Abstract
Recognition of pathogens by Toll-like receptors (TLR) activate multiple signaling cascades and expression of genes tailored to mount a primary immune response, inflammation, cell survival and apoptosis. Although TLR-induced activation of pathways, such as nuclear factor kappaB (NF-κB) and mitogen-activated protein kinases (MAPK), has been well studied, molecular entities controlling quantitative regulation of these pathways during an immune response remain poorly defined. We identified Sam68 as a novel regulator of TLR-induced NF-κB and MAPK activation. We found that TLR2 and TLR3 are totally dependent, whereas TLR4 is only partially dependent on Sam68 to induce the activation of NF-κB c-Rel. Absence of Sam68 greatly decreased TLR2- and TLR3-induced NF-κB p65 activation, whereas TLR4-induced p65 activation in a Sam68-independent manner. In contrast, Sam68 appeared to be a negative regulator of MAPK pathways because absence of Sam68 enhanced TLR2-induced activation of extracellular signal-regulated kinases (ERK) and c-Jun N-terminal kinases (JNK). Interestingly, TLR2- and TLR3-induced gene expression showed a differential requirement of Sam68. Absence of Sam68 impaired TLR2-induced gene expression, suggesting that Sam68 has a critical role in myeloid differentiation primary response gene 88-dependent TLR2 signaling. TLR3-induced gene expression that utilize Toll/Interleukin-1 receptor-domain-containing adapter-inducing beta interferon pathway, depend only partially on Sam68. Our findings suggest that Sam68 may function as an immune rheostat that balances the activation of NF-κB p65 and c-Rel, as well as MAPK, revealing a potential novel target to manipulate TLR signaling.
Similar content being viewed by others
INTRODUCTION
One of the key roles of the immune system is responding to, and limiting infection by, microorganisms including bacteria, viruses, fungi and protozoans. Immunological dogma separates the immune system into two main components: innate and adaptive immunity. Innate immune responses are characterized by early, potent and ‘non-specific’ inflammatory responses in infected tissues meant to limit microbial growth. Adaptive immunity involves the activation of T- and B lymphocytes expressing high-affinity receptors for individual pathogenic antigens that function to drive the protective immune response necessary to clear infection, as well as provide immunological memory to prevent future infections with the same pathogen. The purported ‘non-specific’ nature of the innate immune response is attributed to the innate immune cells expressing receptors termed pattern-recognition receptors (PRRs). PRRs function to recognize conserved microbial structures and/or endogenous host molecules indicative of cellular injury or stress.1 These PRRs can be expressed on the cell surface, in endosomal compartments, or in the cytoplasm of the cell, allowing immune cells to recognize pathogenic infection whether the pathogen resides inside or outside of cells. There are currently four major classes of PRRs: Toll-like receptors (TLRs),2 C-type lectin receptors,3 Nucleotide oligomerization domain-like receptors4, 5 and Retinoic acid-inducible gene 1-like receptors.6 These receptors are capable of recognizing a wide range of molecules, including lipopeptides,7 lipopolysaccharide (LPS),8 flagellin,9, 10, 11 bacterial DNA,12 viral single-stranded/double-stranded RNA,13, 14 fungal components15, 16 and crystalline solids.17, 18 Infectious studies in mice have confirmed the critical role of these PRRs in response to pathogens. TLRs were the first family of these receptors to be discovered and are the most well studied to date.19 This study will focus on regulation of intracellular signaling following TLR engagement.
Upon engagement of TLRs by their cognate ligand(s), multiple intracellular signaling cascades are activated, including nuclear factor kappaB (NF-κB), mitogen-activated protein kinases (MAPK) and interferon regulatory factors (IRFs).20, 21 These cascades result in an altered gene expression and typically lead to the induction of so-called pro-inflammatory cytokines/chemokines, which serve to recruit and activate other immune cells at the site of infection. TLRs are known to activate signaling through the utilization of two adapter proteins, myeloid differentiation primary response gene 88 (MyD88) and Toll/Interleukin-1 receptor-domain-containing adapter-inducing interferon-β (TRIF).2 Most TLRs utilize only one of these adapters to signal, with TLR4 being the lone exception that utilizes both. In general, cell surface TLRs utilize MyD88 and endosomal TLRs utilize TRIF. This was confirmed recently by the discovery that TLR4 signals via MyD88 when at the cell surface, but via TRIF when internalized into an endosome, explaining the previous paradox of TLR4 utilizing both signaling adapters.22 Thus, adapter proteins have a major role in integrating the membrane proximal signaling events by acting as a scaffolding backbone.
