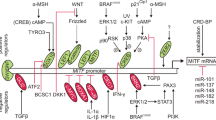
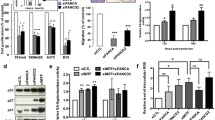
Abstract
Melanoma progression is associated with increased invasion and, often, decreased levels of microphthalmia-associated transcription factor (MITF). Accordingly, downregulation of MITF induces invasion in melanoma cells; however, little is known about the underlying mechanisms. Here, we report for the first time that depletion of MITF results in elevation of intracellular GTP levels and increased amounts of active (GTP-bound) RAC1, RHO-A and RHO-C. Concomitantly, MITF-depleted cells display larger number of invadopodia and increased invasion. We further demonstrate that the gene for guanosine monophosphate reductase (GMPR) is a direct MITF target, and that the partial repression of GMPR accounts mostly for the above phenotypes in MITF-depleted cells. Reciprocally, transactivation of GMPR is required for MITF-dependent suppression of melanoma cell invasion, tumorigenicity and lung colonization. Moreover, loss of GMPR accompanies downregulation of MITF in vemurafenib-resistant BRAFV600E-melanoma cells and underlies the increased invasion in these cells. Our data uncover novel mechanisms linking MITF-dependent inhibition of invasion to suppression of guanylate metabolism.








Similar content being viewed by others
References
Miller AJ, Mihm MC Jr . Melanoma. N Engl J Med 2006; 355: 51–65.
Siegel RL, Miller KD, Jemal A . Cancer statistics, 2015. CA 2015; 65: 5–29.
Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H et al. Distinct sets of genetic alterations in melanoma. N Engl J Med 2005; 353: 2135–2147.
Weaver AM . Invadopodia: specialized cell structures for cancer invasion. Clin Exp Metastasis 2006; 23: 97–105.
Jaffe AB, Hall A . Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol 2005; 21: 247–269.
Halaban R . RAC1 and melanoma. Clin Ther 2015; 37: 682–685.
Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet 2012; 44: 1006–1014.
Bianchi-Smiraglia A, Wawrzyniak JA, Bagati A, Marvin EK, Ackroyd J, Moparthy S et al. Pharmacological targeting of guanosine monophosphate synthase suppresses melanoma cell invasion and tumorigenicity. Cell Death Differ 2015; 22: 1858–1864.
Wawrzyniak JA, Bianchi-Smiraglia A, Bshara W, Mannava S, Ackroyd J, Bagatis A et al. A purine nucloetide biosynthesis enzyme guanosine monophosphate reductase is a suppressor of melanoma invasion. Cell Rep 2013; 5: 493–507.
Hallett MA, Dagher PC, Atkinson SJ . Rho GTPases show differential sensitivity to nucleotide triphosphate depletion in a model of ischemic cell injury. Am J Physiol Cell Physiol 2003; 285: C129–C138.
Mondin M, Moreau V, Genot E, Combe C, Ripoche J, Dubus I . Alterations in cytoskeletal protein expression by mycophenolic acid in human mesangial cells requires Rac inactivation. Biochem Pharmacol 2007; 73: 1491–1498.
Kollareddy M, Dimitrova E, Vallabhaneni KC, Chan A, Le T, Chauhan KM et al. Regulation of nucleotide metabolism by mutant p53 contributes to its gain-of-function activities. Nat Commun 2015; 6: 7389.
Cheli Y, Giuliano S, Botton T, Rocchi S, Hofman V, Hofman P et al. Mitf is the key molecular switch between mouse or human melanoma initiating cells and their differentiated progeny. Oncogene 2011; 30: 2307–2318.
Hoek KS, Goding CR . Cancer stem cells versus phenotype-switching in melanoma. Pigment Cell Melanoma Res 2010; 23: 746–759.
Cheli Y, Ohanna M, Ballotti R, Bertolotto C . Fifteen-year quest for microphthalmia-associated transcription factor target genes. Pigment Cell Melanoma Res 2010; 23: 27–40.
Shibahara S, Takeda K, Yasumoto K, Udono T, Watanabe K, Saito H et al. Microphthalmia-associated transcription factor (MITF): multiplicity in structure, function, and regulation. J Invest Dermatol Symp. Proc. Soc. 2001; 6: 99–104.
McGill GG, Horstmann M, Widlund HR, Du J, Motyckova G, Nishimura EK et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell 2002; 109: 707–718.
