited TNF-α-induced ER stress marker GRP78, p-IRE1α, p-p65 and cartilage matrix degeneration marker MMP13 expression at the protein level, and SA exhibited a similar effect (Fig. 5ciii–vii).
Taken together, these results suggested that SA can directly bind to IRE1α and inhibit IRE1α phosphorylation, which further blocked IRE1α-IκBα-p65 signaling-mediated ER stress.
SA intra-articular injection ameliorates ACLT-induced OA progression by inhibiting IRE1α-mediated ER stress
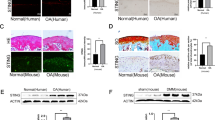
A rat ACLT-induced OA model was constructed for further research. Figure 6a summarizes the protocol of the in vivo study. SA-loaded PLGA scaffolds were intraarticularly injected for controlled release of SA27,28. As shown in Fig. 6bi, no obvious cartilage degeneration was found in the sham group. In the ACLT and vehicle groups, cartilage cell number, cartilage matrix and cartilage thickness decreased significantly compared with those of the sham group, while the SA group showed diminished effects. Knee joint OARSI scoring (Fig. 6bii) and quantitative analysis of the articular cartilage area (Fig. 6biii) showed that SA treatment could reverse ACLT-induced cartilage degeneration.
a Diagram summarizing the animal experiments. ACLT was used to construct the knee OA model. Four weeks later, intra-articular injection of SA-loaded PLGA was performed, and intra-articular injection of PLGA only was used as a control. Four weeks after treatment, rats in each group were sacrificed, and knee joints were subjected to histological analysis. b H&E and safranin O/green staining for each treatment group. H&E (upper panel) and safranin O/green staining (lower panel) showed that ACLT resulted in thicker cartilage and degeneration of the cartilage matrix than sham treatment, and intra-articular injection of SA-loaded PLGA ameliorated this progression compared with vehicle (PLGA only) treatment or sham treatment. OARSI scoring (ii) and cartilage matrix area quantitative analysis (iii) showed that, although OARSI scores in the ACLT, vehicle and SA groups were significantly higher than that of the sham group, the OARSI score in the SA group was significantly lower than those of the ACLT and vehicle groups (n = 5, one-way ANOVA). c Ki67 staining for detecting chondrocyte proliferation. IF staining was used to detect Ki67-positive chondrocytes in each treatment group (i). Quantitative analysis showed that SA dramatically promoted chondrocyte Ki67 expression compared with ACLT (n = 5, one-way ANOVA). d TUNEL staining for detecting chondrocyte apoptosis. TUNEL staining showed that SA treatment reversed ACLT-induced chondrocyte apoptosis (i). Quantitative analysis showed the same trend (ii) (n = 5, one-way ANOVA). ACLT group indicates the group with PBS injection; vehicle group indicates the group injected with only PLGA vehicle; SA group indicates the group injected with SA-loaded PLGA. Dashed lines indicate the cartilage surface. Scale bar 50 μm, data are expressed as the mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, and ns, not significant.
To further prove the mechanisms by which SA inhibits IRE1α-mediated ER stress, we performed IF. Ki67 staining and TUNEL staining were used to evaluate chondrocyte proliferation and apoptosis. The results showed decreased proliferation and increased apoptosis of chondrocytes in both the ACLT and vehicle groups compared with the sham group, while SA treatment reversed the decreased proliferation and increased apoptosis of chondrocytes (Fig. 6c, d). Then, we found that the OA-induced high phosphorylation levels of IRE1α were alleviated by SA treatment (Fig. 7a). Finally, we showed that OA-induced p-p65 nuclear translocation was inhibited by SA treatment (Fig. 7b). These results suggested that SA ameliorates ACLT-induced OA progression by inhibiting IRE1α-mediated ER stress in vivo.
