Abstract
Sexual size dimorphism (SSD), a difference in body size between sexes, is common in many taxa. In insects, females are larger than males in >70% of all taxa in most orders. The fruit fly, Drosophila melanogaster is one prominent model organism to investigate SSD since its clear and representative female-biased SSD and its growth regulation are well studied. Elucidating the number and nature of genetic elements that can potentially influence SSD would be helpful in understanding the evolutionary potential of SSD. Here, we investigated the SSD pattern caused by artificially introduced genetic variation in D. melanogaster, and examined whether variation in SSD was mediated by the sex-specific modification of developmental time. To map the genomic regions that had effects on sexual wing size and/or developmental time differences (SDtD), we reanalyzed previously published genome-wide deficiency mapping data to evaluate the effects of 376 isogenic deficiencies covering a total of ~67% of the genomic regions of the second and third chromosomes of D. melanogaster. We found genetic variation in SSD and SDtD generated by genomic deficiencies, and a negative genetic correlation between size and development time. We also found SSD and SDtD allometries that are not qualitatively congruent, which however overall at best only partly help in explaining the patterns found. We identified several genomic deficiencies with the tendency to either exaggerate or suppress SSD, in agreement with quantitative genetic null expectations of many loci with small effects. These novel findings contribute to a better understanding of the evolutionary potential of sexual dimorphism.
Similar content being viewed by others
Introduction
Body size is one of the key features of organisms, and many ecological traits such as resource acquisition rate, reproductive capability and survival generally show size dependency. Differences in body size between males and females, so-called sexual size dimorphism (SSD), are common in many taxa (Fairbairn, 1997, 2005; Stillwell et al., 2010). In animals, male-biased SSD predominates among birds and mammals while female-biased SSD predominates among poikilothermic vertebrates and invertebrates (Fairbairn, 1997). Key proximate factors behind SSD are sex differences in growth rate and duration (that is, development time), and natural selection that drives the growth of males and females apart will result in changes in adult SSD (Badyaev, 2002).
The pattern of SSD in insects is consistent with that of most other invertebrates: females are larger than males in >70% of the taxa in all major insect orders except Odonata (Stillwell et al., 2010). Both growth rate and developmental time differences between sexes have been shown to contribute quantitatively to SSD in arthropods (Blanckenhorn et al., 2007a). Even though SSD is widespread in insects and developmental processes clearly affect final body size, our general understanding of the causal proximate factors of body size variation is still rudimentary, as detailed genetic and developmental mechanisms underlying SSD have been studied only in few species. The fruit fly, Drosophila melanogaster is one prominent model organism to investigate SSD regulation because it shows clear and representative female-biased SSD and its growth regulation is very well studied. Testa et al. (2013) described complete growth profiles of D. melanogaster males and females, and identified sex-specific growth factors responsible for SSD. They found that growth rate and critical size for pupation significantly contributed to SSD, while developmental time did not. They also found that SSD was lost in insulin-signaling mutants, suggesting that the insulin-signaling pathway plays a critical role in the formation of SSD in D. melanogaster. At the moment, other than this example, little is known about the molecular genetic mechanisms of SSD in insects. Elucidating the number and nature of genetic elements in a genome that can potentially influence the degree and direction of SSD clearly should be helpful in understanding the evolutionary potential of SSD. In the animal kingdom, it is common that male-biased SSD increases and female-biased SSD decreases with body size, an evolutionary pattern termed ‘Rensch’s rule’ (Fairbairn, 1997). How this pattern arises across wide range of taxa is still unclear, but the pattern of genetic variation associated with SSD might help explain it.
In this study, we investigated the SSD pattern caused by artificially introduced genetic variation in D. melanogaster, and examined whether the effect on SSD was mediated by the sex-specific modification of developmental time. To map the genomic regions that had effects on SSD and/or the sexual developmental time difference (SDtD), we here reanalyze the genome-wide deficiency mapping data of Takahashi et al. (2011a,b), and evaluate the effect of 376 isogenic deficiencies covering a total of ~67% of the genomic regions of the second and the third chromosomes of D. melanogaster.
