Abstract
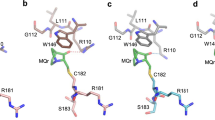
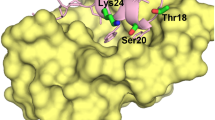
The tumor-suppressor protein p53 is tightly controlled in normal cells by its two negative regulators—the E3 ubiquitin ligase MDM2 and its homolog MDMX. Under stressed conditions such as DNA damage, p53 escapes MDM2- and MDMX-mediated functional inhibition and degradation, acting to prevent damaged cells from proliferating through induction of cell cycle arrest, DNA repair, senescence or apoptosis. Ample evidence suggests that stress signals induce phosphorylation of MDM2 and MDMX, leading to p53 activation. However, the structural basis of stress-induced p53 activation remains poorly understood because of the paucity of technical means to produce site-specifically phosphorylated MDM2 and MDMX proteins for biochemical and biophysical studies. Herein, we report total chemical synthesis, via native chemical ligation, and functional characterization of (24–108)MDMX and its Tyr99-phosphorylated analog with respect to their ability to interact with a panel of p53-derived peptide ligands and PMI, a p53-mimicking but more potent peptide antagonist of MDMX, using FP and surface plasmon resonance techniques. Phosphorylation of MDMX at Tyr99 weakens peptide binding by approximately two orders of magnitude. Comparative X-ray crystallographic analyses of MDMX and of pTyr99 MDMX in complex with PMI as well as modeling studies reveal that the phosphate group of pTyr99 imposes extensive steric clashes with the C-terminus of PMI or p53 peptide and induces a significant lateral shift of the peptide ligand, contributing to the dramatic decrease in the binding affinity of MDMX for p53. Because DNA damage activates c-Abl tyrosine kinase that phosphorylates MDMX at Tyr99, our findings afford a rare glimpse at the structural level of how stress-induced MDMX phosphorylation dislodges p53 from the inhibitory complex and activates it in response to DNA damage.





Similar content being viewed by others
Accession codes
References
Wade M, Li YC, Wahl GM . MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 2013; 13: 83–96.
Levine AJ, Oren M . The first 30 years of p53: growing ever more complex. Nat Rev Cancer 2009; 9: 749–758.
Vogelstein B, Lane D, Levine AJ . Surfing the p53 network. Nature 2000; 408: 307–310.
Vousden KH, Lane DP . p53 in health and disease. Nat Rev Mol Cell Biol 2007; 8: 275–283.
Wade M, Wang YV, Wahl GM . The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol 2010; 20: 299–309.
Jones SN, Roe AE, Donehower LA, Bradley A . Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 1995; 378: 206–208.
Montes de Oca Luna R, Wagner DS, Lozano G . Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 1995; 378: 203–206.
Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet 2001; 29: 92–95.
Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP . Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer 2009; 9: 862–873.
Khoo KH, Verma CS, Lane DP . Drugging the p53 pathway: understanding the route to clinical efficacy. Nat Rev Drug Discov 2014; 13: 217–236.
Popowicz GM, Domling A, Holak TA . The structure-based design of Mdm2/Mdmx-p53 inhibitors gets serious. Angew Chem Int Ed Engl 2011; 50: 2680–2688.
Kruse JP, Gu W . Modes of p53 regulation. Cell 2009; 137: 609–622.
Meek DW . Tumour suppression by p53: a role for the DNA damage response? Nat Rev Cancer 2009; 9: 714–723.
Mayo LD, Turchi JJ, Berberich SJ . Mdm-2 phosphorylation by DNA-dependent protein kinase prevents interaction with p53. Cancer Res 1997; 57: 5013–5016.
Zuckerman V, Lenos K, Popowicz GM, Silberman I, Grossman T, Marine JC et al. c-Abl phosphorylates Hdmx and regulates its interaction with p53. J Biol Chem 2009; 284: 4031–4039.
Chen L, Gilkes DM, Pan Y, Lane WS, Chen J . ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J 2005; 24: 3411–3422.
Maya R, Balass M, Kim ST, Shkedy D, Leal JF, Shifman O et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev 2001; 15: 1067–1077.
Okamoto K, Kashima K, Pereg Y, Ishida M, Yamazaki S, Nota A et al. DNA damage-induced phosphorylation of MdmX at serine 367 activates p53 by targeting MdmX for Mdm2-dependent degradation. Mol Cell Biol 2005; 25: 9608–9620.
Pereg Y, Shkedy D, de Graaf P, Meulmeester E, Edelson-Averbukh M, Salek M et al. Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc Natl Acad Sci USA 2005; 102: 5056–5061.
Cheng Q, Chen L, Li Z, Lane WS, Chen J . ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. EMBO J 2009; 28: 3857–3867.
McCoy MA, Gesell JJ, Senior MM, Wyss DF . Flexible lid to the p53-binding domain of human Mdm2: implications for p53 regulation. Proc Natl Acad Sci USA 2003; 100: 1645–1648.
Showalter SA, Bruschweiler-Li L, Johnson E, Zhang F, Bruschweiler R . Quantitative lid dynamics of MDM2 reveals differential ligand binding modes of the p53-binding cleft. J Am Chem Soc 2008; 130: 6472–6478.