We recently discovered that a bona fide RNA-binding protein called Src-associated substrate in mitosis of 68 kDa (Sam68), acts as a critical adapter in the tumor necrosis factor receptor (TNFR)-signaling complex.23 Sam68 has several structural features that facilitate its function as an adapter protein. It contains a K-homology domain that allows binding to nucleotides, proline-rich regions to bind SH3 domain containing proteins, tyrosine phosphorylation sites that allows binding to SH2 domain containing proteins, as well as arginine methylation sites.24 While studying the role of Sam68 in a TNF signaling pathway, we found that absence of Sam68 leads to impaired activation of the kinases such as IκB kinase and Transforming growth factor beta-activated kinase 1 recruited to the TNFR after stimulation and greatly decreases TNF-induced NF-κB activation.23
Because pro-inflammatory signaling by TNF and TLR share several cellular proteins to activate the NF-κB pathway, we hypothesized that Sam68 is a component of the TLR-induced NF-κB activation pathway. NF-κB is an evolutionarily conserved family of transcription factors that controls both innate and adaptive immune responses, cell survival and apoptosis. NF-κB acts as dimers composed of the combination of the five subunits of the family, RelA (p65), RelB, c-Rel, p50 (and its precursor p105) and p52 (and its precursor p100).25 In the basal state, NF-κB is bound to the inhibitor proteins of kappaB family (IκB) and mostly resides in the cytoplasm. Activation occurs by signal-induced degradation of the IκB proteins and liberation of bound NF-κB, which translocates to the nucleus and activates transcription.
TLR activation has also been shown to activate the MAPK signaling pathways, including extracellular signal-regulated kinases (ERK) and c-Jun N-terminal kinases (JNK) pathways.26 ERK activation is marked by its phosphorylation and it mainly controls cell survival, growth and differentiation.27 JNK activation is associated with the transcription of several genes controlling apoptosis, differentiation and stress response.28
We studied the role of Sam68 in TLR-induced NF-κB and MAPK signaling pathways, using wild-type (WT) and Sam68-deficient macrophages and fibroblasts. Our results show that Sam68 is critical for the nuclear translocation of NF-κB c-Rel following TLR and interleukin (IL)-1R stimulation. This loss of nuclear NF-κB in Sam68-deficient cells results in ablated pro-inflammatory gene induction following TLR2 stimulation, whereas signaling was largely intact following TLR3 activation, indicating a more prominent role for Sam68 in driving gene expression following engagement of MyD88-dependent pathways. We also examined MAPK activation and found that Sam68 KO cells exhibited enhanced ERK and JNK activation, both basally and following stimulation. Together, these data identify Sam68 as a novel regulator of TLR signaling, potentially functioning as a cellular rheostat to fine tune the balance between NF-κB and MAPK signaling, which is critical for the proper immune response during an infection.
MATERIALS AND METHODS
Cells
Mouse embryonic fibroblasts (MEF) and RAW264.7 cells were grown in complete DMEM media (100 U/ml penicillin/streptomycin, 4 mM l-glutamine and 10% fetal bovine serum). For phosphorylation studies, cells were grown overnight in serum-free DMEM. J558L cells were obtained from ATCC and grown in D10 media (complete DMEM supplemented with 10 mM HEPES, 1 × non-essential amino acids (HyClone, Logan, UT, USA), 1 mM sodium pyruvate and 50 μM β-mercaptoethanol) for generation of J558L cultured supernatants.
