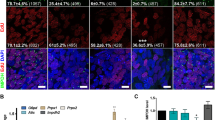
Abstract
This protocol describes how to convert the chromatin structure of sheep and mouse somatic cells into spermatid-like nuclei through the heterologous expression of the protamine 1 gene (Prm1). Furthermore, we also provide step-by-step instructions for somatic cell nuclear transfer (SCNT) of Prm1-remodeled somatic nuclei in sheep oocytes. There is evidence that changing the organization of a somatic cell nucleus with that which mirrors the spermatozoon nucleus leads to better nuclear reprogramming. The protocol may have further potential application in determining the protamine and histone footprints of the whole genome; obtaining 'gametes' from somatic cells; and furthering understanding of the molecular mechanisms regulating the maintenance of DNA methylation in imprinted control regions during male gametogenesis. The protocol is straightforward, and it requires 4 weeks from the establishment of the cell lines to their transfection and the production of cloned blastocysts. It is necessary for researchers to have experience in cell biology and embryology, with basic skills in molecular biology, to carry out the protocol.












Similar content being viewed by others
References
Gurdon, J.B. et al. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature 182, 64–65 (1958).
Wilmut, I. et al. Viable offspring derived from fetal and adult mammalian cells. Nature 385, 810–813 (1997).
Loi, P., Iuso, D., Czernik, M. & Ogura, A. A new, dynamic era for somatic cell nuclear transfer? Trends Biotechnol. Apr 22. pii: S0167-7799(16)30003-8 (2016).
Ogura, A. et al. Recent advancements in cloning by somatic cell nuclear transfer. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20110329 (2013).
Inoue, K. et al. Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science 330, 496–499 (2010).
Kishigami, S. et al. Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem. Biophys. Res. Commun. 340, 183–189 (2006).
Matoba, S. et al. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell 159, 884–895 (2014).
Hosseini, S.M. et al. Epigenetic modification with trichostatin A does not correct specific errors of somatic cell nuclear transfer at the transcriptomic level; highlighting the non-random nature of oocyte-mediated reprogramming errors. BMC Genomics 4, 16 (2016).
Cibelli, J. et al. (eds). Principles 2nd edn. (Academic Press, Elsevier), (2013).
Rathke, C. et al. Chromatin dynamics during spermiogenesis. Biochim. Biophys. Acta 1839, 155–168 (2014).
Gaucher, J. et al. From meiosis to postmeiotic events: the secrets of histone disappearance. FEBS J. 277, 599–604 (2010).
Goudarzi, A. et al. Genome-scale acetylation-dependent histone eviction during spermatogenesis. J. Mol. Biol. 426, 3342–3349 (2014).
Singh, J. & Rao, M.R. Interaction of rat testis protein TP, with nucleosome core particle. Biochem. Int. 17, 701–710 (1988).
Johnson, G.D. et al. The sperm nucleus: chromatin, RNA, and the nuclear matrix. Reproduction 14, 21–36 (2011).
De Vries, M. et al. Chromatin remodelling initiation during human spermiogenesis. Biol. Open. 1, 446–457 (2012).
Palmer, D.K., O'Day, K. & Margolis, R.L. The centromere specific histone CENP-A is selectively retained in discrete foci in mammalian sperm nuclei. Chromosoma 100, 32–36 (1990).
Hammoud, S.S. et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460, 473–478 (2009).
Saitou, M. & Kurimoto, K. Paternal nucleosomes: are they retained in developmental promoters or gene deserts? Dev. Cell 30, 6–8 (2014).
Samans, B. et al. Uniformity of nucleosome preservation pattern in Mammalian sperm and its connection to repetitive DNA elements. Dev. Cell 30, 23–35 (2014).
Van der Heijden, G.W. et al. Asymmetry in histone H3 variants and lysine methylationbetween paternal and maternal chromatin of the early mouse zygote. Mech. Dev. 122, 1008–1022 (2005).
Wu, F. et al. Testis-specific histone variants H2AL1/2 rapidly disappear from paternal heterochromatin after fertilization. J. Reprod. Dev. 54, 413–417 (2008).
Loppin, B. et al. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 437, 1386–1390 (2005).
Okuwaki, M. et al. Function of homo- and hetero-oligomers of human nucleoplasmin/nucleophosmin family proteins NPM1, NPM2 and NPM3 during sperm chromatin remodelling. Nucleic Acids Res. 40, 4861–4878 (2012).
Loi, P. et al. Placental abnormalities associated with post-natal mortality in sheep somatic cell clones. Theriogenology 65, 1110–1121 (2006).
Martínez-Soler, F., Kurtz, K., Ausió, J. & Chiva, M. Transition of nuclear proteins and chromatin structure in spermiogenesis of Sepia officinalis. Mol. Reprod. Dev. 74, 360–370 (2007).
Miller, D., Brinkworth, M. & Iles, D. Paternal DNA packaging in spermatozoa: more than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction 139, 287–301 (2010).
De Mateo, S. et al. Protamine 2 precursors and processing. Protein Pept. Lett. 18, 778–785 (2011).
Iuso, D. et al. Exogenous expression of human protamine 1 (hPrm1) remodels fibroblast nuclei into spermatid-like structures. Cell Rep. 13, 1765–1771 (2015).
Pivot-Pajot, C. et al. Acetylation-dependent chromatin reorganization by BRDT, a testis-specific bromodomain-containing protein. Mol. Cell. Biol. 23, 5354–5365 (2003).
Barckmann, B. & Simonelig, M. Control of maternal mRNA stability in germ cells and early embryos. Biochim. Biophys. Acta 1829, 714–724 (2013).
Loi, P. et al. Nuclei of nonviable ovine somatic cells develop into lambs after nuclear transplantation. Biol. Reprod. 67, 126–132 (2002).
Iuso, D. et al. A simplified approach for oocyte enucleation in mammalian cloning. Cell. Reprogram. 15, 490–494 (2013).
Wakayama, T. & Yanagimachi, R. Mouse cloning with nucleus donor cells of different age and type. Mol. Reprod. Dev. 58, 376–383 (2001).
Miyamoto, K. et al. Effects of synchronization of donor cell cycle on embryonic development and DNA synthesis in porcine nuclear transfer embryos. J. Reprod. Dev. 53, 237–246 (2007).
Wilmut, I. et al. Somatic cell nuclear transfer. Nature 10, 583–586 (2002).
Acknowledgements
This work was funded by grant MIUR/CNR, Program FIRB GA B81J12002520001 (GenHome), and the European Union's Horizon2020 Project "ERAofART" (GA 692185), to P.L. The authors are participating in the COST action FA 1201 'Epiconcept' Epigenetic and Peri-conception Environment. The authors warmly acknowledge L. Palazzese for help with the photos. The authors acknowledge FlowMetric Europe SpA (Italy), an affiliate company of FlowMetric (USA), for cell sorting.
Author information
Authors and Affiliations
Contributions
M.C., D.I., P.T., S.K. and P.L. contributed to the development of the methodology and to the description of the protocol; M.C. and D.I. performed the experiments and prepared the figures. M.C., D.I., S.K. and P.L. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Czernik, M., Iuso, D., Toschi, P. et al. Remodeling somatic nuclei via exogenous expression of protamine 1 to create spermatid-like structures for somatic nuclear transfer. Nat Protoc 11, 2170–2188 (2016). https://doi.org/10.1038/nprot.2016.130
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.130
- Springer Nature Limited
This article is cited by
-
Insights into the roles of sperm in animal cloning
Stem Cell Research & Therapy (2020)