Abstract
Malignant peripheral nerve sheath tumors (MPNSTs) are aggressive, frequently metastatic sarcomas that are associated with neurofibromatosis type 1 (NF1), a prominent inherited genetic disease in humans. Although loss of the NF1 gene predisposes to MPNST induction, relatively long tumor latency in NF1 patients suggests that additional genetic or epigenetic abnormalities are needed for the development of these nerve sheath malignancies. To study the molecular pathways contributing to the formation of MPNSTs in NF1 patients, we used a zebrafish tumor model defined by nf1 loss in a p53-deficient background together with the overexpression of either wild-type or constitutively activated PDGFRA (platelet-derived growth factor receptor-α) under control of the sox10 neural crest-specific promoter. Here we demonstrate the accelerated onset and increased penetrance of MPNST formation in fish overexpressing both the wild-type and the mutant PDGFRA transgenes in cells of neural crest origin. Interestingly, overexpression of the wild-type PDGFRA was even more potent in promoting transformation than the mutant PDGFRA, which is important because ~78% of human MPNSTs have expression of wild-type PDGFRA, whereas only 5% harbor activating mutations of the gene encoding this receptor. Further analysis revealed the induction of cellular senescence in zebrafish embryos overexpressing mutant, but not wild-type, PDGFRA, suggesting a mechanism through which the oncogenic activity of the mutant receptor is tempered by the activation of premature cellular senescence in an NF1-deficient background. Taken together, our study suggests a model in which overexpression of wild-type PDGFRA associated with NF1 deficiency leads to aberrant activation of downstream RAS signaling and thus contributes importantly to MPNST development—a prediction supported by the ability of the kinase inhibitor sunitinib alone and in combination with the MEK inhibitor trametinib to retard MPNST progression in transgenic fish overexpressing the wild-type receptor.
Similar content being viewed by others
Introduction
Malignant peripheral nerve sheath tumors (MPNSTs) are aggressive and often metastatic soft-tissue sarcomas that occur most frequently in children and young adults with neurofibromatosis type 1 (NF1).1 Indeed, about half of all MPNSTs are diagnosed in NF1 patients, and MPNSTs represent one of the most common causes of death in patients who are haploinsufficient for this gene.2 These nerve sheath tumors are thought to arise from cells derived from the neural crest, including progenitors of Schwann cells and perineural cells, which have somatically lost the function of the remaining NF1 allele.3 The prognosis of patients with MPNSTs is very poor, with a high recurrence rate after surgical resection (~40%).4, 5 Currently, complete surgical excision is the only curative therapy for these tumors, but unfortunately they are often not completely resectable. The available chemotherapy regimens are for the most part ineffective, and are often associated with significant toxicity that can severely reduce quality of life. Therefore, improved understanding of MPNST pathogenesis and the identification of new molecular pathways on which these tumors are dependent is urgently needed to improve treatment outcome in NF1 patients who develop malignant nerve sheath tumors. Hopefully, new insights into pathogenesis will implicate potential therapeutic targets for the development of small-molecule drugs with more potent and specific antitumor activity.
Patients with NF1 have only one functional copy of the NF1 gene, which encodes neurofibromin, a protein of more than 2800 amino acids.6 Neurofibromin contains a small region of homology to the RAS GAP protein and is capable of downregulating RAS activity by accelerating the hydrolysis of GTP-bound RAS to its inactive GDP-bound form.7 Thus, when the remaining functional allele of NF1 is lost somatically in nonmyelinating Schwann cell progenitors, the NF1-null progeny acquire aberrant growth properties leading to benign neurofibromas.8, 9 These benign neurofibromas can occur in association with cutaneous nerves in the form of dermal neurofibromas, often causing cosmetic disfiguration. Plexiform neurofibromas are a more serious second type of neurofibroma that form in association with more internal nerve bundles. In this type of neurofibroma, other cell types are recruited, including fibroblasts, perineural cells, endothelial cells and mast cells. Plexiform neurofibromas invade soft-tissue structures and bone, often causing significant functional impairment depending on their location.10 Even though plexiform neurofibromas are benign tumors, they are often unresectable because they involve multiple layers of tissues with essential functions. About 10% of plexiform neurofibromas undergo malignant transformation into MPNSTs,11 often arising from cells with the additional acquired loss of the CDKN2A or TP53 tumor suppressor genes.12
The absence or latency of MPNST development in many NF1 patients suggests that additional gene mutations are needed to induce tumorigenesis. One promising candidate is platelet-derived growth factor receptor-α (PDGFRA), a receptor tyrosine kinase (RTK). Overexpression and chromosomal amplification of the PDGFRA gene have been reported in MPNSTs,13, 14 and this gene was one of the most overexpressed in an analysis of paired benign plexiform neurofibromas and MPNSTs,15 suggesting that this RTK may be upregulated as part of the benign-to-malignant tumor transition. Moreover, PDGFRA is frequently active and phosphorylated in MPNSTs from patients.16, 17 Both PDGFRA and its ligand, PDGF-A, are overexpressed together in human MPNST cells,16 suggesting that PDGF-A ligand acts in an autocrine manner in human MPNSTs to support growth. Paracrine stimulation also supports MPNST growth because Schwann cells derived from nf1−/− mice express growth factor ligands such as PDGF-B, unlike those derived from nf1+/+ mice.18 Overall, both autocrine and paracrine activation of the PDGF receptor seem to be responsible for accelerating human MPNST formation. Taken together, these findings justify a more comprehensive investigation of the role of PDGFRA in the molecular pathogenesis of MPNSTs in patients with loss of the NF1 gene. This imperative gains impetus from reports that RTK inhibitors targeting PDGFRA decrease the growth and invasion rates of MPNST cell lines.16, 19
We previously reported that zebrafish with loss-of-function mutations of p53 develop MPNST,20 reiterating the observation of p53 loss in human MPNST.21 Thus, to discover the molecular pathways contributing to the formation of MPNSTs in NF1 patients, we generated an NF1 mutant zebrafish model and used it to demonstrate that a combination of nf1 loss and p53 deficiency potentiates the penetrance and onset of MPNSTs.22 To better understand the molecular and cellular determinants of NF1-associated MPNST pathogenesis in vivo, we established stable transgenic lines with overexpression of both wild-type and constitutively active mutant of human PDGFRA, using the MPNST-prone zebrafish line with loss of both p53 and nf1. We report here that MPNST onset is accelerated by PDGFRA overexpression. Interestingly, overexpression of wild-type PDGFRA was more active in accelerating MPNST initiation than was mutationally activated PDGFRA in our transgenic zebrafish model, possibly because mutationally active PDGFRA induces senescence in nf1/p53 compound mutant cells. Thus, our study implicates overexpression of wild-type PDGFRA as a key driver in MPNST, which is important because wild-type PDGFRA is frequently documented in human MPNST, whereas mutational activation of the gene is rare. This is a key consideration when implementing targeted therapy, because it indicates that PDGFRA inhibitors are likely to have antiproliferative activity in the large subset of MPNSTs with overexpression of wild-type PDGFRA.