Materials and methods
Datasets
Deficiency strains
To assess the effect of artificially introduced genetic variation on the SSD and SDtD, and in search for genomic regions with effects on SSD, we reanalyzed the deficiency screening data published in Takahashi et al. (2011a, b). These authors measured multiple phenotypic traits for a collection of DrosDel isogenic deficiency strains of D. melanogaster (Ryder et al., 2004) and the corresponding control strain. Because the breakpoints of the deletions in the deficiency strains were determined at a single base-pair resolution, they are a suitable tool for high resolution mapping of the candidate genomic regions (Ryder et al., 2004; Ryder et al., 2007). The control strain (DSK001: w1118iso; 2iso; 3iso), whose X, second and third chromosomes are isogenized, share the same genetic background with the deficiency strains (Ryder et al., 2004; Ryder et al., 2007). Here, we focus on 376 deficiency strains whose trait scores were measured in both studies (Takahashi et al., 2011a, b; Figure 1). The deletions overall covered about 67% of the genomic regions of the second and the third chromosomes, and individual deletions encompassed about 47 genes on average.
Body size and developmental time
Wing size is known to correlate with the sizes of other body parts and is often used as an indicator of body size in D. melanogaster (Gilchrist and Partridge, 1999; Gilchrist et al., 2004). Based on the wing size and thorax length data of 20 species of the genus Drosophila, SSD in wing size highly correlates with SSD in thorax length (correlation coefficient: 0.848, P<0.00001; as calculated with the index described in the following section using data from table 1 in Huey et al., (2006)), indicating that wing size SSD can appropriately represent whole-body SSD. Takahashi et al. (2011b) measured centroid size based on eight landmarks placed on the wing veins. Here, we used these centroid size data to evaluate wing size dimorphism between females and males. To investigate whether genetic variation in SDtD explains the SSD in wing size in D. melanogaster, we additionally considered corresponding developmental time data (measured as days from oviposition to eclosion) from Takahashi et al. (2011a). Because the flies used for the measurement of wing size and developmental time were obtained from the same experiment, the experimental conditions, such as rearing temperature, fly food, larval density and all the experimental equipment, were identical in both studies (Takahashi et al., 2011a, b), so the results are directly comparable. Because of the homozygous lethality of most of the deficiencies, all traits were measured for deficiency-control heterozygotes (Df/+).
Sexual dimorphism
To evaluate SSD, we calculated one of the size dimorphism indices (SDIs) listed in Lovich and Gibbons (1992):

Takahashi et al. (2011a, b) reared 100 eggs per vial and set up five replicate vials for each deficiency-control heterozygote (Df/+) and the control genotype (+/+). We calculated SDIs from the vial-level average centroid size of right and left wings based on up to three females and males that emerged from each replicate vial, and used genotype-level average SDI for correlation analyses. We analogously evaluated SDtD with the same formula and defined it as the developmental time difference index (DtDI).
Statistical analysis
To describe among-genotype SSD and SDtD patterns, we performed major axis regression analyses for female and male wing size and developmental time. We tested whether the regression coefficients significantly differ from unity based on the 95% confidence intervals of the regression coefficient. We then performed multiple regression analyses using wing size as the dependent variable, developmental time, sex and their interaction as fixed independent variables to test the effect of developmental time on wing size and its sex specificity.
To evaluate the effects of deletions on the SDI and DtDI, we performed pairwise comparisons between +/+ and each Df/+ using one-way analysis of variance. To correct for multiple tests with different Df/+ genotypes, we applied the Benjamini–Hochberg procedure to control for the false discovery rate (Benjamini and Hochberg, 1995). In addition, we calculated the effect size (Cohen’s d) for individual comparisons between +/+ and Df/+ to draw a robust conclusion, disregarding sample size variation and the existence of outliers, and to make the results of different tests comparable. Finally, we evaluated with a one-sample t-test whether the deficiencies on average produced biased Cohen’s d scores from zero. All analyses were performed with the statistical software R 2.15.3 (R Foundation for Statistical Computing, Vienna, Austria).
Results
Sexual dimorphism patterns
The estimated regression slope for female on male wing size was significantly smaller than unity (P<0.05; Figure 2a), indicating that deficiencies had size-dependent effects on the degree of SSD, and that there was greater variance in female than in male wing size (variance in females: 0.00077; variance in males: 0.00059). Within the range of wing sizes observed in this study, female-biased SSD increased with wing size (Figure 2a). In the analogous regression of developmental time of females on males, the estimated slope was not significantly different from unity (P>0.05; Figure 2b), indicating equal amounts of variance in female and male developmental time, such that SDtD remains constant over the range of developmental times produced by the deficiencies here.