Generation of bone marrow-derived macrophages
Bone marrow progenitors were collected from femurs and tibias of WT and Sam68 knockout (KO) mice using 25-Gauge syringe and phosphate-buffered saline (PBS). Cells were pelleted and subject to lysis in red blood cell lysis buffer (Biolegend, San Diego, CA, USA) for 10 min on ice. Cells were spun, resuspended in D10 media and were counted on a hemocytometer. Progenitor cells were plated at a density of 2 × 106 cells per 10 cm plate in D10 media containing 15% J558L culture supernatant as a source of granulocyte–macrophage colony-stimulating factor. After 3 days, suspension cells were pelleted and resuspended in fresh D10+J558L and placed back into original 10-cm plates. After an additional 3 days, suspension cells were collected, pelleted, resuspended in fresh C10 (fully supplemented RPMI media)+J558L supernatant and placed into fresh 10-cm plates for continued differentiation into dendritic cells. Fresh D10+J558L media was placed onto adherent cells in original 10-cm plates to continue macrophage differentiation. At day 9 of culture, macrophages and dendritic cells were collected for experimentation.
Reagents and antibodies
Lipofectamine 2000 was obtained from Life Technologies (Carlsbad, CA, USA). Recombinant P3C, FSL-1, poly I:C, LPS, and IL-1β were obtained from Invivogen (San Diego, CA, USA). Antibodies against p65, c-Rel, β-actin, lamin A/C, hNRPA1, Sam68, PLCγ1, JNK1/2 and ERK1/2 were from Santa Cruz Biotechnology (San Diego, CA, USA). Antibodies detecting phosphorylated forms of ERK1/2 and JNK1/2 were from Cell Signaling Technologies (Danvers, MA, USA).
Generation of Cas9-CRISPR knockout RAW macrophages
Sam68-targeted guide RNAs were designed using Zhang lab CRISPR Design and analysis web tool at crispr.mit.edu. All guides were chosen to: minimize off-target effects, target exon 1 to prevent generation of partially truncated proteins, and allow for wobble base-pair mutation in reconstitution plasmids that do not alter amino-acid sequence, but render reconstitution plasmids resistant to Cas9 cleavage. DNA oligos corresponding to guide RNAs were ordered from Integrated DNA Technologies (Coralville, IA, USA) and were cloned into lenti-CRISPR v2 plasmid (Addgene, Cambridge, MA, USA).29 Lentivirus containing Sam68-targeted guides were generated in HEK293T cells as described.29, 30 We used three separate guide constructs to increase likelihood of knockdown and limit off-target effects. RAW macrophages were infected with virus from undiluted culture supernatant overnight and left to recover for an additional day. Cells were then placed on puromycin selection (2 μg/ml) for 48 h followed by 48 h of recovery in fresh media. Puromycin resistant cells were plated at 1000 cells per 15-cm plate and individual colonies grown were picked after 7 days of culture by light trypsinization followed by transfer to 24-well plates. Selected clones were expanded and knockdown of protein was confirmed by western blotting. Five clones lacking Sam68 expression were pooled to minimize clonal variation and used in this study.
Reconstitution of Sam68 in Sam68 KO RAW macrophages
Sam68 KO RAW macrophages were transfected with a pcDNA3-Human Influenza Hemagglutinin (HA)-Sam68 plasmid using lipofectamine reagent following manufacturer’s protocol. Two rounds of transfection were performed to increase the efficiency and expression. Following clonal selection, reconstituted cells were tested for the Sam68 level. Five clones matching the level of endogenous Sam68 expression were pooled and experiments performed.
Ultraviolet radiation treatment of the cells
RAW cells were plated (2.5 × 105 cells per well in 6-cm plates) and treated with 100 mJ/cm2 of 254 nM Ultraviolet C (UVC) radiation using a UV Stratalinker 1800 (Stratagene, La Jolla, CA, USA). Cells were recovered for up to 8 h after the ultraviolet (UV) treatment and then lysed in Triton lysis buffer (10 mM HEPES pH 7.6, 150 mM NaCl, 1 mM EDTA and 1% Triton-X 100). Protein lysate concentrations were determined by bicinchoninic acid (BCA) assay and normalized samples run on 9% SDS–polyacrylamide gel electrophoresis (PAGE) gels for western blot analysis.