Results
I-SceI meganuclease-mediated PDGFRA transgenesis in nf1- and p53-deficient fish
To understand the role of PDGFRA in MPNST pathogenesis, we cloned the cDNA encoding either wild-type or constitutively activated human PDGFRA (exons 8 and 9 deleted)23 under control of the zebrafish sox10 neural crest-specific promoter (Figure 1a). A sox10:mCherry cDNA construct was coinjected with each sox10:PDGFR construct into nf1a+/−; nf1b−/−; p53m/m zebrafish, based on studies showing that three separate transgene constructs will cointegrate into the host genome after coinjection into the one-cell-stage zebrafish embryo.24 Hence, the expression of mCherry serves as a marker for the coexpression of PDGFRA in tissues of the mosaic primary injected animals.
I-SceI meganuclease-mediated human PDGFRA transgenesis in nf1- and p53-deficient fish. (a) Schematic diagram of the DNA constructs used to generate transgenic sox10:mCherry, sox10:wild-type (WT) and constitutively activated (Mut) PDGFRAs zebrafish. I-SceI denotes the I-SceI meganuclease target sequence. (b) Schematic diagram of PCR target of human wild type and constitutively activated mutant PDGFRA (Δ exons 8 and 9) for genotyping. Black arrows represent primer target sites for PCR. (c) PDGFRA wild-type and mutant DNA sequences were detected in genomic DNA of the transgenic zebrafish embryos. Human PDGFRA sequences were confirmed with embryonic DNA of two separate transgenic lines (mC1 and mC2, PW1 and PW2 and PM1 and PM2). Each amplified PCR band of PDGFRA wild type and mutant had sizes of 740 and 497 bp, respectively. Injected plasmids for transgenesis were used as the positive control. (d and e) Sox10 promoter driving mCherry is expressed in cells of neural crest origin during early embryogenesis. Tg (nf1a+/−; nf1b−/−; p53m/m; sox10:mCherry) zebrafish embryo at 3 d.p.f. showed mCherry expression throughout otic vesicle (OV), branchial arches (BA), oligodendrocytes (O), jaw cartilage (JC) and pectoral fins (PF).
At maturity 3 months after injection, the primary injected animals were outbred to establish stable transgenic lines with overexpression of either wild-type or mutant PDGFRA in nf1a+/−; nf1b−/−; p53m/m zebrafish. To confirm the presence of the human PDGFRA gene in zebrafish DNA, we performed PCR with the extracted total DNA from individual embryos of the stable lines, detecting the human wild-type PDGFRA (740 bp) and mutant (497 bp) PCR bands (Figures 1b and c). In the stable transgenic lines, the mCherry fluorescence, driven by the sox10 promoter, is expressed in cells of neural crest origin during early embryogenesis (Figures 1d and e).25
Hyperplasia of oligodendrocyte progenitor cells (OPCs) and decreased lateral stripe melanophores have been reported in our previous study with nf1 mutant zebrafish.22 We examined whether PDGFRA overexpression affects these cells (Supplementary Figure S1). In these results, 6 days post fertilization (d.p.f.) embryos from both wild-type and mutant PDGFRA transgenic lines demonstrated increased numbers of sox10:mCherry-positive OPCs in the spinal cord. Embryos transgenic for mutant PDGFRA had the most marked increase in OPCs, with approximately twice the number of sox10:mCherry-positive OPCs compared with the mCherry control transgenic larvae in the nf1a+/+; nf1b−/−; p53m/m background. Decreased lateral stripe melanophores were identified in our current study in the nf1a+/+; nf1b−/−; p53m/m background, similar to results we reported previously,22 and there was no detectable difference among the mCherry control, wild-type and mutant PDGFRA transgenic zebrafish. We did not observe any additional developmental defects in the PDGFRA transgenic zebrafish embryos.
Both wild-type and constitutively activated mutant forms of PDGFRA accelerate MPNST tumorigenicity in nf1 - and p53-deficient fish
We performed tumor watches using both mosaic primary injected fish and established stable transgenic lines, and found that both wild-type and constitutively activated mutant PDGFRA transgenes accelerated MPNST tumorigenesis and increased tumor penetrance in nf1a+/−; nf1b−/−; p53m/m zebrafish (Figures 2a and b). In primary injectants, both wild-type and mutant PDGFRA-injected zebrafish developed mCherry-positive tumors much earlier and more frequently than in mCherry control fish (Figure 2a). In PDGFRA mutant injectants, tumor onset began at 20 weeks post fertilization (w.p.f.), whereas in those PDGFRA injected with wild-type, they began to form at 25 w.p.f., and mCherry-only control transgenic fish developed tumors from 31 weeks. Interestingly, both wild-type and PDGFRA mutant injectants showed higher penetrance rates than did mCherry control zebrafish (71.4% and 55.6% vs 27.3% at 40 w.p.f). These results in the primary injectants show that both wild-type and constitutively activated PDGFRA overexpression cooperate with loss of nf1 and p53 function to accelerate the onset and increase the penetrance of tumor formation.
Both wild-type and constitutively activated mutant forms of PDGFRA accelerate MPNST tumorigenicity in nf1- and p53-deficient fish. (a and b) Kaplan–Meier analysis of tumorigenesis in fish with either mosaic or stable expression of the PDGFRA transgene. Onset of MPNSTs in nf1- and p53-deficient zebrafish (nf1a+/–; nf1b–/–; p53m/m) injected with the following DNA constructs: (1) sox10:mCherry alone (mCherry, red); (2) sox10:PDGFRA wild-type and sox10:mCherry (PDGFRA wild-type, yellow); or (3) sox10:PDGFRA mutant and sox10:mCherry (PDGFRA mutant, blue). Note that wild-type PDGFRA overexpression accelerated the onset of MPNSTs more rapidly than constitutively activated mutant PDGFRA. (c–h) Representative images of the sox10 promoter driving mCherry-positive tumors under different conditions. Transgenic sox10:mCherry (c and d), sox10:mcherry/PDGFRA wild-type (e and f) and sox10:mCherry/PDGFRA CA mutant (g and h) in the nf1a+/−; nf1b−/−; p53m/m background induced tumors that strongly expressed mCherry protein (>30 w.p.f., scale bar=10 mm). (i) Western blot analysis for PDGFRA in protein lysates prepared from tumors of mCherry control, PDGFRA wild-type and PDGFRA mutant fish. PDGFRA wild-type, mutant and their phosphorylated (active) forms were detected. β-Tubulin was used an internal control for equal loading. Arrow denotes protein size (kDa).