Allometric regression plots of mean male on mean female natural-log-transformed (a) wing size and (b) developmental time for 376 Df/+ and +/+ genotypes. Broken line represents Y=X; solid line represents the estimated regression line. Slope and intercept estimates are given with upper and lower limits of 95% confidence intervals that are asymmetric around the estimates in parentheses.
Multiple regression analysis showed that both developmental time and sex had significant effects on wing size (developmental time: P<0.0001, sex: P=0.021), while their interaction was not significant (P=0.378), indicating that the effect of developmental time on wing size was consistent for the sexes and did not differentially affect the general SSD pattern (Figure 3). Developmental time was negatively related to wing size, that is, longer developmental time resulted in smaller wing size (Figure 3).
Mean developmental time and wing size of 376 Df/+ and +/+ genotypes for females (○) and males (Δ). Solid line represents the estimated regression line for the larger females and broken line represents the estimated line for males. Slope and intercept estimates are given with 95% confidence intervals in parentheses.
Effect of deficiencies on sexual dimorphism
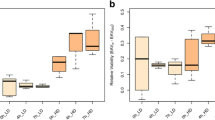
The control genotype (+/+) showed significantly female-biased SSD (mean centroid size±s.d. for male: 3.121±0.046, for female: 3.528±0.026; P<0.0001 using a t-test, SDI=−0.116), while its SDtD was also slightly female biased but not so significantly (mean developmental time±s.d. for male: 13.455±0.245, for female: 13.501±0.126; P=0.724, DtDI=−0.003). Cohen’s d, a measure of effect size of individual deficiencies calculated for both traits showed unimodal distributions centered around zero (Cohen’s d for SDI: mean −0.698, range −4.010 to 2.870; Cohen’s d for DtDI: mean −0.450, range −4.592 to 2.859), indicating that most deficiencies had small effects on SDI and DtDI (Figure 4). Nevertheless, for both SDI and DtDI, Cohen’s d was significantly biased in the negative direction (SDI: t=−16.261, P<0.0001; DtDI: t=−8.630, P<0.0001), indicating that the deficiencies tended to reduce SSD and SDtD on average. Effect size for SDI was significantly negatively correlated with that for DtDI (correlation coefficient: −0.261, P<0.0001), suggesting a genetic correlation between SSD and SDtD (Figure 5).
SDI of Df/+ genotypes with extreme top and bottom 2.5% effect sizes plus the SDI of +/+ controls are shown in Figure 6. Some of the Df/+ showed significantly larger or smaller SDI compared to +/+ when tested individually, but after adjustment of P-values for multiple comparisons, no Df/+ differed significantly from +/+ (Figure 6).
Size dimorphism index (SDI) scores of Df/+ genotypes with top and bottom 2.5% effect size (solid bars) relative to the control +/+ (open bar). Error bars represent standard errors. Asterisks denote statistically significant differences between the +/+ and each Df/+ genotype: *P<0.05, **P<0.01 and ***P<0.001.
