Abstract
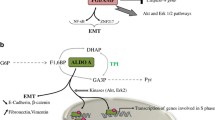
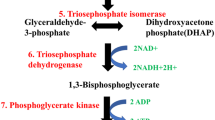
Cancer cells reprogram their metabolism to meet the requirement for survival and rapid growth. One hallmark of cancer metabolism is elevated aerobic glycolysis and reduced oxidative phosphorylation. Emerging evidence showed that most glycolytic enzymes are deregulated in cancer cells and play important roles in tumorigenesis. Recent studies revealed that all essential glycolytic enzymes can be translocated into nucleus where they participate in tumor progression independent of their canonical metabolic roles. These noncanonical functions include anti-apoptosis, regulation of epigenetic modifications, modulation of transcription factors and co-factors, extracellular cytokine, protein kinase activity and mTORC1 signaling pathway, suggesting that these multifaceted glycolytic enzymes not only function in canonical metabolism but also directly link metabolism to epigenetic and transcription programs implicated in tumorigenesis. These findings underscore our understanding about how tumor cells adapt to nutrient and fuel availability in the environment and most importantly, provide insights into development of cancer therapy.



Similar content being viewed by others
References
Vander Heiden MG, Cantley LC, Thompson CB . Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009; 324: 1029–1033.
Dang CV, Semenza GL . Oncogenic alterations of metabolism. Trends Biochem Sci 1999; 24: 68–72.
Kennedy KM, Scarbrough PM, Ribeiro A, Richardson R, Yuan H, Sonveaux P et al. Catabolism of exogenous lactate reveals it as a legitimate metabolic substrate in breast cancer. PloS One 2013; 8: e75154.
Suda T, Takubo K, Semenza GL . Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell 2011; 9: 298–310.
Amelio I, Cutruzzola F, Antonov A, Agostini M, Melino G . Serine and glycine metabolism in cancer. Trends Biochem Sci 2014; 39: 191–198.
Stincone A, Prigione A, Cramer T, Wamelink MM, Campbell K, Cheung E et al. The return of metabolism: biochemistry and physiology of the pentose phosphate pathway. Biol Rev Camb Philos Soc 2015; 90: 927–963.
Lincet H, Icard P . How do glycolytic enzymes favour cancer cell proliferation by nonmetabolic functions? Oncogene 2015; 34: 3751–3759.
Patra KC, Wang Q, Bhaskar PT, Miller L, Wang Z, Wheaton W et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell 2013; 24: 213–228.
Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA 3rd et al. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 2012; 337: 975–980.
Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 2011; 334: 1278–1283.
Gruning NM, Rinnerthaler M, Bluemlein K, Mulleder M, Wamelink MM, Lehrach H et al. Pyruvate kinase triggers a metabolic feedback loop that controls redox metabolism in respiring cells. Cell Metab 2011; 14: 415–427.
Li S, Swanson SK, Gogol M, Florens L, Washburn MP, Workman JL et al. Serine and SAM Responsive Complex SESAME Regulates Histone Modification Crosstalk by Sensing Cellular Metabolism. Mol Cell 2015; 60: 408–421.
Kim JW, Dang CV . Multifaceted roles of glycolytic enzymes. Trends Biochem Sci 2005; 30: 142–150.
Pastorino JG, Hoek JB . Hexokinase II: the integration of energy metabolism and control of apoptosis. Curr Med Chem 2003; 10: 1535–1551.
Haga A, Funasaka T, Niinaka Y, Raz A, Nagase H . Autocrine motility factor signaling induces tumor apoptotic resistance by regulations Apaf-1 and Caspase-9 apoptosome expression. Int J Cancer 2003; 107: 707–714.
Colell A, Ricci JE, Tait S, Milasta S, Maurer U, Bouchier-Hayes L et al. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell 2007; 129: 983–997.
Tarze A, Deniaud A, Le Bras M, Maillier E, Molle D, Larochette N et al. GAPDH, a novel regulator of the pro-apoptotic mitochondrial membrane permeabilization. Oncogene 2007; 26: 2606–2620.
Fan J, Krautkramer KA, Feldman JL, Denu JM . Metabolic regulation of histone post-translational modifications. ACS Chem Biol 2015; 10: 95–108.
Lu C, Thompson CB . Metabolic regulation of epigenetics. Cell Metab 2012; 16: 9–17.
Ryall JG, Cliff T, Dalton S, Sartorelli V . Metabolic Reprogramming of Stem Cell Epigenetics. Cell Stem Cell 2015; 17: 651–662.
Li S, Kong L, Yu X, Zheng Y . Host-virus interactions: from the perspectives of epigenetics. Rev Med Virol 2014; 24: 223–241.