a P-IRE1α in chondrocytes in each treatment group. IF was used to detect p-IRE1α in each treatment group. p-IRE1α was highly expressed in the cytoplasm of chondrocytes in the ACLT and vehicle groups, and SA dramatically decreased the expression of p-IRE1α in the cytoplasm of chondrocytes (i). Quantitative analysis showed that ACLT-induced high expression of p-IRE1α was significantly reversed by SA treatment (ii) (n = 5, one-way ANOVA). b P-p65 nuclear translocation in each treatment group. ACLT-induced p-p65 nuclear translocation was ameliorated by SA treatment (i). Quantitative analysis of each treatment group showed the same trend (ii). Scale bar, 50 μm; the ACLT group indicates the group injected with PBS; the vehicle group indicates the group injected with only PLGA vehicle; the SA group indicates the group injected with SA-loaded PLGA. Dashed lines indicate the cartilage surface. The data are expressed as the mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, and ns, not significant. c A proposed model of action. SA binding to IRE1α blocked IRE1α phosphorylation and inhibited IRE1α-mediated ER stress via IRE1α-IκBα-p65 signaling.
In summary, our in vitro and in vivo tests showed that SA binds to IRE1α and blocks IRE1α phosphorylation, which inhibits IRE1α-mediated ER stress by IRE1α-IκBα-p65 signaling (Fig. 7c).
Discussion
With the aging population, the morbidity of OA is increasing year by year33. DMOADs have been identified as potential treatments to address the increasing OA prevalence2,34. In this study, we found that SA not only inhibits TNF-α-induced chondrocyte inflammatory factor expression and extracellular matrix degeneration but also enhances chondrocyte proliferation and inhibits chondrocyte apoptosis. Mechanistically, we clarified that SA may regulate OA progression by binding with IRE1α and regulating IRE1α-mediated ER stress.
As the most common chemically standardized willow bark extract, SA is a nonsteroidal anti-inflammatory drug (NSAID), which may cause gastrointestinal, renal, and cardiovascular toxicity6,34. As reported by Schmid et al., oral ingestion of 13.97 mmol of pure SA resulted in a peak serum concentration of 800 µM, which indicated that oral ingestion of SA could lead to side effects or low concentrations of SA in the knee joint7. As a natural small molecular drug, SA can cross the cell membrane and bind with intracellular targets, thus avoiding the serious side effects caused by oral ingestion. By directly adding SA to the chondrocyte medium and intra-articular injection of SA-loaded PLGA, we demonstrated the anti-inflammatory, pro-proliferative, and antiapoptotic effects of SA in the progression of OA. These results indicate the advantage and efficiency of intra-articular use of SA.
Recently, in addition to the anti-inflammatory, antipyretic, antirheumatic, and antiseptic properties of SA10, Gao et al.12 found that SA prevents collagen type II degradation by inhibiting the activation of the NF-κB proinflammatory pathway. However, the underlying detailed mechanisms are still unclear. In this study, we obtained similar results by detecting cartilage matrix degeneration and inflammatory markers. Furthermore, DEGs found by RNA sequencing and KEGG enrichment analysis showed that ER stress was highly involved in the protective effects of SA on cartilage degeneration. Next, molecular docking and DARTS analysis showed how SA can occupy the phosphorylation sites of IRE1α and block IRE1α phosphorylation-mediated p65 nuclear translocation and downstream gene expression. Thus, this study helped elucidate the SA-mediated protective effects on cartilage degeneration.
