Abstract
We performed a genome-wide association study for primary open-angle glaucoma (POAG) in 1,007 cases with high-pressure glaucoma (HPG) and 1,009 controls from southern China. We observed genome-wide significant association at multiple SNPs near ABCA1 at 9q31.1 (rs2487032; P = 1.66 × 10−8) and suggestive evidence of association in PMM2 at 16p13.2 (rs3785176; P = 3.18 × 10−6). We replicated these findings in a set of 525 HPG cases and 912 controls from Singapore and a further set of 1,374 POAG cases and 4,053 controls from China. We observed genome-wide significant association with more than one SNP at the two loci (P = 2.79 × 10−19 for rs2487032 representing ABCA1 and P = 5.77 × 10−10 for rs3785176 representing PMM2). Both ABCA1 and PMM2 are expressed in the trabecular meshwork, optic nerve and other ocular tissues. In addition, ABCA1 is highly expressed in the ganglion cell layer of the retina, a finding consistent with it having a role in the development of glaucoma.


Similar content being viewed by others
Accession codes
Change history
09 September 2014
In the version of this article initially published online, there were separate errors in the affiliations and in the Acknowledgments. In the affiliations, Chiea Chuen Khor and Chi Pui Pang are not affiliated with Sichuan Translational Medicine Hospital, Chinese Academy of Sciences, Chengdu, China, and the last four authors (Chiea Chuen Khor, Chi Pui Pang, Xinghuai Sun and Zhenglin Yang) jointly directed the work. In the Acknowledgments, two grants from the Biomedical Research Council, Singapore, were incorrectly assigned. Tien Yin Wong is the recipient of BMRC 09/1/35/19/616, and Eranga N. Vithana and Chiea Chuen Khor are the joint recipients of BMRC 10/1/35/19/675. These errors have been corrected for the print, PDF and HTML versions of this article.
References
Wang, Y.X., Xu, L., Yang, H. & Jonas, J.B. Prevalence of glaucoma in North China: the Beijing Eye Study. Am. J. Ophthalmol. 150, 917–924 (2010).
He, M. et al. Prevalence and clinical characteristics of glaucoma in adult Chinese: a population-based study in Liwan District, Guangzhou. Invest. Ophthalmol. Vis. Sci. 47, 2782–2788 (2006).
Stone, E.M. et al. Identification of a gene that causes primary open angle glaucoma. Science 275, 668–670 (1997).
Rezaie, T. et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science 295, 1077–1079 (2002).
Footz, T.K. et al. Glaucoma-associated WDR36 variants encode functional defects in a yeast model system. Hum. Mol. Genet. 18, 1276–1287 (2009).
Fingert, J.H. et al. Copy number variations on chromosome 12q14 in patients with normal tension glaucoma. Hum. Mol. Genet. 20, 2482–2494 (2011).
Pasutto, F. et al. Variants in ASB10 are associated with open-angle glaucoma. Hum. Mol. Genet. 21, 1336–1349 (2012).
Fingert, J.H. et al. Analysis of myocilin mutations in 1703 glaucoma patients from five different populations. Hum. Mol. Genet. 8, 899–905 (1999).
Alward, W.L. et al. Evaluation of optineurin sequence variations in 1,048 patients with open-angle glaucoma. Am. J. Ophthalmol. 136, 904–910 (2003).
Hauser, M.A. et al. Distribution of WDR36 DNA sequence variants in patients with primary open-angle glaucoma. Invest. Ophthalmol. Vis. Sci. 47, 2542–2546 (2006).
Allingham, R.R., Liu, Y. & Rhee, D.J. The genetics of primary open-angle glaucoma: a review. Exp. Eye Res. 88, 837–844 (2009).
Fingert, J.H. Primary open-angle glaucoma genes. Eye (Lond.) 25, 587–595 (2011).
Thorleifsson, G. et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat. Genet. 42, 906–909 (2010).
Burdon, K.P. et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat. Genet. 43, 574–578 (2011).
Wiggs, J.L. et al. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet. 8, e1002654 (2012).
Nakano, M. et al. Three susceptible loci associated with primary open-angle glaucoma identified by genome-wide association study in a Japanese population. Proc. Natl. Acad. Sci. USA 106, 12838–12842 (2009).
Nakano, M. et al. Common variants in CDKN2B-AS1 associated with optic-nerve vulnerability of glaucoma identified by genome-wide association studies in Japanese. PLoS ONE 7, e33389 (2012).
Macgregor, S. et al. Genome-wide association identifies ATOH7 as a major gene determining human optic disc size. Hum. Mol. Genet. 19, 2716–2724 (2010).
Ramdas, W.D. et al. A genome-wide association study of optic disc parameters. PLoS Genet. 6, e1000978 (2010).
Khor, C.C. et al. Genome-wide association study identifies susceptibility loci for dengue shock syndrome at MICB and PLCE1. Nat. Genet. 43, 1139–1141 (2011).