Boukouris AE, Zervopoulos SD, Michelakis ED . Metabolic Enzymes Moonlighting in the Nucleus: Metabolic Regulation of Gene Transcription. Trends Biochem Sci 2016; 41: 712–730.
Huang X, Holden HM, Raushel FM . Channeling of substrates and intermediates in enzyme-catalyzed reactions. Ann Rev Biochem 2001; 70: 149–180.
Matsuda S, Adachi J, Ihara M, Tanuma N, Shima H, Kakizuka A et al. Nuclear pyruvate kinase M2 complex serves as a transcriptional coactivator of arylhydrocarbon receptor. Nucleic Acids Res 2016; 44: 636–647.
Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson TH, Haromy A et al. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell 2014; 158: 84–97.
Ventura M, Mateo F, Serratosa J, Salaet I, Carujo S, Bachs O et al. Nuclear translocation of glyceraldehyde-3-phosphate dehydrogenase is regulated by acetylation. Int J Biochem Cell Biol 2010; 42: 1672–1680.
Berger F, Lau C, Dahlmann M, Ziegler M . Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J Biol Chem 2005; 280: 36334–36341.
Castonguay Z, Auger C, Thomas SC, Chahma M, Appanna VD . Nuclear lactate dehydrogenase modulates histone modification in human hepatocytes. Biochem Biophys Res Commun 2014; 454: 172–177.
Zheng L, Roeder RG, Luo Y . S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 2003; 114: 255–266.
Katoh Y, Ikura T, Hoshikawa Y, Tashiro S, Ito T, Ohta M et al. Methionine adenosyltransferase II serves as a transcriptional corepressor of Maf oncoprotein. Mol Cell 2011; 41: 554–566.
Kera Y, Katoh Y, Ohta M, Matsumoto M, Takano-Yamamoto T, Igarashi K . Methionine adenosyltransferase II-dependent histone H3K9 methylation at the COX-2 gene locus. J Biol Chem 2013; 288: 13592–13601.
Li S, Shogren-Knaak MA . Cross-talk between histone H3 tails produces cooperative nucleosome acetylation. Proc Natl Acad Sci USA 2008; 105: 18243–18248.
Li S, Shogren-Knaak MA . The Gcn5 bromodomain of the SAGA complex facilitates cooperative and cross-tail acetylation of nucleosomes. J Biol Chem 2009; 284: 9411–9417.
Seligson DB, Horvath S, McBrian MA, Mah V, Yu H, Tze S et al. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol 2009; 174: 1619–1628.
Friis RM, Wu BP, Reinke SN, Hockman DJ, Sykes BD, Schultz MC . A glycolytic burst drives glucose induction of global histone acetylation by picNuA4 and SAGA. Nucleic Acids Res 2009; 37: 3969–3980.
McBrian MA, Behbahan IS, Ferrari R, Su T, Huang TW, Li K et al. Histone acetylation regulates intracellular pH. Mol Cell 2013; 49: 310–321.
Liu XS, Little JB, Yuan ZM . Glycolytic metabolism influences global chromatin structure. Oncotarget 2015; 6: 4214–4225.
Cai L, Sutter BM, Li B, Tu BP . Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell 2011; 42: 426–437.
Thangaraju M, Gopal E, Martin PM, Ananth S, Smith SB, Prasad PD et al. SLC5A8 triggers tumor cell apoptosis through pyruvate-dependent inhibition of histone deacetylases. Cancer Res 2006; 66: 11560–11564.
Latham T, Mackay L, Sproul D, Karim M, Culley J, Harrison DJ et al. Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucleic Acids Res 2012; 40: 4794–4803.
Su X, Wellen KE, Rabinowitz JD . Metabolic control of methylation and acetylation. Curr Opin Chem Biol 2016; 30: 52–60.
Ringel AE, Ryznar R, Picariello H, Huang KL, Lazarus AG, Holmes SG . Yeast Tdh3 (glyceraldehyde 3-phosphate dehydrogenase) is a Sir2-interacting factor that regulates transcriptional silencing and rDNA recombination. PLoS Genet 2013; 9: e1003871.
Xu YF, Zhao X, Glass DS, Absalan F, Perlman DH, Broach JR et al. Regulation of yeast pyruvate kinase by ultrasensitive allostery independent of phosphorylation. Mol Cell 2012; 48: 52–62.
Wilson MJ, Shivapurkar N, Poirier LA . Hypomethylation of hepatic nuclear DNA in rats fed with a carcinogenic methyl-deficient diet. Biochem J 1984; 218: 987–990.
Tanaka Y, Yano H, Ogasawara S, Yoshioka S, Imamura H, Okamoto K et al. Mild Glucose Starvation Induces KDM2A-Mediated H3K36me2 Demethylation through AMPK To Reduce rRNA Transcription and Cell Proliferation. Mol Cell Biol 2015; 35: 4170–4184.
Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB . Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 2015; 518: 413–416.
Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell 2012; 150: 685–696.
Goel A, Mathupala SP, Pedersen PL . Glucose metabolism in cancer. Evidence that demethylation events play a role in activating type II hexokinase gene expression. J Biol Chem 2003; 278: 15333–15340.
Johnson C, Warmoes MO, Shen X, Locasale JW . Epigenetics and cancer metabolism. Cancer Lett 2015; 356: 309–314.
Galdieri L, Zhang T, Rogerson D, Vancura A . Reduced histone expression or a defect in chromatin assembly induces respiration. Mol Cell Biol 2016; 36: 1064–1077.
Mehrotra S, Galdieri L, Zhang T, Zhang M, Pemberton LF, Vancura A . Histone hypoacetylation-activated genes are repressed by acetyl-CoA- and chromatin-mediated mechanism. Biochim Biophys Acta 2014; 1839: 751–763.
Keller KE, Doctor ZM, Dwyer ZW, Lee YS . SAICAR induces protein kinase activity of PKM2 that is necessary for sustained proliferative signaling of cancer cells. Mol Cell 2014; 53: 700–709.
Gao X, Wang H, Yang JJ, Liu X, Liu ZR . Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell 2012; 45: 598–609.
Feo S, Arcuri D, Piddini E, Passantino R, Giallongo A . ENO1 gene product binds to the c-myc promoter and acts as a transcriptional repressor: relationship with Myc promoter-binding protein 1 (MBP-1). FEBS Lett 2000; 473: 47–52.
Lee J, Kim HK, Han YM, Kim J . Pyruvate kinase isozyme type M2 (PKM2) interacts and cooperates with Oct-4 in regulating transcription. Int J Biochem Cell Biol 2008; 40: 1043–1054.
Luo W, Hu H, Chang R, Zhong J, Knabel M, O’Meally R et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 2011; 145: 732–744.
Enzo E, Santinon G, Pocaterra A, Aragona M, Bresolin S, Forcato M et al. Aerobic glycolysis tunes YAP/TAZ transcriptional activity. EMBO J 2015; 34: 1349–1370.
Lucarelli G, Rutigliano M, Sanguedolce F, Galleggiante V, Giglio A, Cagiano S et al. Increased expression of the autocrine motility factor is associated with poor prognosis in patients with clear cell-renal cell carcinoma. Medicine 2015; 94: e2117.
Fairbank M, St-Pierre P, Nabi IR . The complex biology of autocrine motility factor/phosphoglucose isomerase (AMF/PGI) and its receptor, the gp78/AMFR E3 ubiquitin ligase. Mol Biosyst 2009; 5: 793–801.
Araki K, Shimura T, Yajima T, Tsutsumi S, Suzuki H, Okada K et al. Phosphoglucose isomerase/autocrine motility factor promotes melanoma cell migration through ERK activation dependent on autocrine production of interleukin-8. J Biol Chem 2009; 284: 32305–32311.
Niinaka Y, Harada K, Fujimuro M, Oda M, Haga A, Hosoki M et al. Silencing of autocrine motility factor induces mesenchymal-to-epithelial transition and suppression of osteosarcoma pulmonary metastasis. Cancer Res 2010; 70: 9483–9493.
Kho DH, Zhang T, Balan V, Wang Y, Ha SW, Xie Y et al. Autocrine motility factor modulates EGF-mediated invasion signaling. Cancer Res 2014; 74: 2229–2237.
Kho DH, Nangia-Makker P, Balan V, Hogan V, Tait L, Wang Y et al. Autocrine motility factor promotes HER2 cleavage and signaling in breast cancer cells. Cancer Res 2013; 73: 1411–1419.
Fu M, Li L, Albrecht T, Johnson JD, Kojic LD, Nabi IR . Autocrine motility factor/phosphoglucose isomerase regulates ER stress and cell death through control of ER calcium release. Cell Death Differ 2011; 18: 1057–1070.
Sun S, Liang X, Zhang X, Liu T, Shi Q, Song Y et al. Phosphoglycerate kinase-1 is a predictor of poor survival and a novel prognostic biomarker of chemoresistance to paclitaxel treatment in breast cancer. Br J Cancer 2015; 112: 1332–1339.
Li X, Jiang Y, Meisenhelder J, Yang W, Hawke DH, Zheng Y et al. Mitochondria-translocated PGK1 functions as a protein kinase to coordinate glycolysis and the TCA cycle in tumorigenesis. Mol Cell 2016; 61: 705–719.
He CL, Bian YY, Xue Y, Liu ZX, Zhou KQ, Yao CF et al. Pyruvate kinase M2 Activates mTORC1 by phosphorylating AKT1S1. Sci Rep 2016; 6: 21524.