Cellular fractionations for western blotting
MEFs and RAW macrophages were collected from plates using PBS+10 mM EDTA and were pelleted at 1000g in a refrigerated centrifuge. Cells were then resuspended in cytoplasmic lysis buffer plus 1 × protease inhibitor cocktail (10 mM HEPES pH 7.6, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA and 1 mM DTT) for 15 min on ice. A final concentration of 0.625% NP40 was added to the lysate and vortexed immediately for 10 s to disrupt remaining cellular membrane. Lysates were spun at 10 000g for 30 s at 4 °C and supernatants were used as cytoplasmic lysate. Pellets were washed once in 3 × original volume of cytoplasmic buffer without NP40 and spun at 10 000g for 5 s at 4 °C to remove any remaining cytoplasmic proteins. Following aspiration of supernatant, pellets were lysed for 30 min on ice in nuclear lysis buffer plus 1 × protease inhibitor cocktail (20 mM HEPES pH 7.6, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA and 1 mM DTT). Pellets were vortexed for 5 s, three times during lysis to ensure complete disruption. Lysates were spun at 10 000g for 10 min at 4 °C and supernatants were used as nuclear lysates. Protein levels were normalized using BCA assay (Thermo Scientific, Rockford, IL, USA). For phosphorylation studies, MEFs and RAWs were collected as before and pelleted. Pellets were resuspended in phospho-lysis buffer plus 1 × protease inhibitor cocktail (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton-X 100, 25 μM sodium pyrophosphate, 1 mM β-glycerophosphate, 5 mM iodoacetamide and 5 mM N-ethyl maleimide). Lysates were centrifuged at 10 000g for 10 min at 4 °C. Protein concentrations of supernatants were determined by BCA assay. After normalization of protein levels, appropriate volumes of 4 × Laemmli sample buffer were added to dilute to 1 × and samples boiled for 5 min at 95 °C. For western blot analysis, cytoplasmic/nuclear lysates were resolved through 9% SDS–PAGE gels and lysates to examine phospho-proteins were resolved through 10% SDS–PAGE gels. Proteins from the gel were transferred onto nitrocellulose membranes, probed using relevant antibodies and visualized by enhanced chemiluminescence assay.
Quantitative real-time PCR
To directly quantify gene expression, quantitative real-time PCR analysis was performed. Total RNA was isolated from stimulated RAW cells using the DNAaway RNA miniprep kit (BioBasic, Markham, ON, CANADA). RNA yields were quantified by NanoDrop spectrophotometer (Thermo, Wilmington, DE, USA) and 1 μg of total RNA converted to complementary DNA (cDNA) using the qScript cDNA Supermix (Quanta BioSciences, Gaithersburg, MD, USA). A measure of 40 ng of cDNA was then run in duplicate for genes of interest (Supplementary Table S1). Triplicate samples were run for all stimulation conditions. Gene expression is quantified as fold induction over mock treatment, using the ΔΔCt method. All values are normalized to the housekeeping gene L32.