Wellbrock C, Rana S, Paterson H, Pickersgill H, Brummelkamp T, Marais R . Oncogenic BRAF regulates melanoma proliferation through the lineage specific factor MITF. PLoS ONE 2008; 3: e2734.
Arozarena I, Bischof H, Gilby D, Belloni B, Dummer R, Wellbrock C . In melanoma, beta-catenin is a suppressor of invasion. Oncogene 2011; 30: 4531–4543.
Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS et al. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev 2006; 20: 3426–3439.
Cheli Y, Giuliano S, Fenouille N, Allegra M, Hofman V, Hofman P et al. Hypoxia and MITF control metastatic behaviour in mouse and human melanoma cells. Oncogene 2012; 31: 2461–2470.
Javelaud D, Alexaki VI, Pierrat MJ, Hoek KS, Dennler S, Van Kempen L et al. GLI2 and M-MITF transcription factors control exclusive gene expression programs and inversely regulate invasion in human melanoma cells. Pigment Cell Melanoma Res 2011; 24: 932–943.
Vachtenheim J, Ondrusova L . Microphthalmia-associated transcription factor expression levels in melanoma cells contribute to cell invasion and proliferation. Exp Dermatol 2015; 24: 481–484.
Hotary KB, Allen ED, Brooks PC, Datta NS, Long MW, Weiss SJ . Membrane type I matrix metalloproteinase usurps tumor growth control imposed by the three-dimensional extracellular matrix. Cell 2003; 114: 33–45.
Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011; 364: 2507–2516.
Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci USA 2008; 105: 3041–3046.
Muller J, Krijgsman O, Tsoi J, Robert L, Hugo W, Song C et al. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat Commun 2014; 5: 5712.
Ji Z, Erin Chen Y, Kumar R, Taylor M, Jenny Njauw CN, Miao B et al. MITF modulates therapeutic resistance through EGFR signaling. J Invest Dermatol 2015; 135: 1863–1872.
Konieczkowski DJ, Johannessen CM, Abudayyeh O, Kim JW, Cooper ZA, Piris A et al. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov 2014; 4: 816–827.
Paraiso KH, Das Thakur M, Fang B, Koomen JM, Fedorenko IV, John JK et al. Ligand-independent EPHA2 signaling drives the adoption of a targeted therapy-mediated metastatic melanoma phenotype. Cancer Discov 2015; 5: 264–273.
O'Connell MP, Marchbank K, Webster MR, Valiga AA, Kaur A, Vultur A et al. Hypoxia induces phenotypic plasticity and therapy resistance in melanoma via the tyrosine kinase receptors ROR1 and ROR2. Cancer Discov 2013; 3: 1378–1393.
Hoek KS, Schlegel NC, Eichhoff OM, Widmer DS, Praetorius C, Einarsson SO et al. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res 2008; 21: 665–676.
Salti GI, Manougian T, Farolan M, Shilkaitis A, Majumdar D, Das Gupta TK . Micropthalmia transcription factor: a new prognostic marker in intermediate-thickness cutaneous malignant melanoma. Cancer Res 2000; 60: 5012–5016.
Bauer NN, Chen YW, Samant RS, Shevde LA, Fodstad O . Rac1 activity regulates proliferation of aggressive metastatic melanoma. Exp Cell Res 2007; 313: 3832–3839.
Bartolome RA, Galvez BG, Longo N, Baleux F, Van Muijen GN, Sanchez-Mateos P et al. Stromal cell-derived factor-1alpha promotes melanoma cell invasion across basement membranes involving stimulation of membrane-type 1 matrix metalloproteinase and Rho GTPase activities. Cancer Res 2004; 64: 2534–2543.
Wang J, Huang SK, Marzese DM, Hsu SC, Kawas NP, Chong KK et al. Epigenetic changes of EGFR have an important role in BRAF inhibitor-resistant cutaneous melanomas. J Invest Dermatol 2015; 135: 532–541.
Zubrilov I, Sagi-Assif O, Izraely S, Meshel T, Ben-Menahem S, Ginat R et al. Vemurafenib resistance selects for highly malignant brain and lung-metastasizing melanoma cells. Cancer Lett 2015; 361: 86–96.
Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci USA 2009; 106: 1814–1819.
Friedl P, Alexander S . Cancer invasion and the microenvironment: plasticity and reciprocity. Cell 2011; 147: 992–1009.
Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 2005; 436: 117–122.
Wellbrock C, Marais R . Elevated expression of MITF counteracts B-RAF-stimulated melanocyte and melanoma cell proliferation. J Cell Biol 2005; 170: 703–708.
Golan T, Messer AR, Amitai-Lange A, Melamed Z, Ohana R, Bell RE et al. Interactions of melanoma cells with distal keratinocytes trigger metastasis via notch signaling inhibition of MITF. Mol Cell 2015; 59: 664–676.
Felicetti F, Errico MC, Bottero L, Segnalini P, Stoppacciaro A, Biffoni M et al. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res 2008; 68: 2745–2754.
Kamai T, Tsujii T, Arai K, Takagi K, Asami H, Ito Y et al. Significant association of Rho/ROCK pathway with invasion and metastasis of bladder cancer. Clin Cancer Res 2003; 9: 2632–2641.
Arozarena I, Sanchez-Laorden B, Packer L, Hidalgo-Carcedo C, Hayward R, Viros A et al. Oncogenic BRAF induces melanoma cell invasion by downregulating the cGMP-specific phosphodiesterase PDE5A. Cancer Cell 2011; 19: 45–57.
Franklin TJ, Twose PA . Reduction in beta-adrenergic response of cultured glioma cells following depletion of intracellular GTP. Eur J Biochem 1977; 77: 113–117.
Imamura H, Nhat KP, Togawa H, Saito K, Iino R, Kato-Yamada Y et al. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc Natl Acad Sci USA 2009; 106: 15651–15656.
van Horssen R, Janssen E, Peters W, van de Pasch L, Lindert MM, van Dommelen MM et al. Modulation of cell motility by spatial repositioning of enzymatic ATP/ADP exchange capacity. J Biol Chem 2009; 284: 1620–1627.
Mannava S, Moparthy KC, Wheeler LJ, Natarajan V, Zucker SN, Fink EE et al. Depletion of deoxyribonucleotide pools is an endogenous source of DNA damage in cells undergoing oncogene-induced senescence. Am J Pathol 2013; 182: 142–151.
Fink EE, Mannava S, Bagati A, Bianchi-Smiraglia A, Nair JR, Moparthy K et al. Mitochondrial thioredoxin reductase regulates major cytotoxicity pathways of proteasome inhibitors in multiple myeloma cells. Leukemia 2015; 30: 104–111.
Acknowledgements
We are grateful to Dr Dominic Smiraglia (Roswell Park Cancer Institute) for critical reading of the manuscript and to the Pathology Resource Network and the Clinical Data Network at Roswell Park Cancer Institute for providing human specimens. This work has been supported by NIH grants CA120244 and CA190533 (MAN), CA083081 (DSS), Ruth L Kirschstein National Research Service Award F32CA189622 (AB-S); American Cancer Society grant RSG-10-121-01 (MAN) and Jennifer Linscott Tietgen Foundation (MAN). This work was also supported in part by NCI Cancer Center Support Grant CA016056 to the Roswell Park Cancer Institute.
Author contributions
AB-S, AB and MAN designed the experiments and wrote the manuscript; AB-S and AB performed most of the experiments and analyzed the data; SM, EEF, JAW, EKM, PJ, MK, CEF, AEB, NIK, BCL, LMR, SB and JA performed some of the experiments; WB performed and scored the IHC staining; DSS supervised part of the study; MAN conceived the initial hypothesis and supervised the study. All authors discussed the results and commented on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Bianchi-Smiraglia, A., Bagati, A., Fink, E. et al. Microphthalmia-associated transcription factor suppresses invasion by reducing intracellular GTP pools. Oncogene 36, 84–96 (2017). https://doi.org/10.1038/onc.2016.178
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.178
- Springer Nature Limited
This article is cited by
-
Leveraging a disulfidptosis/ferroptosis-based signature to predict the prognosis of lung adenocarcinoma
Cancer Cell International (2023)
-
Deficiency of gluconeogenic enzyme PCK1 promotes metabolic-associated fatty liver disease through PI3K/AKT/PDGF axis activation in male mice
Nature Communications (2023)
-
A Controlled Transcription-Driven Light-Up Aptamer Amplification for Nucleoside Triphosphate Detection
BioChip Journal (2023)
-
A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy
Cell Research (2023)
-
Signal pathways of melanoma and targeted therapy
Signal Transduction and Targeted Therapy (2021)