Kong L, Li S, Yu X, Fang X, Xu A, Huang M et al. Hepatitis C virus and its protein NS4B activate the cancer-related STAT3 pathway via the endoplasmic reticulum overload response. Arch Virol 2016; 161: 2149–2159.
Adams V, Griffin LD, Gelb BD, McCabe ER . Protein kinase activity of rat brain hexokinase. Biochem Biophys Res Commun 1991; 177: 1101–1106.
Faivre S, Kroemer G, Raymond E . Current development of mTOR inhibitors as anticancer agents. Nat Rev Drug Discov 2006; 5: 671–688.
Lee MN, Ha SH, Kim J, Koh A, Lee CS, Kim JH et al. Glycolytic flux signals to mTOR through glyceraldehyde-3-phosphate dehydrogenase-mediated regulation of Rheb. Mol Cell Biol 2009; 29: 3991–4001.
Zhang JY, Zhang F, Hong CQ, Giuliano AE, Cui XJ, Zhou GJ et al. Critical protein GAPDH and its regulatory mechanisms in cancer cells. Cancer Biol Med 2015; 12: 10–22.
Zhao Y, Butler EB, Tan M . Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis 2013; 4: e532.
Gong L, Wei Y, Yu X, Peng J, Leng X . 3-Bromopyruvic acid, a hexokinase II inhibitor, is an effective antitumor agent on the hepatoma cells: in vitro and in vivo findings. Anti-Cancer Agents Med Chem 2014; 14: 771–776.
Ros S, Schulze A . Balancing glycolytic flux: the role of 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatases in cancer metabolism. Cancer Metab 2013; 1: 8.
Doherty JR, Cleveland JL . Targeting lactate metabolism for cancer therapeutics. J Clin Invest 2013; 123: 3685–3692.
Khalid MH, Shibata S, Hiura T . Effects of clotrimazole on the growth, morphological characteristics, and cisplatin sensitivity of human glioblastoma cells in vitro. J Neurosurg 1999; 90: 918–927.
Penso J, Beitner R . Clotrimazole and bifonazole detach hexokinase from mitochondria of melanoma cells. Eur J Pharmacol 1998; 342: 113–117.
Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol 2012; 8: 839–847.
Choi SY, Xue H, Wu R, Fazli L, Lin D, Collins CC et al. The MCT4 gene: a novel, potential target for therapy of advanced prostate cancer. Clin. Cancer Res. 2016; 22: 2721–2733.
Olson KA, Schell JC, Rutter J . Pyruvate and metabolic flexibility: illuminating a path toward selective cancer therapies. Trends Biochem Sci 2016; 41: 219–230.
Gancedo JM . Yeast carbon catabolite repression. Microbiol Mol Biol Rev 1998; 62: 334–361.
Popanda O, Fox G, Thielmann HW . Modulation of DNA polymerases alpha, delta and epsilon by lactate dehydrogenase and 3-phosphoglycerate kinase. Biochim Biophys Acta 1998; 1397: 102–117.
Jiang Y, Li X, Yang W, Hawke DH, Zheng Y, Xia Y et al. PKM2 regulates chromosome segregation and mitosis progression of tumor cells. Mol Cell 2014; 53: 75–87.
Grosse F, Nasheuer HP, Scholtissek S, Schomburg U . Lactate dehydrogenase and glyceraldehyde-phosphate dehydrogenase are single-stranded DNA-binding proteins that affect the DNA-polymerase-alpha-primase complex. Eur J Biochem/FEBS 1986; 160: 459–467.
Acknowledgements
We apologize to colleagues whose work cannot be cited here because of space limitation. We thank members of Li laboratory for critical reading of this manuscript. This work was funded by grants from National Nature Science Foundation of China (No. 31671335 to S Li; No. 31600046 to X Yu).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Yu, X., Li, S. Non-metabolic functions of glycolytic enzymes in tumorigenesis. Oncogene 36, 2629–2636 (2017). https://doi.org/10.1038/onc.2016.410
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.410
- Springer Nature Limited
This article is cited by
-
Tyrosine hydroxylase inhibits HCC progression by downregulating TGFβ/Smad signaling
European Journal of Medical Research (2024)
-
Nuclear GAPDH in cortical microglia mediates cellular stress-induced cognitive inflexibility
Molecular Psychiatry (2024)
-
SHMT2 regulates esophageal cancer cell progression and immune Escape by mediating m6A modification of c-myc
Cell & Bioscience (2023)
-
Glc7/PP1 dephosphorylates histone H3T11 to regulate autophagy and telomere silencing in response to nutrient availability
Cell Discovery (2023)
-
Identification of ENO1 as a prognostic biomarker and molecular target among ENOs in bladder cancer
Journal of Translational Medicine (2022)