Discussion
In this study, a collection of 376 isogenic deficiencies revealed a significant positive genetic correlation between male and female wing sizes in D. melanogaster, as can be generally expected (Fairbairn et al., 2007). However, the slope of the regression of male on female wing size was significantly less than unity (that is, hypoallometric, <1), such that the degree of female-biased SSD increased with body size over the body size range observed here (Figure 2a). This within-species SSD pattern is opposite to the generally hyperallometric slope (>1) obtained among a wide range of Drosophila species, which in general display female-biased SSD regardless of the measures of body size used (Huey et al., 2006; Blanckenhorn et al., 2007b). The potential genetic variation in SSD found here for our 376 isogenic deficiency heterozygotes thus is opposite to what is predicted by Rensch’s rule (Rensch, 1960). This is equivalent to a pattern of (phylo)genetic variation in male body size being generally greater than that of females (Blanckenhorn et al., 2007b). In contrast, our results here rather agree with opposite patterns of phenotypic variance found in insects (Teder and Tammaru, 2005). Blanckenhorn et al. (2007b) also found incongruent SSD patterns among species (according to Rensch’s rule) vs among populations and among families within species (inconsistent with Rensch’s rule) for sepsid flies with female-biased SSD, but not for scathophagid flies with male-biased SSD. If the genetic variation in SSD we documented here reflects the general pattern in Drosophila, the SSD variation with body size observed among Drosophila species (Blanckenhorn et al., 2007b; Huey et al., 2006) is unlikely to be a mere by-product of body size evolution within species; instead our results suggest that natural selection might directly act on dimorphism itself, as intra-specific patterns, and thus presumably mechanisms, do not predict inter-specific patterns of SSD. The problem is akin to the relationship between ontogenetic and static allometry, which do not necessarily have to be congruent (Cheverud, 1982; Pelabon et al., 2013). However, as genetic variation in the current study was artificially introduced by using a collection of isogenic deficiencies, we cannot be sure that it actually reflects natural genetic variation in Drosophila. In each Df/+ genotype in the current study, we expected a 50% reduction in the gene expression level of the genes encompassed by any deficiency compared to the control genotype. In total, 8783 genes were encompassed by the 376 deficiencies in the current study, and some of them might not show genetic variation in expression level in wild fly populations. Hence, what we uncovered in the current study must be qualified as potential genetic variation in SSD, and further genetic study of natural populations is necessary to understand the evolutionary potential of SSD in Drosophila spp.
Our study also revealed the expected genetic correlation between male and female developmental times (Figure 2b), but the slope of this regression did not significantly differ from unity. The hypoallometric pattern for SSD and isometric pattern for SDtD are thus qualitatively inconsistent, and therefore the sex differences, as well as the SSD allometry not following Rensch’s rule displayed in Figure 2, are only distinct for wing size (and not developmental time), which is also true across Drosophila species (Blanckenhorn et al., 2007a). Overall, therefore, sex differences in developmental time cannot explain sex differences in body size, and SDtD cannot explain the allometric pattern of SSD in any simple way, confirming similar conclusions of previous studies (Blanckenhorn et al., 2007a; Testa et al., 2013). Contrary to expectation, we here also found a significant negative genetic correlation between wing size and developmental time that was equal for both sexes (Figure 3). Again, this lack of interaction between developmental time and sex in mediating wing size cannot help explain the differential body size allometry of males and females opposite to Rensch’s rule found here. Whereas Nunney (1996) found a strongly positive within-species genetic correlation between body size and developmental time in an artificial selection study of D. melanogaster, the two traits are typically correlated negatively across environments (for example, food restriction produces smaller flies that take longer to develop: Blanckenhorn, 1999) or across (clinal) populations (James et al., 1995). Whether and why this confers an adaptive advantage in nature is not completely clear, but life history optimality models generally predict the above-mentioned response to food limitation (Berrigan and Koella, 1994; Stearns and Koella, 1986). In the current dataset, deleterious effects of some of the deficiencies might be manifested as slow development (that is, long developmental time) and small body size, while advantageous effects of other deficiencies might be opposite. Alternatively, it is conceivable that most deficiencies produced less fit deviations in terms of developmental time and body size from a possibly existing optimal phenotype, in which case mostly ‘poor’ genotypes were produced that mirror the effect typically produced by ‘poor’ environments, thus explaining the obtained negative correlation (Stearns and Koella, 1986), but this remains a conjecture.
When examining the effect of individual deficiencies on SDI and DtDI, effect size values showed bell-shape distributions centering around zero in both cases (Figure 4). These results indicate that most of the deficiencies had little effect on SDI and/or DtDI, and suggest that SSD and SDtD are under robust genetic control of many loci with small effect in D. melanogaster, confirming the null expectation from quantitative genetic theory (Falconer, 1989; Roff, 1997; Lynch and Walsh, 1998). Nevertheless, the effect of the deficiencies was significantly biased toward negative deviations, indicating that they tend to reduce SSD and SDtD on average. The relationship between the effect sizes for DtDI and SDI also showed a significantly negative correlation (Figure 5), indicating that the deficiencies that influenced DtDI positively tended to influence SDI negatively. Thus there is a negative genetic correlation between SSD and SDtD in D. melanogaster. The developmental mechanism mediating this negative genetic correlation is still unknown at the moment, but at least one of our results suggests that SDtD and SSD may be causally linked. Modification of developmental time by the deficiencies combined with the negative genetic correlation between wing size and developmental time might cause the negative genetic correlation between SSD and SDtD.