In these stable transgenic lines derived from outbreeding the primary injectants, including Tg(nf1a+/−; nf1b−/−; p53m/m; sox10: wild-type PDGFRA; sox10:mCherry), Tg(nf1a+/−; nf1b−/−; p53m/m; sox10:PDGFRA mutant; sox10:mCherry), and Tg(nf1a+/−; nf1b−/−; p53m/m; sox10:mCherry) also developed tumors (Figure 2b). In the stable fish lines, the tumor onset times and penetrance patterns associated with both the wild-type and mutant forms of PDGFRA were similar to those of the mosaic PDGFRA wild-type and mutant injectants, reflecting a faster growth rate of tumors overexpressing wild-type compared with constitutively activated PDGFRA. Tumors arose at 18 w.p.f. in the wild-type PDGFRA-overexpressing fish, with 89.3% of this group developing MPNSTs by 30 w.p.f. Transgenic PDGFRA mutant zebrafish began to show tumors by 20 w.p.f., with 48.3% developing tumors by 30 w.p.f. This contrasts with the lack of onset of tumors in the mCherry control zebrafish until 24 w.p.f., and a tumor penetrance rate of 22.8% at 30 w.p.f.
These findings reinforce the status of PDGFRA as a key driver of tumorigenesis in MPNST. The faster development of tumors in PDGFRA wild-type overexpressors compared with PDGFRA mutant fish was surprising, likely reflecting optimal PDGFRA tyrosine kinase activity when the wild-type receptor is overexpressed as opposed to constitutive activation of the PDGFRA gene. Apparently, the constitutively activated mutant PDGFRA is so active in an nf1/p53-deficient background that it actually reduces tumor cell growth by inducing cellular senescence (see below and Figures 4d–l). This interpretation also agrees with reports that human MPNST samples often show high levels of PDGFRA expression without any mutation in the PDGFRA coding sequence.16
The sox10 promoter is highly expressed in MPNST,26, 27 and thus transgenic sox10:mCherry (Figures 2c and d), sox10:mcherry/PDGFRA wild-type (Figures 2e and f) and sox10:mcherry/PDGFRA mutant (Figures 2g and h) zebrafish developed tumors with strong mCherry expression. Evaluation of the induced zebrafish MPNSTs for PDGFRA expression demonstrated expression of human PDGFRAs in protein lysates prepared from tumors of fish transgenic for wild-type and mutant PDGFRAs (Figure 2i). Their phosphorylated (active) forms were also detected. Interestingly, a phosphoprotein with the same mobility as human PDGFRA was detected in mCherry control tumor lysates, suggesting that the endogenous zebrafish receptor tyrosine kinase is activated during the molecular pathogenesis of zebrafish MPNST.
PDGFRA-induced zebrafish MPNSTs exhibit similar histology to that of human MPNSTs
To examine the histology of the induced tumors in our transgenic zebrafish, we performed hematoxylin and eosin (H&E) staining. The results were consistent with the diagnostic histologic features of human MPNST: spindle-shaped cells with long serpentine-like nuclei that stack into short fascicles, typically organized into whorls (Figures 3a–c).
PDGFRA-induced zebrafish MPNST tumors exhibit similar histology to that of human MPNSTs. (a–c) The histopathology after H&E staining of the zebrafish MPNSTs was identical in the nf1a+/−; nf1b−/−; p53m/m zebrafish when either PDGFRA wild-type or mutant proteins were overexpressed. In each genotype, the zebrafish MPNST histopathology was very similar to that of human MPNSTs. The zebrafish MPNSTs comprise spindle cells that stack into short fascicles, typically with a whirling organization pattern. MPNST cells have long serpentine-like nuclei and are spindle-shaped. (d–i) Markers of cells of neural crest origin were expressed by the zebrafish MPNST tumors, such as S100 (d–f) and Sox10 (g–i). S100 was detected in the cell membrane and nucleus, whereas Sox10 was detected in the nucleus. Scale bar=200 μm.
We also examined the expression of two neural crest cell lineage markers, S100 and Sox10, in our zebrafish MPNST tumors, as these are used diagnostically to distinguish human MPNST from other types of solid tumors.26, 27 Both S100 (Figures 3d–f) and Sox10 (Figures 3g–i) were expressed by tumor cells in samples of tumors arising in each of our transgenic lines. S100 is localized in the cell membrane and nucleus, whereas Sox10 is only detected in the nucleus. The histopathologic features were identical to those in MPNSTs arising in the nf1a+/−; nf1b−/−; p53m/m line expressing mCherry, and in fish with the same background mutations but expressing the wild-type or mutant PDGFRA transgene.
Additionally, we performed immunohistochemical staining to identify which cells and tissues expressed PDGFRA in adult zebrafish (Supplementary Figure S2). Endogenous PDGFRA was expressed in many adult zebrafish tissues such as brain, gill and oocytes. MPNST tumor cells from sox10:mCherry, wild-type and mutant PDGFRA zebrafish lines each also expressed detectable PDGFRA. The mutant PDGFRA protein is predominately expressed in the cytosol, as noted previously.23
Overexpression of the PDGFRA wild-type gene activates AKT and ERK at levels optimal for tumorigenicity
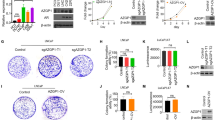
We expected that PDGFRA overexpression and activation of its tyrosine kinase domain would activate phosphatidylinositol-4,5-bisphosphate 3 kinase and Ras downstream signals in neural crest origin cells, and consequently would accelerate MPNST tumorigenicity. To test this prediction, we performed western blot analysis for AKT and ERK1/2 activation in protein lysates prepared from tumors of mCherry control, wild-type PDGFRA and PDGFRA mutant fish. Both AKT and ERK1/2 were phosphorylated in all types of primary MPNST lysates (Figure 4a). However, the phospho-AKT and phospho-ERK1/2 levels in the lysate of the PDGFRA mutant tumor were higher compared with those of the mCherry control and PDGFRA wild-type tumors. The mean values for the phosphorylated (p)-AKT/AKT and p-ERK/ERK ratios in the PDGFRA mutant were 3.68- and 3.10-fold higher, respectively, compared with that for the mCherry control (Figures 4b and c), whereas the phosphorylation levels of AKT and ERK1/2 in the lysates of mCherry and PDGFRA wild-type tumors were similar. Because the PDGFRA mutant tumors grow more slowly than PDGFRA wild-type tumors, our findings suggest that the wild-type PDGFRA may activate Ras downstream signals at levels optimal for MPNST tumorigenicity, whereas the PDGFRA mutant may transmit a stronger than optimal signal.
Overexpression of the PDGFRA wild-type gene activates AKT and ERK at levels optimal for tumorigenicity. (a) Western blot analysis for AKT and ERK1/2 activation in protein lysates prepared from tumors of mCherry control, PDGFRA wild-type, and PDGFRA mutant fish. Similar levels of p-AKT and p-ERK1/2 were detected in PDGFRA wild-type tumors as compared with mCherry control tumors; however, tumors induced by the PDGFRA mutant showed increased levels of p-AKT and p-ERK1/2. Proteins were detected by stripping the membrane and reprobing. Arrows denote protein size (kDa). (b and c) Statistical analysis of mean±s.d. of p-AKT/total AKT and p-ERK/total AKT using ImageJ. Asterisks indicate statistical significance (**P<0.005). (d, g and j) Representative images of SA-β galactosidase-stained transgenic fish embryos of mCherry control (n=12), PDGFRA wild-type (n=36) and PDGFRA mutant (n=32) in the nf1a+/−; nf1b−/−; p53m/m background at 4 d.p.f. The boxed areas were magnified at the right bottom corner of each panel. (e, h and k) Transverse cryosections through the spinal cord of embryos at 4 d.p.f., with mCherry and DAPI (4',6-diamidino-2-phenylindole; f, i and l).