The ER is a multifunctional organelle where protein folding occurs prior to transport to the extracellular surface or different intracellular sites35,36,37,38. Three ER transmembrane proteins mediate the unfolded protein response (UPR), including IRE1, pancreatic endoplasmic reticulum kinase (PERK), and activating transcription factor 6 (ATF6)36,37. These three ER transmembrane proteins regulate thousands of genes involved in controlling the function of the ER while also regulating signal transduction of ER stress39,40. Among them, IRE1 is the most conserved gene from yeast to humans and has two subtypes: IRE1α and IRE1β. IRE1α is commonly expressed in most cells and tissues, while IRE1β is limited to gastrointestinal epithelial cells41. Notably, IRE1α plays a key role in chondrocyte proliferation, ECM production, etc42., which indicates the regulatory function of IRE1α-mediated ER stress in chondrocyte proliferation and degeneration. Jacqueline et al.43 found that the two top risk factors for OA, age and obesity, were highly associated with ER stress, and resveratrol mitigated early joint degeneration by inhibiting ER stress. Benedetta et al.44 found that chronic ER stress decreased chondrocyte proliferation. Kung et al.45,46 reported that hypertrophic chondrocytes have limited potential to cope with increased ER stress, and they also found that increased ER stress is sufficient to reduce chondrocyte proliferation. Rajpar et al.47 showed that ER stress plays a direct role in cartilage pathology. It is widely accepted that ER stress is directly associated with chondrocyte apoptosis and death48,49,50. Moreover, recent studies have confirmed that OA inflammation is related to ER stress51,52,53,54,55. In this study, we first detected the expression levels of PERK, p-PERK, IRE1α, p-IRE1α, and AFT6 after stimulation with TNF-α in the presence of SA and found that SA did not influence TNF-α-induced PERK phosphorylation or ATF6 expression. However, SA dramatically rescued TNF-α-induced IRE1-α phosphorylation. Furthermore, we showed that by directly binding with IRE1α, SA inhibited IRE1α-mediated ER stress and subsequently promoted chondrocyte proliferation, decreased inflammatory factor expression, and inhibited chondrocyte apoptosis through both in vitro and in vivo tests.
Recently, ER stress regulators were identified as new drug targets for several diseases35,36,37,56. During the progression of OA, chondrocytes are responsible for the biogenesis and maintenance of cartilage ECM. Several cellular stresses, including hypoxia, oxidative stress, nutrient deprivation, aging, injury, etc., can cause excessive unfolding or misfolded proteins on the ER and trigger ER stress43,50. Huang et al.42 reported that IRE1α regulates chondrocyte apoptosis by activating NF-κB signaling. Ye et al.57 reported that phosphorylated IRE1α activates NF-κB signaling by releasing IκBα. Released NF-κB dimers translocate to the nucleus and bind κB sites in the promoters or enhancers of target genes58. However, IRE1α can also activate RNase to splice XBP1 mRNA and produce the homeostatic transcription factor XBP1s, which mainly participates in ER chaperone expression, lipid biosynthesis, hexosamine biosynthesis and proinflammatory cytokine expression16,40,59. We showed that instead of influencing the expression of XBP1u and XPB1s at the mRNA level or influencing the expression of XPB1s at the protein level, SA regulated ER stress by regulating p-IRE1α and p-IκBα and activating p-p65 nuclear translocation. Furthermore, molecular docking and DARTS analysis showed that SA can bind the phosphorylation site of IRE1α and block IRE1α phosphorylation, which is followed by the decreased expression of p-κBα and p-p65 nuclear translocation. Therefore, as one of the key ER stress regulators, IRE1α is a potential drug target for OA treatment.
As a whole joint disease, OA, especially late-stage OA, is characterized by dramatic synovium, cartilage, and subchondral bone pathologies1,60,61. However, when early-stage OA featured cartilage pathology62,63, we found that topical use of SA promoted chondrocyte proliferation and inhibited cartilage matrix degeneration. Therefore, SA shows potential as an effective drug for modifying early-stage OA. More studies that focus on the effect of SA on other cell types, such as synoviocytes and immune cells, are needed and will contribute to the further clinical translation of SA.
This study demonstrated that SA directly binds to IRE1α and blocks IRE1α phosphorylation, which inhibits IκBα phosphorylation and p65 nuclear translocation. Subsequently, these events inhibit chondrocyte apoptosis, promote chondrocyte proliferation, and decrease the expression of inflammatory factors. Thus, SA is a potential drug for the clinical modification of OA progression by intra-articular injection.