Matthijs, G. et al. Mutations in PMM2, a phosphomannomutase gene on chromosome 16p13, in carbohydrate-deficient glycoprotein type I syndrome (Jaeken syndrome). Nat. Genet. 16, 88–92 (1997).
Jensen, H., Kjaergaard, S., Klie, F. & Moller, H.U. Ophthalmic manifestations of congenital disorder of glycosylation type 1a. Ophthalmic Genet. 24, 81–88 (2003).
Thompson, D.A. et al. Retinal on-pathway deficit in congenital disorder of glycosylation due to phosphomannomutase deficiency. Arch. Ophthalmol. 130, 712–719 (2012).
Fukumoto, H., Deng, A., Irizarry, M.C., Fitzgerald, M.L. & Rebeck, G.W. Induction of the cholesterol transporter ABCA1 in central nervous system cells by liver X receptor agonists increases secreted Aβ levels. J. Biol. Chem. 277, 48508–48513 (2002).
Oram, J.F. & Lawn, R.M. ABCA1. The gatekeeper for eliminating excess tissue cholesterol. J. Lipid Res. 42, 1173–1179 (2001).
Brooks-Wilson, A. et al. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat. Genet. 22, 336–345 (1999).
Cheung, M.C., Mendez, A.J., Wolf, A.C. & Knopp, R.H. Characterization of apolipoprotein A-I– and A-II–containing lipoproteins in a new case of high density lipoprotein deficiency resembling Tangier disease and their effects on intracellular cholesterol efflux. J. Clin. Invest. 91, 522–529 (1993).
Pressly, T.A., Scott, W.J., Ide, C.H., Winkler, A. & Reams, G.P. Ocular complications of Tangier disease. Am. J. Med. 83, 991–994 (1987).
Chen, W. et al. Genetic variants near TIMP3 and high-density lipoprotein–associated loci influence susceptibility to age-related macular degeneration. Proc. Natl. Acad. Sci. USA 107, 7401–7406 (2010).
Willer, C.J. et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40, 161–169 (2008).
Guay, S.P. et al. ABCA1 gene promoter DNA methylation is associated with HDL particle profile and coronary artery disease in familial hypercholesterolemia. Epigenetics 7, 464–472 (2012).
Helgadottir, A. et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 316, 1491–1493 (2007).
Scott, L.J. et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316, 1341–1345 (2007).
Lettre, G. et al. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genet. 7, e1001300 (2011).
Howell, G.R. et al. Molecular clustering identifies complement and endothelin induction as early events in a mouse model of glaucoma. J. Clin. Invest. 121, 1429–1444 (2011).
Yeghiazaryan, K. et al. An enhanced expression of ABC 1 transporter in circulating leukocytes as a potential molecular marker for the diagnostics of glaucoma. Amino Acids 28, 207–211 (2005).
Kielar, D. et al. Rapid quantification of human ABCA1 mRNA in various cell types and tissues by real-time reverse transcription–PCR. Clin. Chem. 47, 2089–2097 (2001).
Zhang, X.J. et al. Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat. Genet. 41, 205–210 (2009).
Klein, R.J. et al. Complement factor H polymorphism in age-related macular degeneration. Science 308, 385–389 (2005).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Shi, Y. et al. Genetic variants at 13q12.12 are associated with high myopia in the Han Chinese population. Am. J. Hum. Genet. 88, 805–813 (2011).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Han, J.W. et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet. 41, 1234–1237 (2009).
Liu, X. et al. Association study of complement factor H, C2, Cfb, and C3 and age-related macular degeneration in a Han Chinese population. Retina 30, 1177–1184 (2010).
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913 (2007).
Willer, C.J., Li, Y. & Abecasis, G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Mogilenko, D.A. et al. Endogenous apolipoprotein A-I stabilizes ATP-binding cassette transporter A1 and modulates Toll-like receptor 4 signaling in human macrophages. FASEB J. 26, 2019–2030 (2012).
Sieg, S., Smith, D., Yildirim, Z. & Kaplan, D. Fas ligand deficiency in HIV disease. Proc. Natl. Acad. Sci. USA 94, 5860–5865 (1997).