Worrall EG, Worrall L, Blackburn E, Walkinshaw M, Hupp TR . The effects of phosphomimetic lid mutation on the thermostability of the N-terminal domain of MDM2. J Mol Biol 2010; 398: 414–428.
Zhan C, Varney K, Yuan W, Zhao L, Lu W . Interrogation of MDM2 phosphorylation in p53 activation using native chemical ligation: the functional role of Ser17 phosphorylation in MDM2 reexamined. J Am Chem Soc 2012; 134: 6855–6864.
Goldberg Z, Vogt Sionov R, Berger M, Zwang Y, Perets R, Van Etten RA et al. Tyrosine phosphorylation of Mdm2 by c-Abl: implications for p53 regulation. EMBO J 2002; 21: 3715–3727.
Waning DL, Lehman JA, Batuello CN, Mayo LD . c-Abl phosphorylation of Mdm2 facilitates Mdm2-Mdmx complex formation. J Biol Chem 2011; 286: 216–222.
Popowicz GM, Czarna A, Holak TA . Structure of the human Mdmx protein bound to the p53 tumor suppressor transactivation domain. Cell Cycle 2008; 7: 2441–2443.
Dawson PE, Kent SB . Synthesis of native proteins by chemical ligation. Annu Rev Biochem 2000; 69: 923–960.
Dawson PE, Muir TW, Clark-Lewis I, Kent SB . Synthesis of proteins by native chemical ligation. Science 1994; 266: 776–779.
Pazgier M, Liu M, Zou G, Yuan W, Li C, Li C et al. Structural basis for high-affinity peptide inhibition of p53 interactions with MDM2 and MDMX. Proc Natl Acad Sci USA 2009; 106: 4665–4670.
Li C, Pazgier M, Li C, Yuan W, Liu M, Wei G et al. Systematic mutational analysis of peptide inhibition of the p53-MDM2/MDMX interactions. J Mol Biol 2010; 398: 200–213.
Zhan C, Zhao L, Wei X, Wu X, Chen X, Yuan W et al. An ultrahigh affinity d-peptide antagonist Of MDM2. J Med Chem 2012; 55: 6237–6241.
Tarrant MK, Cole PA . The chemical biology of protein phosphorylation. Annu Rev Biochem 2009; 78: 797–825.
Kent SB . Total chemical synthesis of proteins. Chem Soc Rev 2009; 38: 338–351.
Muir TW, Sondhi D, Cole PA . Expressed protein ligation: a general method for protein engineering. Proc Natl Acad Sci USA 1998; 95: 6705–6710.
Schwarzer D, Cole PA . Protein semisynthesis and expressed protein ligation: chasing a protein's tail. Curr Opin Chem Biol 2005; 9: 561–569.
Noren CJ, Anthony-Cahill SJ, Griffith MC, Schultz PG . A general method for site-specific incorporation of unnatural amino acids into proteins. Science 1989; 244: 182–188.
Lu W, Gong D, Bar-Sagi D, Cole PA . Site-specific incorporation of a phosphotyrosine mimetic reveals a role for tyrosine phosphorylation of SHP-2 in cell signaling. Mol Cell 2001; 8: 759–769.
Lu W, Shen K, Cole PA . Chemical dissection of the effects of tyrosine phosphorylation of SHP-2. Biochemistry 2003; 42: 5461–5468.
Kallen J, Goepfert A, Blechschmidt A, Izaac A, Geiser M, Tavares G et al. Crystal structures of human MdmX (HdmX) in complex with p53 peptide analogues reveal surprising conformational changes. J Biol Chem 2009; 284: 8812–8821.
Schnolzer M, Alewood P, Jones A, Alewood D, Kent SB . In situ neutralization in Boc-chemistry solid phase peptide synthesis. Rapid, high yield assembly of difficult sequences. Int J Pept Protein Res 1992; 40: 180–193.
Otwinowski Z, Minor W . Processing of X-ray diffraction data collected in oscillation mode. Carter CW Jr, Sweet RM . Methods in Enzymology: Macromolecylar Crystallography, Part A. Academic Press: New York, 1997; 276: 307–326.
Murshudov GN, Vagin AA, Dodson EJ . Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 1997; 53: 240–255.
Emsley P, Cowtan K . Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60: 2126–2132.
Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 2010; 66: 12–21.
Acknowledgements
This work was partially supported by the US National Institutes of Health grant CA167296 and the Overseas Scholars Collaborative Research grant 81128015 from the National Natural Science Foundation of China (WL), and by the National Basic Research Program of China (973 Program, 2013CB932500) (W-YL). XC was the recipient of a scholarship from the China Scholarship Council. Portions of this research were carried out at the University of Maryland X-ray Crystallography Shared Service and the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program (P41RR001209), and the National Institute of General Medical Sciences.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Chen, X., Gohain, N., Zhan, C. et al. Structural basis of how stress-induced MDMX phosphorylation activates p53. Oncogene 35, 1919–1925 (2016). https://doi.org/10.1038/onc.2015.255
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.255
- Springer Nature Limited