PDGFRA mutant overexpressors showed senescence-associated β-galactosidase activity
We hypothesized that excessive activation of Ras signaling by the mutant PDGFRA receptor might induce senescence in a subset of the MPNST cells, thus delaying the initiation of MPNST tumorigenesis during development. To test this hypothesis, we checked senescence-associated β-galactosidase activity of the cells in our zebrafish model at 4 d.p.f. In this result, the PDGFRA mutant overexpressors in nf1- and p53-deficient background showed higher levels senescence-associated β-galactosidase activity than did the wild-type PDGFRA overexpressors or controls (Figures 4d, g and j). In transverse sections of the embryos, senescence-associated β-galactosidase-stained cells coincided with or were adjacent to Sox10-expressing mCherry-positive cells within the ventral neural tube of transgenic fish overexpressing the PDGFRA mutant cDNA (Figures 4e, f, h, i, k and l). This result suggests that overexpression of the PDGFRA mutant in cells of neural crest origin may induce senescence through hyperstimulation of Ras signaling. Senescent cells become postmitotic, so this process would delay the onset and retard the growth of MPNST tumors in the PDGFRA mutant animals.
Furthermore, to detect senescence in MPNSTs derived from wild-type or mutant PDGFRA transgenic zebrafish, we performed β-galactosidase staining of the cryosectioned tumor tissues. We also examined fibrosis in both groups of tumors with the trichrome stain and antibodies to detect collagen I expression. Additionally, CD31 expression levels were assessed in the cryosectioned tumor tissues. In these experiments, we did not observe any differences in senescence, fibrosis or vascularity among the transgenic and mCherry control tumors (Supplementary Figures S4).
Sunitinib retards primary MPNST progression in wild-type PDGFRA transgenic fish
To test whether inhibition of PDGFRA and other tyrosine kinase receptors would affect tumor cell growth and survival, we treated sox10:PDGFRA wild-type transgenic MPNST fish with the RTK inhibitor, sunitinib for 10 days (2 μm sunitinib or dimethyl sulfoxide (DMSO) in fish water). In the adults, the drug was effectively absorbed by the gills and transported to the tumor cells and normal cells through the blood. Comparison of mCherry expression by the tumor pretreatment and post-treatment tumor area indicated retarded tumor cell growth in the sunitinib-treated fish (Figures 5a–l). Sunitinib-treated PDGFRA wild-type transgenic fish showed an average tumor growth of 11% over 10 days, compared with 300% growth over 10 days in the DMSO-treated fish (Figure 5m).
Sunitinib retards the progression of primary MPNSTs in transgenic fish with wild-type PDGFRA. (a–f) Images of Tg (sox10:PDGFRA wild-type; sox10:mCherry; nf1a+/−; nf1b−/−; p53m/m) zebrafish with primary MPNSTs before drug treatment. mCherry driven by the sox10 promoter is expressed by the tumor cells. Fish with MPNSTs were incubated in 2 μm of sunitinib (n=3) or DMSO (n=3) in the fish water for 10 days. (g–l) After drug treatment, tumor images were taken under the same condition as before treatment. (m) Comparison of cross-sectional areas of MPNSTs after sunitinib treatment and the DMSO control shows that the tumors increase in size when treated with the DMSO vehicle, and that this growth appears to be retarded by sunitinib treatment (P<0.05).
Sunitinib promotes apoptosis of MPNST cells of PDGFRA wild-type transgenic fish
To study how sunitinib treatment leads to tumor shrinkage, we first examined the histopathology of the DMSO- or sunitinib-treated fish after H&E staining (Figures 6a–h). The sunitinib-treated tumors showed areas of necrosis (Figures 6c, d and g, h), with the necrotic centers surrounded by MPNST cells, whereas areas of necrosis were not detected in the DMSO control tumors (Figures 6a, b and e, f). We also performed proliferating cell nuclear antigen and cleaved caspase-3 immunostaining to assess proliferation and apoptosis, respectively (Figures 6i–l and m–p). Positive staining for proliferating cell nuclear antigen was observed in both the DMSO- and the sunitinib-treated MPNSTs at about the same levels, with the positive immunostaining of cleaved caspase-3 observed only in the cell nuclei of tumors in the sunitinib-treated MPNSTs. This result suggests that sunitinib retards the progression of primary MPNSTs by inducing apoptotic cell death rather than by simply inhibiting cell proliferation.
Sunitinib promotes apoptosis of primary MPNST progression in transgenic fish expressing wild-type PDGFRA. (a–d) Histopathology after H&E staining of the DMSO- (a and b) or sunitinib- (c and d) treated MPNSTs. Black arrowheads show areas of MPNSTs (black scale bar=5 mm). Proliferating cell nuclear antigen (e–h) and cleaved caspase-3 (i–l) stains of each condition for proliferation and apoptosis analysis, respectively. The right upper corner of each panel shows magnified stained areas.
Sunitinib activity is enhanced when given in combination with the MEK inhibitor trametinib in an in vivo assay using implanted zebrafish MPNST cells
MEK inhibitors have been reported to show activity against human MPNST cell lines.28 To test whether sunitinib activity is enhanced when given in combination with a MEK inhibitor in vivo, we used the Food and Drug Administration-approved MEK inhibitor trametinib for this experiment. We transplanted primary MPNST cells of wild-type PDGFRA transgenic fish in the nf1a+/−; nf1b−/−; p53m/m background into 2-day-old embryos. Because thymocytes have not begun to develop at this stage of embryogenesis, injected tumor cells from a donor fish with MPNST will expand in Casper recipient zebrafish embryos without rejection, even up to 8 weeks of age (Figures 7a–f). After cell implantation at 2 d.p.f., embryos were incubated for 24 h before drug treatment. Successfully implanted embryos were incubated with drugs for 3 days (Figure 7g). Both trametinib and sunitinib alone were able to significantly suppress MPNST cell growth compared with the DMSO control group. Our results show marked potentiation of cell killing in embryos receiving sunitinib plus trametinib, compared with sunitinib alone (Figures 7i–n). These results support the concept that RTK-RAS-MAPK signaling is hyperactivated in nf1-deficient zebrafish, and thus direct inhibition of MEK is able to synergize with the RTK inhibitory activity of sunitinib to improve the response of MPNST tumor cells in vivo.