RESULTS
Sam68 differentially regulates P3C and IL-1-induced NF-κB and MAPK pathways in fibroblasts
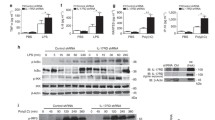
Our prior work has demonstrated that Sam68 is a critical component of pro-inflammatory signaling by TNF, where it acts as an adapter that links multiple proteins recruited to the TNF receptor complex leading to NF-κB activation.23 Because pro-inflammatory signaling by TLR is also capable of inducing NF-κB activation, we investigated the role of Sam68 in TLR signaling. We stimulated WT and Sam68 KO MEF with the diacylated lipopeptide FSL-1 or the triacylated lipopeptide Pam3CSK4 (P3C) to stimulate TLR2/6 and TLR2/1, respectively.31, 32 FSL-1 and P3C stimulation resulted in nuclear translocation of both NF-κB p65 and NF-κB c-Rel in WT MEF cells (Figure 1a). We found that Sam68 deficiency differentially affects the nuclear translocation of the two NF-κB subunits, p65 and c-Rel, which control the activation of innate immune response genes. Although Sam68 deficiency resulted in only a minor decrease in nuclear translocation of p65, TLR2- mediated nuclear translocation of c-Rel was greatly diminished following FSL-1 stimulation for 1 h and P3C stimulation for 1 and 3 h. Notably, Sam68 KO cells showed a weaker accumulation of nuclear c-Rel at 3 h following FSL-1 stimulation. This suggests either Sam68 is essential only for the early activation of c-Rel by FSL-1 or the existence of a Sam68-independent mechanism for the delayed FSL-1-induced c-Rel nuclear translocation (Figure 1a). Because we observed that Sam68 KO MEF cells expressed lower levels of c-Rel, we confirmed that Sam68 deficiency indeed decreases nuclear c-Rel levels by calculating the nuclear/cytoplasmic ratio of c-Rel using densitometric analysis (Uppsala, SWE). This further demonstrated that nuclear c-Rel was substantially diminished following FSL-1 treatment at 1 h and P3C treatment at 1 and 3 h (Supplementary Figure 1a). These findings were reproduced in response to IL-1R activation as well, with Sam68 deficiency resulting in slightly diminished IL-1-induced p65 nuclear translocation and almost complete inhibition of c-Rel nuclear translocation (Figure 1b). Given that P3C is present in a range of bacterial species and that it induced stronger NF-κB activation than FSL-1, we chose to use P3C as our TLR2 agonist for this study.
Sam68 is critical for TLR-induced signaling in fibroblasts. (a) WT and Sam68 KO MEFs (1 × 105) were stimulated with FSL-1 (300 ng/ml) or Pam3CSK4 (200 ng/ml) for 1 and 3 h. (b) WT and Sam68 KO MEFs (1 × 105) were stimulated with IL-1β (100 ng/ml) for 15–120 min. (c) WT and Sam68 KO MEFs (2 × 105) were stimulated with Pam3CSK4 (200 ng/ml) for the indicated time points. Cells were collected, and cytoplasmic and nuclear fractions were prepared. Western blots were probed to identify nuclear NF-κB translocation of NF-κB p65 and c-Rel. Actin and lamin were used as loading controls for cytoplasm and nucleus, respectively, and PLCγ1 was used as fractionation purity control to show the lack of cytoplasmic contamination in the nucleus. (d, e) WT and Sam68 KO MEFs (2 × 105) were serum starved (0% FBS) overnight and stimulated with Pam3CSK4 or IL-1β for the indicated time points. Cells were collected and lysed in buffer containing phosphatase inhibitors. Protein samples were probed for p-ERK1, p-ERK2, p-JNKp54, p-JNKp46 and total ERK/JNK. b represents one of 2 independent experiments and a, c, d and e are representative of ⩾3 independent experiments. ERK, extracellular signal-regulated kinases; IL, interleukin; JNK, c-Jun N-terminal kinases; KO, knockout; MEF, Mouse embryonic fibroblasts; NF-κB, nuclear factor kappaB; TLR, Toll-like receptor; WT, wild type.
To delineate whether the altered TLR2-induced NF-κB nuclear translocation in Sam68 KO cells was a temporal defect, we performed a kinetic analysis of P3C stimulation of WT and Sam68 KO MEFs. Cells were stimulated with P3C from 15 min through 6 h. Nuclear NF-κB p65 levels were decreased at 15 and 30 min, but it resolved to comparable levels in WT and Sam68 KO cells after 6 h of treatment, indicating that Sam68 deficiency affects only transient p65 activation (Figure 1c). By contrast, nuclear translocation of NF-κB c-Rel was greatly decreased in the Sam68 KO MEFs. Interestingly, at 1 h, following P3C stimulation we observed a modest increase in nuclear NF-κB c-Rel (Figures 1a and c and higher exposure of Figure 1c in Supplementary Figure 1b), which indicates the presence of a Sam68-independent transient c-Rel activation mechanism. P3C stimulation-induced elevation of p65 levels both in WT and Sam68 KO cells, and significant enhancement of c-Rel levels in the cytoplasm of WT cells. Interestingly, the early increase in P3C-induced cytoplasmic c-Rel was markedly reduced in the absence of Sam68, although long treatment for 6 h resulted in accumulation of cytoplasmic c-Rel in Sam68 KO cells. However, this enhanced cytoplasmic c-Rel was found not sufficient to cause its nuclear translocation, suggesting that Sam68 affects both P3C-induced synthesis and nuclear translocation of c-Rel (Figure 1c).