References
Hunter, D. J., March, L. & Chew, M. Osteoarthritis in 2020 and beyond: a Lancet Commission. Lancet 396, 1711–1712 (2020).
Hunter, D. J. Pharmacologic therapy for osteoarthritis–the era of disease modification. Nat. Rev. Rheumatol. 7, 13–22 (2011).
Roemer, F. W., Kwoh, C. K., Hayashi, D., Felson, D. T. & Guermazi, A. The role of radiography and MRI for eligibility assessment in DMOAD trials of knee OA. Nat. Rev. Rheumatol. 14, 372–380 (2018).
Fiebich, B. L. & Appel, K. Anti-inflammatory effects of willow bark extract. Clin. Pharmacol. Ther. 74, 96 (2003). author reply 96-97.
Vlachojannis, J. E., Cameron, M. & Chrubasik, S. A systematic review on the effectiveness of willow bark for musculoskeletal pain. Phytother. Res. 23, 897–900 (2009).
Willow, B. Drugs and Lactation Database (LactMed) Bethesda (MD) (National Library of Medicine, 2006).
Schmid, B., Kotter, I. & Heide, L. Pharmacokinetics of salicin after oral administration of a standardised willow bark extract. Eur. J. Clin. Pharmacol. 57, 387–391 (2001).
Kong, C. S. et al. Salicin, an extract from white willow bark, inhibits angiogenesis by blocking the ROS-ERK pathways. Phytother. Res. 28, 1246–1251 (2014).
Guo, F., Wu, R. & Xu, J. Salicin prevents TNF-alpha-induced cellular senescence in human umbilical vein endothelial cells (HUVECs). Artif. Cells Nanomed. Biotechnol. 47, 2618–2623 (2019).
Adamiak, K., Lewandowska, K. & Sionkowska, A. The infuence of salicin on rheological and film-forming properties of collagen. Molecules 26, 1661 (2021).
Marz, R. W. & Kemper, F. [Willow bark extract–effects and effectiveness. Status of current knowledge regarding pharmacology, toxicology and clinical aspects]. Wien. Med. Wochenschr. 152, 354–359 (2002).
Gao, F. & Zhang, S. Salicin inhibits AGE-induced degradation of type II collagen and aggrecan in human SW1353 chondrocytes: therapeutic potential in osteoarthritis. Artif. Cells Nanomed. Biotechnol. 47, 1043–1049 (2019).
Biegert, C. et al. Efficacy and safety of willow bark extract in the treatment of osteoarthritis and rheumatoid arthritis: results of 2 randomized double-blind controlled trials. J. Rheumatol. 31, 2121–2130 (2004).
Gosset, M., Berenbaum, F., Thirion, S. & Jacques, C. Primary culture and phenotyping of murine chondrocytes. Nat. Protoc. 3, 1253–1260 (2008).
Mi, B. et al. Osteoblast/osteoclast and immune cocktail therapy of an exosome/drug delivery multifunctional hydrogel accelerates fracture repair. ACS Nano https://pubs.acs.org/doi/10.1021/acsnano.1c08284 (2022).
Wang, L. et al. Divergent allosteric control of the IRE1alpha endoribonuclease using kinase inhibitors. Nat. Chem. Biol. 8, 982–989 (2012).
Xu, K., Gao, Y., Yang, L., Liu, Y. & Wang, C. Magnolin exhibits anti-inflammatory effects on chondrocytes via the NF-kappaB pathway for attenuating anterior cruciate ligament transection-induced osteoarthritis. Connect. Tissue Res. 62, 475–484 (2020).
Hu, G. et al. MicroRNA-145 attenuates TNF-alpha-driven cartilage matrix degradation in osteoarthritis via direct suppression of MKK4. Cell Death Dis. 8, e3140 (2017).