Sunitinib activity is enhanced when given in combination with the MEK inhibitor trametinib in an assay using implanted zebrafish MPNST cells. (a–f) Photomicrographs of mCherry expressing transplanted MPNST cells. (a) Primary zebrafish MPNSTs were harvested from wild-type PDGFRA transgenic fish, and (b) ~100–120 tumor cells were implanted into the yolk sac of a 2 d.p.f. embryo. (c) Illustration of our implantation. Red arrow represents direction of the microneedle for cell injection. (d) Representative image of implanted MPNST embryo at 3 d.p.f. (e) Injected MPNST cells were expanded and (f) formed tumor mass in embryos. (g) Schematic diagram of our implantation and drug treatment assay. MPNST cells were transplanted into 2 d.p.f. embryos and treated with drugs in the fish water from 3 to 6 d.p.f. (h–m) Representative fish images at 6 d.p.f. after drug treatment. Because autofluorescence was expressed from the embryo gut (j), mCherry expressed areas in yolk sac were only analyzed for this assay. (n) MPNST tumor cell growth in the yolk sac of the implanted embryos. These embryos were treated with vehicle control, trametinib, sunitinib or combinations of trametinib and sunitinib (n=20 per condition). The asterisks indicate the range of the different P-values of Student's t-test (*P<0.05; **P<0.01; ***P<0.001; ****P<0.0001). Black bar in each column scatter plot represents the median value.
Discussion
Here we compare the consequences of overexpression of wild-type and mutant PDGFRA on the onset and penetrance of MPNSTs arising in our nf1- and p53-deficient zebrafish model. Relying on the sox10 promoter to overexpress wild-type and mutant PDGFRA in cells of neural crest origin, we demonstrate that PDGFRA overexpression accelerates MPNST tumorigenicity in zebrafish. Surprisingly, overexpression of wild-type PDGFRA was even more active in promoting the development of MPNST than was overexpression of a constitutively activated mutant PDGFRA in the nf1- and p53- deficient background (see Figures 2a and b). This observation has considerable clinical importance, because PDGFRA is frequently expressed at high levels in primary human MPNSTs, but it is only rarely mutated in these tumors.16 Our results implicate this high level of expression of the wild-type receptor in pathogenesis, which is difficult to infer in the absence of evidence from an animal model. In addition, our studies show that inhibition of the tyrosine kinase activity with sunitinib in tumors arising in animals transgenic for the wild-type receptor is active in inducing apoptosis and delaying tumor growth, suggesting dependence on this pathway for maintenance as well as for the initiation of MPNST, and suggesting a role for this drug in the management of tumors that overexpress the wild-type receptor.
Mutational activation of PDGFRA is common in many types of cancers, such as gastrointestinal stromal tumors,29, 30 myeloproliferative malignancies associated with hypereosinophilia31, 32, 33 and glioblastoma.23, 34 Most of these mutations act by promoting dimerization of the receptors in the absence of ligand, resulting in activation of the receptor’s intracellular tyrosine kinase domain.35 However, PDGFRA mutations in human MPNSTs are relatively rare, despite evidence of high levels of expression of the receptor and increased DNA copy number for the region surrounding the location of the gene.14, 16 Our studies suggest that this is because overexpression of constitutively active mutant PDGFRA induces excessive levels of oncogenic stress in NF1-deficient Schwann cell precursors, resulting in premature cell senescence.36, 37, 38 It would appear that high levels of wild-type PDGFRA expression are sufficient to promote dimerization of a subset of the receptors on the cell surface by mass action, and that this relatively modest level of tyrosine kinase signaling is optimal to promote malignant transformation of NF1 defiant Schwann cells.
A second possibility is that other oncogenic signals activated by wild-type PDGFRA may be involved in MPNST development. Clarke et al.23 reported that wild-type PDGFRA and exon 8- and 9-deleted mutant forms were differently localized in cells, with the mutants mainly expressed in cell cytosol and the wild-type form in the cell membrane. Because many cell signals are precisely regulated through receptors and transporters on the cell membrane, changes in the localization of PDGFRA may affect the outcome of RTK-related cell signaling such as calcium regulation, endocytosis and focal adhesion.39, 40, 41 Thus, signaling through the correctly localized wild-type PDGFRA may produce optimal signals for MPNST initiation.
The Sox10 protein is a neural crest transcription factor that is active in the differentiation of Schwann cell precursors.42 In our previous study, we confirmed that GFP driven by the sox10 promoter is strongly expressed in OPCs and Schwann cells,22 so that this promoter is ideal for driving gene expression in this lineage. Moreover, the tumor cells themselves express Sox10 (Shin et al.22 and Figures 3g–i) and fluorescent protein driven by the sox10 promoter is readily apparent in the primary MPNSTs of adult fish under the fluorescent microscope (Figures 2c–h), providing a useful marker for monitoring MPNST growth in living adult tumor bearing fish.
The constitutively active form of PDGFRA that we used for this study was first identified in human high-grade gliomas,23 and possesses an in-frame deletion (exons 8 and 9) in the extracellular domain. We previously reported two cases of high-grade gliomas in the nf1a+/−; nf1b−/−; p53m/m-stable zebrafish line, and these tumor cells expressed Sox10.22 In our current study, we identified one high-grade glioma in a fish transgenic for the sox10:PDGFRA mutant (Supplementary Figure S3), but we did not observe accelerated onset or increased penetrance of high-grade gliomas in the PDGFRA transgenic zebrafish. We are not sure why this promoter–receptor combination was not active in promoting the pathogenesis of high-grade gliomas in the zebrafish model. The sox10-mCherry expression is clearly visible in the brains of the transgenic fish, but it is possible that the sox10 promoter is not expressed early enough in the central nervous system glial lineage to drive the transformation of glial stem and progenitor cells. Alternatively, the central nervous system glial cells may be more susceptible to cell senescence induced by the PDGFRA transgenes, although we do not observe increased β-gal staining in the central nervous system of these fish.
In a previous study, PDGFRA was expressed in 75% of human primary MPNSTs and MPNST cell lines,16 and overexpression of epidermal growth factor receptor together with p53 or pten mutations increased MPNST development in mouse models.43, 44 Our study supports the hypothesis that wild-type PDGFRA overexpression accelerates the onset of MPNST development and would be sensitive to inhibition in MPNST patients. In human MPNSTs, Zietsch et al.13 reported that the cell line S462 harbors the 4q12 amplicon (PDGFRA, KIT and KDR genes) and is sensitive to sunitinib, with an IC50 (half-maximal inhibitory concentration) below 1.0 μm. Moreover, sunitinib induced apoptosis and prevented PDGFRA downstream signaling in response to the ligand, PDGF-AA. Holtkamp et al.16 also reported that PDGFRA in the S462 cell line is phosphorylated in response PDGF-AA. In addition, Aoki et al.19 has shown that cell invasion by the human MPNST cell lines, FU-SFT8611 and FU-SFT9817, is increased in response to the PDGF-AA and PDGF-BB ligands. Based on these results, we believe the role of PDGFRA in MPNST growth is evolutionarily conserved and that our zebrafish model faithfully represents human MPNSTs. Our results showing measurable activities of sunitinib and trametinib appear to accurately reflect the sensitivity of the human disease. In future studies with our MPNST model, it will be important to test other RTK inhibitors such as imatinib, pazopanib and sorafenib. The ideal properties of the zebrafish system for evaluating the activity of drugs administered into the fish water or delivered by daily oral gavage suggest that this system may be ideal for identifying drugs that can synergize with sunitinib to yield much more substantial levels of tumor cell killing. For example, others have reported activity of MEK28 and BET45 inhibitors in human MPNSTs and further investigation revealed that the MEK inhibitor trametinib enhanced antitumor effects of sunitinib in vivo (see Figure 7). Assays in the zebrafish may be one of the best ways to test combinations of three or more drugs in vivo at multiple dose levels in preclinical studies to guide the development of clinical trials for this disease.