Acknowledgements
We thank all the individuals with POAG and their families for participating in this study. The samples from Fudan University used for the analyses described in this manuscript were obtained from the EENT Hospital Biobank. We also thank B. Zhao and J. Sang of Beijing Tongren Hospital for sample collection. This research project was supported by the National Natural Science Foundation of China (81025006 (Z.Y.), 81170883 (Z.Y.), 81430008 (Z.Y.), 81200723 (Y. Chen), 81271007 (X. Zhu) and 81271005 (N.W.)); the National Basic Research Program of China (973 Program, 2011CB504604 to Z.Y.); the Department of Science and Technology of Sichuan Province, China (2014SZ0169 (Z.Y.), 2012SZ0219 (Z.Y.) and 2011jtd0020 (Z.Y.)); the Special Scientific Research Project of Health Professions (201302015 (X.S.)); research grants 467708 (C.P.P. and C.C.Y.T.) and 468810 (C.P.P. and C.K.S.L.) from the General Research Fund, Hong Kong; the National Medical Research Council, Singapore (NMRC/TCR/002-SERI/2008 (R626/47/2008TCR (T.A.)), CSA R613/34/2008 (T.A.), NMRC 0796/2003 (T.Y.W.) and STaR/0003/2008 (T.Y.W.)); the National Research Foundation of Singapore, Biomedical Research Council, Singapore (BMRC 09/1/35/19/616 (T.Y.W.), 08/1/35/19/550 (T.A.) and 10/1/35/19/675 (E.N.V. and C.C.K.)); and the Genome Institute of Singapore (GIS/12-AR2105 (C.C.K.)).
Author information
Authors and Affiliations
Contributions
Z.Y., X.S., C.P.P. and Y. Chen designed the study. Y. Chen, Y.L., E.N.V., L.J., T.Y.W., L.J.C., P.O.S.T., S.Q., Z.L., B.M., Q.L., C.G., C.K.S.L., W.C., C.C.Y.T., Y. Cheng, N.W., T.A., C.C.K., C.P.P., X.S. and Z.Y. recruited the participants. C.C.K., Y.L., X. Li and Z.Y. performed the genotyping. X. Zuo, Y. Chen, Y.L., C.C.K., X. Liu, X. Zhang, Q.Y. and Z.Y. performed the statistical analysis. B.G., X. Zhu, X. Liu and Z.Y. performed the immunohistochemistry and gene expression studies. Z.Y. wrote the initial draft, with edits from C.C.K. Q.Y. and C.P.P. corrected the English spelling and grammar. All authors critically revised and gave final approval of this manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 2 Principal-components analysis (PCA) for the GWAS samples.
PCA was performed on the 1,971 GWAS samples (966 cases and 1,005 controls).
Supplementary Figure 3 Quantile-quantile plots of observed P values (−log10 P) for association.
The genomic inflation factor (λ) in the GWAS was 1.016.
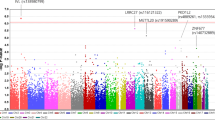
Supplementary Figure 4 Association results for genotyped SNPs at the discovery stage.
The genome-wide distribution of −log10 P values from the unadjusted Cochran-Armitage trend test is shown across the chromosomes. Values are shown for 870,261 SNPs that were of sufficient quality, after quality control filtering, in 966 unrelated Han Chinese cases with HPG and in 1,005 unrelated Han Chinese controls, after genetic matching and correction for the inflation factor of 1.016. Each chromosome is depicted in a different color. −log10 (UNADJ), the −log10 P value of the Cochran-Armitage trend test, was not adjusted for multiple testing.
Supplementary Figure 5 LD block analysis of the POAG-associated SNPs at the chromosome 9 ABCA1 locus (top) and the chromosome 16 PMM2 locus (bottom).
Numbers within the filled diamonds reflect the pairwise LD value between the markers using the r2 algorithm.
Supplementary Figure 6 LD block analysis of the disease-associated SNPs in the ABCA1 gene and their relationship with previously described markers also mapping to ABCA1 showing association with HDL and CAD.
LD block analysis was performed using CHB genotyping data from the dbSNP database. Previous studies indicated that rs4149274 in intron 5 of the ABCA1 gene was significantly associated with HDL and CAD, and rs4149274 and rs1883025 in intron 2 of the ABCA1 gene were shown to be significantly associated with AMD. rs2164560, rs2472459 and rs2472519 are associated with POAG in this study. Values for r2 (left) and D′ (right) are shown in the block.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Supplementary Tables 1–7 and Supplementary Note. (PDF 3242 kb)
Rights and permissions
About this article
Cite this article
Chen, Y., Lin, Y., Vithana, E. et al. Common variants near ABCA1 and in PMM2 are associated with primary open-angle glaucoma. Nat Genet 46, 1115–1119 (2014). https://doi.org/10.1038/ng.3078
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3078
- Springer Nature America, Inc.
This article is cited by
-
Profile of Dr. Zhenglin Yang
Science China Life Sciences (2023)
-
Primary open-angle glaucoma risk prediction with ABCA1 and LOC102723944 variants and their genotype–phenotype correlations in southern Chinese population
Molecular Genetics and Genomics (2023)
-
Cholesterol homeostasis regulated by ABCA1 is critical for retinal ganglion cell survival
Science China Life Sciences (2023)
-
Genetic variants associated with glaucomatous visual field loss in primary open-angle glaucoma
Scientific Reports (2022)
-
TIPARP is involved in the regulation of intraocular pressure
Communications Biology (2022)