Zhao, Y. et al. Cortistatin binds to TNF-alpha receptors and protects against osteoarthritis. EBioMedicine 41, 556–570 (2019).
Wang, C. et al. Ursolic acid protects chondrocytes, exhibits anti-inflammatory properties via regulation of the NF-kappaB/NLRP3 inflammasome pathway and ameliorates osteoarthritis. Biomed. Pharmacother. 130, 110568 (2020).
Wang, C. et al. Safflower yellow alleviates osteoarthritis and prevents inflammation by inhibiting PGE2 release and regulating NF-kappaB/SIRT1/AMPK signaling pathways. Phytomedicine 78, 153305 (2020).
Dai, G. et al. LncRNA H19 regulates BMP2-induced hypertrophic differentiation of mesenchymal stem cells by promoting Runx2 phosphorylation. Front. Cell Dev. Biol. 8, 580 (2020).
Xiao, H. et al. BMP9 exhibits dual and coupled roles in inducing osteogenic and angiogenic differentiation of mesenchymal stem cells (MSCs). Biosci. Rep. 40, BSR20201262 (2020).
Liao, J. et al. lncRNA H19 mediates BMP9-induced osteogenic differentiation of mesenchymal stem cells (MSCs) through Notch signaling. Oncotarget 8, 53581–53601 (2017).
Lomenick, B., Jung, G., Wohlschlegel, J. A. & Huang, J. Target identification using drug affinity responsive target stability (DARTS). Curr. Protoc. Chem. Biol. 3, 163–180 (2011).
Cui, Z. et al. Halofuginone attenuates osteoarthritis by inhibition of TGF-beta activity and H-type vessel formation in subchondral bone. Ann. Rheum. Dis. 75, 1714–1721 (2016).
Jin, S. et al. Recent advances in PLGA-based biomaterials for bone tissue regeneration. Acta Biomater. 127, 56–79 (2021).
Cheng, G. et al. Controlled co-delivery of growth factors through layer-by-layer assembly of core-shell nanofibers for improving bone regeneration. ACS Nano 13, 6372–6382 (2019).
Liao, J. et al. Sox9 potentiates BMP2-induced chondrogenic differentiation and inhibits BMP2-induced osteogenic differentiation. PLoS ONE 9, e89025 (2014).
Gerwin, N., Bendele, A. M., Glasson, S. & Carlson, C. S. The OARSI histopathology initiative – recommendations for histological assessments of osteoarthritis in the rat. Osteoarthritis Cartilage 18, S24–S34 (2010).
Lin, X. et al. The E3 ubiquitin ligase Itch limits the progression of post-traumatic osteoarthritis in mice by inhibiting macrophage polarization. Osteoarthritis Cartilage 29, 1225–1236 (2021).
Lazic, S. E., Clarke-Williams, C. J. & Munafo, M. R. What exactly is ‘N’ in cell culture and animal experiments? PLoS Biol. 16, e2005282 (2018).
Global Burden of Disease Study 2013 Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 386, 743–800 (2015).
Drugs for osteoarthritis. JAMA 325, 581–582 (2021).
Kovaleva, V. & Saarma, M. Endoplasmic reticulum stress regulators: new drug targets for Parkinson’s disease. J. Parkinsons Dis. 11, S219–S228 (2021).
Mustapha, S. et al. Potential roles of endoplasmic reticulum stress and cellular proteins implicated in diabesity. Oxid. Med. Cell. Longev. 2021, 8830880 (2021).
Peng, M., Chen, F., Wu, Z. & Shen, J. Endoplasmic reticulum stress, a target for drug design and drug resistance in parasitosis. Front. Microbiol. 12, 670874 (2021).
Hu, P., Han, Z., Couvillon, A. D., Kaufman, R. J. & Exton, J. H. Autocrine tumor necrosis factor alpha links endoplasmic reticulum stress to the membrane death receptor pathway through IRE1alpha-mediated NF-kappaB activation and down-regulation of TRAF2 expression. Mol. Cell Biol. 26, 3071–3084 (2006).