Materials and methods
Zebrafish lines
The nf1a+/−; nf1b−/−; p53m/m zebrafish line22 was used for injections to generate the sox10:PDGFRA wild-type and mutant and sox10:mCherry transgenic lines. Zebrafish were maintained under standard conditions as described previously.46 All experiments involving zebrafish were approved by the Institutional Animal Care and Use Committee of the Dana-Farber Cancer Institute.
DNA constructs for transgenesis
The 7 kb promoter region of the sox10 gene was amplified by PCR from a zebrafish promoter construct kindly provided by Dr Robert N Kelsh,25 and subcloned into vectors to drive the expression of several genes, including Tg(sox10:mCherry), Tg(sox10:PDGFRA wild-type) and Tg(sox10:PDGFRA mutant) in tissues normally expressing the sox10 gene. The wild-type and mutant PDGFRA cDNAs were provided by Dr Eric C Holland.47 Embryos were injected with these DNA constructs at the one-cell stage and grown to adulthood. Fin clips from the offspring were genotyped for the stable integration and germline transmission of the transgenes. The Tg(sox10:mCherry), Tg(sox10:PDGFRA wild-type) and Tg(sox10:PDGFRA mutant) zebrafish lines are designated the ‘mCherry', ‘wild-type PDGFRA’ and ‘PDGFRA mutant’ transgenic line in this article, respectively.
Genotyping assays
To extract genomic DNA, embryos or tail clips were placed in clean PCR tubes and 40 μl of lysis buffer (10 mm Tris (pH 8.3), 50 mm KCl, 0.3% Tween-20, 0.3% NP40, 1 mg/ml proteinase K) was added to each tube. The PCR tubes incubated at 55 °C for 2 h to overnight, depending on the size of the sample, and then the tubes were heated to 95 °C for 15 min for the proteinase K inactivation. Using 2 μl of the lysate as a PCR template, PCR reaction was performed with NEB Taq DNA polymerase (NEB, Ipswich, MA, USA; M0273) according to the manufacturer's instructions. The primers used for human PDGFRA amplification were as follows: (forward) 5′-ATCAAACCCACCTTCAGCCA-3′ and (reverse) 5′-TCAATGACCCTCCAGCGAAT-3′. PCR was started at 95 °C for 2 min and then underwent 40 cycles of 95 °C for 30 s, 55 °C for 30 s and 68 °C for 45 s. For nf1a, nf1b and p53, we used protocols described previously.20, 22, 48
Tumor watch of transgenic fish
mCherry, wild-type and mutant PDGFRA heterozygous transgenic fish were crossed with nf1a+/−; nf1b−/−; p53m/m, and offsprings were screened every week starting from 12 w.p.f. for fluorescent mCherry-expressing cell masses indicative of tumors. In addition, for Figure 2a, either human wild-type or mutant PDGFRAs were overexpressed in nf1- and p53-deficient fish as mosaics by coinjecting the following constructs into the one-cell stage of nf1- and p53-deficient embryos: (1) sox10:PDGFRA wild-type with sox10:mCherry; (2) sox10:PDGFRA mutant with sox10:mCherry; or (3) sox10:mCherry alone. The primary injectants were raised and monitored for the onset of tumorigenesis as described above. Fish with tumors were separated and analyzed further by H&E staining and immunohistochemical assays.
Western blotting
Protein lysates were prepared with mCherry expressed tumors from nf1a+/−; nf1b−/−; p53m/m fish. Briefly, mCherry-positive tumor mass were harvested from tricaine-anesthetized tumor fish, and were lysed on ice in 1 × RIPA (Cell Signaling, Danvers, MA, USA) containing, PhosSTOP phosphatase inhibitor cocktail tablet (Roche, Indianapolis, IN, USA) and complete protease inhibitor tablet (Roche). The inhibitors were prepared following the manufacturer’s recommendation. Protein lysates were separated by gel electrophoresis, transferred to PVDF membranes and probed overnight at 4 °C with the following primary antibodies: anti-PDGFRA (Cell Signaling; 5241; 1:500), anti-p-tyrosine (EMD Millipore, Billerica, MA, USA; 05-321; 1:1000), anti-β-tubulin (Cell Signaling 2146; 1:2000), anti-AKT (Cell Signaling; 9272; 1:1000), anti-p-AKT (Cell Signaling; 4060; 1:1000), anti-p-ERK1/2 (Cell Signaling; 4377;1:1000) and anti-ERK1/2 (Cell Signaling; 9102; 1:1000). Primary antibody binding was visualized on X-ray film using anti-mouse-HRP (Cell Signaling; 7076; 1:10 000) or anti-rabbit-HRP (Cell Signaling; 7074; 1:10 000) secondary antibodies along with SuperSignal West Dura or Femto (Thermo Fisher Scientific, Waltham, MA, USA) chemiluminescent substrates. Stripping was performed using Restore PLUS Western Blot Stripping Buffer (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Senescence-associated β-galactosidase staining
The Senescence-Associated β-Galactosidase Detection Kit (CS0030; Sigma-Aldrich, St Louis, MO, USA) was used for fish embryo senescence detection according to the manufacturer’s protocol. Briefly, fish embryos at 4 d.p.f. were fixed with 4% paraformaldehyde for 24 h at 4 °C, and were washed three times in phosphate-buffered saline (pH 7.4) for 1 h each. After washing, the embryos were incubated with the senescence staining mixture for 24 h at 37 °C. The experiment was performed in duplicate.
Paraffin sectioning, cryosectioning and immunostaining
Fish were fixed with 4% paraformaldehyde and embedded in paraffin blocks for paraffin sectioning. Sections were immunostained by conventional protocols49 using antibodies against S100 (Dako, Carpinteria, CA, USA; Z0311; 1:1000), Sox10 (GeneTex, Irvine, CA, USA; GTX128374; 1:1000), proliferating cell nuclear antigen (EMD Millipore; NA03; 1:50) and cleaved caspase-3 (Cell Signaling 9664; 1:125). Cryosectioning of β-galactosidase-stained embryos was performed as described previously.50
Drug treatment
Wild-type PDGFRA expressed tumor fish were exposed to 2 μm sunitinib (LC Laboratories, Woburn, MA, USA; S-8803) or DMSO added to the fish water. The tumor fish were fed with dry food (GEMMA Micro 300; Skretting, Tooele, UT, USA) every other day. The fish water was exchanged with fresh water (containing sunitinib or DMSO) 2 h after feeding. After 10 days of treatment, quantitative assessment of the remaining tumor was measured using the ImageJ software (NIH, Bethesda, MD, USA) for each treated tumor fish. Embryos were treated with sunitinib, trametinib (Selleck, Houston, TX, USA; GSK1120212) or a combination of both for 3 days under dark conditions.