The extreme deficiencies within the top or bottom 2.5% effect sizes (20 deficiencies in total; Figure 6) showed relatively clear effects on SDI that were statistically significant when tested individually, but not after adjusting for multiple comparisons, and they were not distinguishable from random noise. That is, an approximately equal number of deficiencies with strong effect exaggerated SSD while others suppressed SSD relative to the control treatment. Again, this suggests that there are multiple genetic factors with mostly small effects that can potentially influence the degree and direction of SSD in D. melanogaster. Among the 20 deficiencies with top or bottom 2.5% effect size on SDI, only one deficiency, Df(2 L)ED105, showed a strong pleiotropic effect on DtDI, again not more than expected by chance. Despite our conclusion above that the genetic correlation between body size and development time is a general feature of the genetic architecture underlying SSD and SDtD, it therefore nevertheless seems that many genetic factors influence body size independently of development time, and SSD independently of SDtD.
In conclusion, we found robust genetic regulation of SSD in D. melanogaster affected by many loci of small effect, confirming a null expectation from quantitative genetic theory. Genetic variation in SSD generated by genomic deficiencies violated Rensch’s rule, such that the within-species allometric pattern is opposite to the among-species pattern in Drosophila (Huey et al., 2006; Blanckenhorn et al., 2007b). Although sex differences in development time are principally expected to produce corresponding sex differences in body size (Blanckenhorn et al., 2007a; Teder, 2013), the SSD and SDtD allometry patterns found here (Figure 2) do not agree qualitatively, thus failing to provide explanatory power regarding their relationship. The lack of a direct connection may be mediated by the negative genetic correlation between wing size and development time we found (Figure 3), which was clearly determined by the induced genetic effects (Figure 5). This negative correlation is unexpected within species because, typically, it takes more time to get large, predicting a positive association, but is predicted by life history theory to occur across environments (Stearns and Koella, 1986; Nunney, 1996; Blanckenhorn, 1999). In addition, a negative genetic correlation between SSD and SDtD was revealed by the genomic deficiencies, but again, the developmental mechanism mediating the negative genetic correlation is still unknown. Lastly, we identified several genomic deficiencies with tendency to either exaggerate or suppress SSD, probably not more than expected assuming many loci of small effect distributed randomly over the genome. These novel findings contribute to a better understanding of the evolutionary potential of sexual dimorphism.
Data Archiving
There were no data to deposit.
References
Badyaev AV . (2002). Growing apart: an ontogenetic perspective on the evolution of sexual size dimorphism. Trends Ecol Evol 17: 369–378.
Benjamini Y, Hochberg Y . (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57: 289–300.
Berrigan D, Koella JC . (1994). The evolution of reaction norms: simple models for age and size at maturity. J Evol Biol 7: 549–566.
Blanckenhorn WU . (1999). Different growth responses to food shortage and temperature in three insect species with similar life histories. Evol Ecol 13: 395–409.
Blanckenhorn WU, Dixon AF, Fairbairn DJ, Foellmer MW, Gibert P, van der Linde K et al. (2007a). Proximate causes of Rensch's rule: does sexual size dimorphism in arthropods result from sex differences in development time? Am Nat 169: 245–257.
Blanckenhorn WU, Meier R, Teder T . (2007b). Rensch's rule in insects: patterns among and within species. In: Fairbairn DJ, Blanckenhorn WU, Székely T (eds). Sex, Size and Gender Roles. Oxford University Press: New York.
Cheverud JM . (1982). Relationships among ontogenetic, static, and evolutionary allometry. Am J Phys Anthropol 59: 139–149.
Fairbairn DJ . (1997). Allometry for sexual size dimorphism: pattern and process in the coevolution of body size in males and females. Annu Rev Ecol Syst 28: 659–687.
Fairbairn DJ . (2005). Allometry for sexual size dimorphism: testing two hypotheses for Rensch's rule in the water strider Aquarius remigis. Am Nat 166: S69–S84.
Fairbairn DJ, Blanckenhorn WU, Székely T . (2007) Sex, Size and Gender Roles: Evolutionary Studies of Sexual Size Dimorphism. Oxford University Press: New York.