Hetz, C., Zhang, K. & Kaufman, R. J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 21, 421–438 (2020).
Marciniak, S. J., Chambers, J. E. & Ron, D. Pharmacological targeting of endoplasmic reticulum stress in disease. Nat. Rev. Drug Discov. 21, 115–140 (2022).
Sepulveda, D. et al. Interactome screening identifies the ER luminal chaperone Hsp47 as a regulator of the unfolded protein response transducer IRE1alpha. Mol. Cell 69, 238–252.e237 (2018).
Huang, R. et al. IRE1 signaling regulates chondrocyte apoptosis and death fate in the osteoarthritis. J. Cell. Physiol. 237, 118–127 (2021).
Hecht, J. T. et al. Primary osteoarthritis early joint degeneration induced by endoplasmic reticulum stress is mitigated by resveratrol. Am. J. Pathol. 91, 1624–1637 (2021).
Gualeni, B. et al. A novel transgenic mouse model of growth plate dysplasia reveals that decreased chondrocyte proliferation due to chronic ER stress is a key factor in reduced bone growth. Dis. Model Mech. 6, 1414–1425 (2013).
Kung, L. H., Rajpar, M. H., Preziosi, R., Briggs, M. D. & Boot-Handford, R. P. Increased classical endoplasmic reticulum stress is sufficient to reduce chondrocyte proliferation rate in the growth plate and decrease bone growth. PLoS ONE 10, e0117016 (2015).
Kung, L. H., Rajpar, M. H., Briggs, M. D. & Boot-Handford, R. P. Hypertrophic chondrocytes have a limited capacity to cope with increases in endoplasmic reticulum stress without triggering the unfolded protein response. J. Histochem. Cytochem. 60, 734–748 (2012).
Rajpar, M. H. et al. Targeted induction of endoplasmic reticulum stress induces cartilage pathology. PLoS Genet. 5, e1000691 (2009).
Briggs, M. D., Bell, P. A. & Pirog, K. A. The utility of mouse models to provide information regarding the pathomolecular mechanisms in human genetic skeletal diseases: the emerging role of endoplasmic reticulum stress (Review). Int. J. Mol. Med. 35, 1483–1492 (2015).
Hughes, A., Oxford, A. E., Tawara, K., Jorcyk, C. L. & Oxford, J. T. Endoplasmic reticulum stress and unfolded protein response in cartilage pathophysiology; contributing factors to apoptosis and osteoarthritis. Int. J. Mol. Sci. 18, 665 (2017).
Rellmann, Y., Eidhof, E. & Dreier, R. Review: ER stress-induced cell death in osteoarthritic cartilage. Cell. Signal. 78, 109880 (2021).
de Seny, D. et al. Proteins involved in the endoplasmic reticulum stress are modulated in synovitis of osteoarthritis, chronic pyrophosphate arthropathy and rheumatoid arthritis, and correlate with the histological inflammatory score. Sci. Rep. 10, 14159 (2020).
Kusaczuk, M., Naumowicz, M., Kretowski, R., Cukierman, B. & Cechowska-Pasko, M. Molecular and cellular effects of chemical chaperone-TUDCA on ER-stressed NHAC-kn human articular chondrocytes cultured in normoxic and hypoxic conditions. Molecules 26, 878 (2021).
Kim, T. J. et al. Valdecoxib improves lipid-induced skeletal muscle insulin resistance via simultaneous suppression of inflammation and endoplasmic reticulum stress. Biochem. Pharmacol. 188, 114557 (2021).
Xie, C. L. et al. Vitexin alleviates ER-stress-activated apoptosis and the related inflammation in chondrocytes and inhibits the degeneration of cartilage in rats. Food Funct. 9, 5740–5749 (2018).