Embryonic implantation assay
Primary MPNST cells derived from wild-type PDGFRA expressed tumor fish were injected into the yolk sac of each embryo (roy and nacre double homozygous mutant; casper line) at 2 d.p.f. Successfully implanted embryos were collected and randomly distributed into the well of a 12-well plate, and incubated with either DMSO or drugs for 3 days at 28 °C. After drug treatment for 3 days, each embryo was imaged, and tumor area was quantitated using the ImageJ software (NIH). For further details about the implantation, see Supplementary Information.
Statistical analysis
Statistical analysis was performed with Prism 5 software (GraphPad, La Jolla, CA, USA). Kaplan–Meier methods and the log-rank test were applied to assess the rate of tumor development in Figure 2. The exact sample size is indicated in each figure and legend. A two-tailed unpaired t-test was used for the analysis in Figure 4 and data were plotted along with the standard deviation. A two-tailed unpaired t-test was also used for the analysis in Figure 5. For the experiment shown in Figure 7, the power to detect a 50% decrease in tumor cells with an agent compared with DMSO is 91% when nine embryos are tested per condition, assuming that if the mean signal is reduced by 50%, the standard deviation is also reduced by 50%, so at least nine embryos were tested at each condition. The largest and smallest values for each condition were removed from analysis as outliers according to the pre-established criteria, and the median values are shown in Figure 7. A two-tailed unpaired t-test was used for the analysis of significance in Figure 7. After drug treatment, digital images were recorded of the fluorescence signal for every embryo and the area of the fluorescence signal was measured quantitatively using the ImageJ software (NIH). The variance of the area of mCherry expression in the embryos was calculated for each group and shown to be within 30% of the DMSO control embryos. If the standard deviation for specific compounds varies by >30% from the DMSO control embryos, and then we would test for significance using the Welch t-test. Embryos were injected without randomization for this experiment. The investigators were not blinded to the experimental group.
References
Ferrari A, Bisogno G, Carli M . Management of childhood malignant peripheral nerve sheath tumor. Paediatr Drugs 2007; 9: 239–248.
Evans DG, O'Hara C, Wilding A, Ingham SL, Howard E, Dawson J et al. Mortality in neurofibromatosis 1: in North West England: an assessment of actuarial survival in a region of the UK since 1989. Eur J Hum Genet 2011; 19: 1187–1191.
Thway K, Fisher C . Malignant peripheral nerve sheath tumor: pathology and genetics. Ann Diagn Pathol 2013; 18: 109–116.
Kourea HP, Bilsky MH, Leung DH, Lewis JJ, Woodruff JM . Subdiaphragmatic and intrathoracic paraspinal malignant peripheral nerve sheath tumors: a clinicopathologic study of 25 patients and 26 tumors. Cancer 1998; 82: 2191–2203.
Wong WW, Hirose T, Scheithauer BW, Schild SE, Gunderson LL . Malignant peripheral nerve sheath tumor: analysis of treatment outcome. Int J Radiat Oncol Biol Phys 1998; 42: 351–360.
Cichowski K, Jacks T . NF1 tumor suppressor gene function: narrowing the GAP. Cell 2001; 104: 593–604.
Martin GA, Viskochil D, Bollag G, McCabe PC, Crosier WJ, Haubruck H et al. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell 1990; 63: 843–849.
Knudson AG . Two genetic hits (more or less) to cancer. Nat Rev Cancer 2001; 1: 157–162.
Rubin JB, Gutmann DH . Neurofibromatosis type 1—a model for nervous system tumour formation? Nat Rev Cancer 2005; 5: 557–564.
Fortman BJ, Kuszyk BS, Urban BA, Fishman EK . Neurofibromatosis type 1: a diagnostic mimicker at CT. Radiographics 2001; 21: 601–612.
Mautner VF, Friedrich RE, von Deimling A, Hagel C, Korf B, Knofel MT et al. Malignant peripheral nerve sheath tumours in neurofibromatosis type 1: MRI supports the diagnosis of malignant plexiform neurofibroma. Neuroradiology 2003; 45: 618–625.
Lee W, Teckie S, Wiesner T, Ran L, Prieto Granada CN, Lin M et al. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat Genet 2014; 46: 1227–1232.
Zietsch J, Ziegenhagen N, Heppner FL, Reuss D, von Deimling A, Holtkamp N . The 4q12 amplicon in malignant peripheral nerve sheath tumors: consequences on gene expression and implications for sunitinib treatment. PLoS One 2010; 5: e11858.
Mantripragada KK, Spurlock G, Kluwe L, Chuzhanova N, Ferner RE, Frayling IM et al. High-resolution DNA copy number profiling of malignant peripheral nerve sheath tumors using targeted microarray-based comparative genomic hybridization. Clin Cancer Res 2008; 14: 1015–1024.
Holtkamp N, Mautner VF, Friedrich RE, Harder A, Hartmann C, Theallier-Janko A et al. Differentially expressed genes in neurofibromatosis 1-associated neurofibromas and malignant peripheral nerve sheath tumors. Acta Neuropathol 2004; 107: 159–168.
Holtkamp N, Okuducu AF, Mucha J, Afanasieva A, Hartmann C, Atallah I et al. Mutation and expression of PDGFRA and KIT in malignant peripheral nerve sheath tumors, and its implications for imatinib sensitivity. Carcinogenesis 2006; 27: 664–671.
Perrone F, Da Riva L, Orsenigo M, Losa M, Jocolle G, Millefanti C et al. PDGFRA, PDGFRB, EGFR, and downstream signaling activation in malignant peripheral nerve sheath tumor. Neuro-oncology 2009; 11: 725–736.
Mashour GA, Ratner N, Khan GA, Wang HL, Martuza RL, Kurtz A . The angiogenic factor midkine is aberrantly expressed in NF1-deficient Schwann cells and is a mitogen for neurofibroma-derived cells. Oncogene 2001; 20: 97–105.
Aoki M, Nabeshima K, Koga K, Hamasaki M, Suzumiya J, Tamura K et al. Imatinib mesylate inhibits cell invasion of malignant peripheral nerve sheath tumor induced by platelet-derived growth factor-BB. Lab Invest 2007; 87: 767–779.
Berghmans S, Murphey RD, Wienholds E, Neuberg D, Kutok JL, Fletcher CD et al. Tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci USA 2005; 102: 407–412.
Legius E, Dierick H, Wu R, Hall BK, Marynen P, Cassiman JJ et al. TP53 mutations are frequent in malignant NF1 tumors. Genes Chromosomes Cancer 1994; 10: 250–255.
Shin J, Padmanabhan A, de Groh ED, Lee JS, Haidar S, Dahlberg S et al. Zebrafish neurofibromatosis type 1 genes have redundant functions in tumorigenesis and embryonic development. Dis Models Mech 2012; 5: 881–894.
Clarke ID, Dirks PB . A human brain tumor-derived PDGFR-alpha deletion mutant is transforming. Oncogene 2003; 22: 722–733.