Falconer DS . (1989). Introduction to quantitative genetics. Longman Scientific: Harlow.
Gilchrist AS, Partridge L . (1999). A comparison of the genetic basis of wing size divergence in three parallel body size clines of Drosophila melanogaster. Genetics 153: 1775–1787.
Gilchrist GW, Huey RB, Balanya J, Pascual M, Serra L . (2004). A time series of evolution in action: a latitudinal cline in wing size in South American Drosophila subobscura. Evolution 58: 768–780.
Huey RB, Moreteau B, Moreteau JC, Gibert P, Gilchrist GW, Ives AR et al. (2006). Sexual size dimorphism in a Drosophila clade, the D. obscura group. Zoology (Jena) 109: 318–330.
James AC, Azevedo RB, Partridge L . (1995). Cellular basis and developmental timing in a size cline of Drosophila melanogaster. Genetics 140: 659–666.
Lovich JE, Gibbons JW . (1992). A review of techniques for quantifying sexual size dimorphism. Growth Dev Aging 56: 269–281.
Lynch M, Walsh B . (1998) Genetics and Analysis of Quantitative Traits. Sinauer Associates: Sunderland, MA.
Nunney L . (1996). The response to selection for fast larval development in Drosophila melanogaster and its effect on adult weight: an example of a fitness trade-off. Evolution 50: 1193–1204.
Pelabon C, Bolstad GH, Egset CK, Cheverud JM, Pavlicev M, Rosenqvist G . (2013). On the relationship between ontogenetic and static allometry. Am Nat 181: 195–212.
Rensch B . (1960) Evolution Above the Species Level. Columbia University Press: New York.
Roff DA . (1997) Evolutionary Quantitative Genetics. Chapman and Hall: New York.
Ryder E, Ashburner M, Bautista-Llacer R, Drummond J, Webster J, Johnson G et al. (2007). The DrosDel deletion collection: a Drosophila genomewide chromosomal deficiency resource. Genetics 177: 615–629.
Ryder E, Fiona B, Michael A, Rosa B-L, Darin C, Jenny D et al. (2004). The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167: 797–813.
Stearns SC, Koella J . (1986). The evolution of phenotypic plasticity in life history traits: predictions of reaction norms for age and size at maturity. Evolution 40: 893–914.
Stillwell RC, Blanckenhorn WU, Teder T, Davidowitz G, Fox CW . (2010). Sex differences in phenotypic plasticity affect variation in sexual size dimorphism in insects: from physiology to evolution. Annu Rev Entomol 55: 227–245.
Takahashi KH, Okada Y, Teramura K . (2011a). Genome-wide deficiency mapping of the regions responsible for temporal canalization of the developmental processes of Drosophila melanogaster. J Hered 102: 448–457.
Takahashi KH, Okada Y, Teramura K, Tsujino M . (2011b). Deficiency mapping of the genomic regions associated with effects on developmental stability in Drosophila melanogaster. Evolution 65: 3565–3577.
Teder T . (2014). Sexual size dimorphism requires a corresponding sex difference in development time: a meta-analysis in insects. Funct Ecol 28: 479–486.
Teder T, Tammaru T . (2005). Sexual size dimorphism within species increases with body size in insects. Oikos 108: 321–334.
Testa ND, Ghosh SM, Shingleton AW . (2013). Sex-specific weight loss mediates sexual size dimorphism in Drosophila melanogaster. PLoS ONE 8: e58936.
Acknowledgements
This work was financially supported by Special Coordination Funds for Promoting Sciences and Technology of The Ministry of Education, Sport, Culture, Science and Technology of Japan, and a Grant-in-Aid for Scientific Research (KAKENHI 25440212) to KHT.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Takahashi, K., Blanckenhorn, W. Effect of genomic deficiencies on sexual size dimorphism through modification of developmental time in Drosophila melanogaster. Heredity 115, 140–145 (2015). https://doi.org/10.1038/hdy.2015.1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2015.1
- Springer Nature Switzerland AG
This article is cited by
-
The sex-limited effects of mutations in the EGFR and TGF-β signaling pathways on shape and size sexual dimorphism and allometry in the Drosophila wing
Development Genes and Evolution (2016)