Li, K. et al. DEPTOR prevents osteoarthritis development via interplay with TRC8 to reduce endoplasmic reticulum stress in chondrocytes. J. Bone Miner. Res. 36, 400–411 (2021).
de Seabra Rodrigues Dias, I. R. et al. Potential therapeutic compounds from traditional Chinese medicine targeting endoplasmic reticulum stress to alleviate rheumatoid arthritis. Pharmacol. Res. 170, 105696 (2021).
Ye, L. et al. Endoplasmic reticulum stress is involved in ventilator-induced lung injury in mice via the IRE1alpha-TRAF2-NF-kappaB pathway. Int. Immunopharmacol. 78, 106069 (2020).
Ghosh, S. & Hayden, M. S. New regulators of NF-kappaB in inflammation. Nat. Rev. Immunol. 8, 837–848 (2008).
Madhavan, A. et al. Pharmacologic IRE1/XBP1s activation promotes systemic adaptive remodeling in obesity. Nat. Commun. 13, 608 (2022).
Hunter, D. J. & Bierma-Zeinstra, S. Osteoarthritis. Lancet 393, 1745–1759 (2019).
Sharma, L. Osteoarthritis of the Knee. N. Engl. J. Med. 384, 51–59 (2021).
Berenbaum, F. & Walker, C. Osteoarthritis and inflammation: a serious disease with overlapping phenotypic patterns. Postgrad. Med. 132, 377–384 (2020).
Breedveld, F. C. Osteoarthritis–the impact of a serious disease. Rheumatology 43, i4–i8 (2004).
Acknowledgements
We are grateful for the support of the Key Laboratory of Biorheological Science and Technology, College of Bioengineering, Chongqing University. The authors would like to thank Dr. Tingting Peng from the Chongqing BI Academy for technical and scientific support. The authors are also grateful to T.-C. He and Annie Wang of The University of Chicago Medical Center for critical reading of the manuscript. The reported work was supported by the National Natural Science Foundation of China (NSFC) (#81972069 and #82002312). This project was also supported by the Innovation Project from Chongqing Municipal Education Commission (#CYB21169), the Science and Technology Research Program of Chongqing Education Commission (#KJQN202100431 and #KJZD-M202100401), the Cultivating Program and Candidate of Tip-Top Talent of The First Affiliated Hospital of Chongqing Medical University (#2018PYJJ-11 and # BJRC2021-04), and the Cultivating Program of Postdoctoral Research of The First Affiliated Hospital of Chongqing Medical University (CYYY-BSHPYXM-202202). J.Y. was supported by a postdoctoral fellowship from Chongqing Medical University. Funding sources were not involved in designing the study; in collecting, analyzing and interpreting data; in writing the report; or in the decision to submit the paper for publication.
Author information
Authors and Affiliations
Contributions
Conception and design: W.H. and J.L.; analysis and interpretation of the data: Z.Z., S.G., C.C., W.X., P.X., C.D., B.C., and Y.G.; drafting of the article: J.L. and Z.Z.; critical revision of the article for important intellectual content: J.L. and W.H.; provision of study materials: J.L., C.W. and W.H.; statistical expertize: Z.Z.; funding: J.L., Z.Z., and W.H.; Administrative, technical, or logistic support: J.L. and W.H.; collection and assembly of data: Z.Z., J.L., Z.C., and C.W.; final approval of the article: all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhu, Z., Gao, S., Chen, C. et al. The natural product salicin alleviates osteoarthritis progression by binding to IRE1α and inhibiting endoplasmic reticulum stress through the IRE1α-IκBα-p65 signaling pathway. Exp Mol Med 54, 1927–1939 (2022). https://doi.org/10.1038/s12276-022-00879-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s12276-022-00879-w
- Springer Nature Limited
This article is cited by
-
Endoplasmic reticulum stress: a novel targeted approach to repair bone defects by regulating osteogenesis and angiogenesis
Journal of Translational Medicine (2023)