Langenau DM, Keefe MD, Storer NY, Jette CA, Smith AC, Ceol CJ et al. Co-injection strategies to modify radiation sensitivity and tumor initiation in transgenic Zebrafish. Oncogene 2008; 27: 4242–4248.
Dutton JR, Antonellis A, Carney TJ, Rodrigues FS, Pavan WJ, Ward A et al. An evolutionarily conserved intronic region controls the spatiotemporal expression of the transcription factor Sox10. BMC Dev Biol 2008; 8: 105.
Nonaka D, Chiriboga L, Rubin BP . Sox10: a pan-schwannian and melanocytic marker. Am J Surg Pathol 2008; 32: 1291–1298.
Karamchandani JR, Nielsen TO, van de Rijn M, West RB . Sox10 and S100 in the diagnosis of soft-tissue neoplasms. Appl Immunohistochem Mol Morphol 2012; 20: 445–450.
Jessen WJ, Miller SJ, Jousma E, Wu J, Rizvi TA, Brundage ME et al. MEK inhibition exhibits efficacy in human and mouse neurofibromatosis tumors. J Clin Invest 2013; 123: 340–347.
Toffalini F, Demoulin JB . New insights into the mechanisms of hematopoietic cell transformation by activated receptor tyrosine kinases. Blood 2010; 116: 2429–2437.
Corless CL, Schroeder A, Griffith D, Town A, McGreevey L, Harrell P et al. PDGFRA mutations in gastrointestinal stromal tumors: frequency, spectrum and in vitro sensitivity to imatinib. J Clin Oncol 2005; 23: 5357–5364.
Demoulin JB, Montano-Almendras CP . Platelet-derived growth factors and their receptors in normal and malignant hematopoiesis. Am J Blood Res 2012; 2: 44–56.
Medves S, Demoulin JB . Tyrosine kinase gene fusions in cancer: translating mechanisms into targeted therapies. J Cell Mol Med 2012; 16: 237–248.
Cools J, DeAngelo DJ, Gotlib J, Stover EH, Legare RD, Cortes J et al. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med 2003; 348: 1201–1214.
Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010; 17: 98–110.
Velghe AI, Van Cauwenberghe S, Polyansky AA, Chand D, Montano-Almendras CP, Charni S et al. PDGFRA alterations in cancer: characterization of a gain-of-function V536E transmembrane mutant as well as loss-of-function and passenger mutations. Oncogene 2014; 33: 2568–2576.
Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW . Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997; 88: 593–602.
Dimauro T, David G . Ras-induced senescence and its physiological relevance in cancer. Curr Cancer Drug Targets 2010; 10: 869–876.
Downward J . Ras signalling and apoptosis. Curr Opin Genet Dev 1998; 8: 49–54.
Baker SA, Hennig GW, Salter AK, Kurahashi M, Ward SM, Sanders KM . Distribution and Ca(2+) signalling of fibroblast-like (PDGFR(+)) cells in the murine gastric fundus. J Physiol 2013; 591: 6193–6208.
Alexander A . Endocytosis and intracellular sorting of receptor tyrosine kinases. Front Biosci 1998; 3: d729–d738.
Riemenschneider MJ, Mueller W, Betensky RA, Mohapatra G, Louis DN . In situ analysis of integrin and growth factor receptor signaling pathways in human glioblastomas suggests overlapping relationships with focal adhesion kinase activation. Am J Pathol 2005; 167: 1379–1387.
Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA et al. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev 2001; 15: 66–78.
Keng VW, Watson AL, Rahrmann EP, Li H, Tschida BR, Moriarity BS et al. Conditional inactivation of Pten with EGFR overexpression in schwann cells models sporadic MPNST. Sarcoma 2012; 2012: 620834.
Rahrmann EP, Watson AL, Keng VW, Choi K, Moriarity BS, Beckmann DA et al. Forward genetic screen for malignant peripheral nerve sheath tumor formation identifies new genes and pathways driving tumorigenesis. Nat Genet 2013; 45: 756–766.
De Raedt T, Beert E, Pasmant E, Luscan A, Brems H, Ortonne N et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature 2014; 514: 247–251.
Westerfield M . The Zebrafish Book. A Guide for the Laboratory use of Zebrafish (Danio rerio) 4th edn. University of Oregon Press, 2000.
Ozawa T, Brennan CW, Wang L, Squatrito M, Sasayama T, Nakada M et al. PDGFRA gene rearrangements are frequent genetic events in PDGFRA-amplified glioblastomas. Genes Dev 2010; 24: 2205–2218.
Lee JS, Padmanabhan A, Shin J, Zhu S, Guo F, Kanki JP et al. Oligodendrocyte progenitor cell numbers and migration are regulated by the zebrafish orthologs of the NF1 tumor suppressor gene. Hum Mol Genet 2010; 19: 4643–4653.
Macdonald R . Zebrafish immunohistochemistry. Methods Mol Biol 1999; 127: 77–88.
Tao T, Shi H, Guan Y, Huang D, Chen Y, Lane DP et al. Def defines a conserved nucleolar pathway that leads p53 to proteasome-independent degradation. Cell Res 2013; 23: 620–634.
Acknowledgements
This work was supported by the Latsis family fellowship from the Boston Children’s Hospital Neurofibromatosis program, a grant from Department of Defense (W81XWH-12-1-0125), a Drug Discovery Initiative Award from the Children’s Tumor Foundation and the NF1 Research Consortium Fund. We thank John Gilbert for editorial review and critical comments, and Dr Donna Neuberg for biostatistical advice regarding the sample size, statistical power and most appropriate statistical tests. We are grateful to Hillary Layden and Shawna Chamberlin for their expert assistance with zebrafish husbandry and to Dr Eric Holland for providing the wild-type and mutant PDGFRA cDNAs.
Author contributions
DHK, SH and ATL conceived and designed the experiments; DHK performed the experiments; SR performed and analyzed the histologic analysis and immunostaining; DHK, SH and ATL analyzed the data and wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Ki, D., He, S., Rodig, S. et al. Overexpression of PDGFRA cooperates with loss of NF1 and p53 to accelerate the molecular pathogenesis of malignant peripheral nerve sheath tumors. Oncogene 36, 1058–1068 (2017). https://doi.org/10.1038/onc.2016.269
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.269
- Springer Nature Limited
This article is cited by
-
Malignant Peripheral Nerve Sheath Tumor, a Heterogeneous, Aggressive Cancer with Diverse Biomarkers and No Targeted Standard of Care: Review of the Literature and Ongoing Investigational Agents
Targeted Oncology (2024)
-
The therapeutic potential of neurofibromin signaling pathways and binding partners
Communications Biology (2023)
-
Malignant peripheral nerve sheath tumor: models, biology, and translation
Oncogene (2022)
-
Enhancer reprogramming in PRC2-deficient malignant peripheral nerve sheath tumors induces a targetable de-differentiated state
Acta Neuropathologica (2021)
-
Curse of the devil: molecular insights into the emergence of transmissible cancers in the Tasmanian devil (Sarcophilus harrisii)
Cellular and Molecular Life Sciences (2020)