Our previous work has revealed that TNF-induced MAPK activation was enhanced in Sam68 KO cells.23 We next sought to determine whether TLR-induced MAPK activation was dependent on Sam68. We found that absence of Sam68 greatly enhanced ERK activation, as indicated by phosphorylation of ERK1 (top band) and ERK2 (bottom band) following P3C stimulation (Figure 1d). We also examined JNK activation and found that Sam68 KO MEFs had enhanced basal phosphorylation of JNKp54 (top) and JNKp46 (bottom band) compared with WT cells, with moderate increases in P3C-induced JNKp46 phosphorylation (Figure 1d). IL-1β stimulation also resulted in enhanced ERK1 and ERK2 activation, and pronounced JNKp46 activation in Sam68 KO cells (Figure 1e). The relatively low levels of MAPK activation, which we observed are likely due to the use of low doses of ligands (200 ng/ml compared with 1 μg/ml as previously reported33) that we determined as the threshold required for NF-κB activation. These data suggests that Sam68 deficiency results in enhanced MAPK activation and provide the first experimental evidence that Sam68 functions as a dual regulator of NF-κB and MAPK signaling in response to TLR signaling.
Sam68 is a selective regulator of TLR-induced NF-κB activation in macrophages
To determine the role of Sam68 in TLR-induced NF-κB activation in an immune cell, we generated RAW264.7 macrophages that lack Sam68 by CRISPR-Cas9-mediated gene editing (Supplementary Figure 2). To study TLR signaling, we pooled five Sam68 knockdown clones to minimize effects of variant viral integration. We studied three TLR ligands, P3C, poly (I:C) and LPS, which utilize MyD88, TRIF or both MyD88 and TRIF, respectively, in signaling. First, we studied stimulation of RAW macrophages using P3C that utilizes TLR2/MyD88 for signaling. We found that Sam68 deficiency completely blocks P3C-induced nuclear translocation of NF-κB c-Rel. However, lack of Sam68 did not completely block nuclear translocation of p65, suggesting that P3C may activate NF-κB p65 in a Sam68-independent manner (Figure 2a). In order to determine the optimal dose of poly (I:C) to be used, we stimulated WT and Sam68 KO RAWs with 5 and 10 μg/ml, and found that 10 μg/ml was necessary to induce potent NF-κB nuclear translocation (Supplementary Figure 3). We found that the role of Sam68 in TLR signaling is not limited to MyD88-dependent pathways as poly (I:C) treatment also resulted in complete absence of nuclear NF-κB c-Rel and substantially decreased p65 nuclear translocation in Sam68 KO RAWs (Figure 2b). Interestingly, LPS treatment did not show any significant changes in p65 nuclear translocation, although it only partially decreased c-Rel levels compared with WT cells (Figure 2b). This indicates the existence of a Sam68-independent LPS-induced c-Rel activation pathway. Taken together, comparative study of three TLR pathways suggest a differential requirement of Sam68 in MyD88-dependent, TRIF-dependent and MyD88/TRIF-dependent signaling pathways.
TLR-induced activation of NF-κB in macrophages requires Sam68. (a–b) WT and Sam68 KO RAW264.7 macrophages (1 × 106) were stimulated with (a) Pam3CSK4 (200 ng/ml), (b) poly I:C (10 μg/ml) and LPS (1 μg/ml) for up to 2 h. Cytoplasmic and nuclear fractions were analyzed as indicated to identify nuclear NF-κB translocation. (c) WT, Sam68 KO and Sam68+ RAW264.7 macrophages (1 × 106) were stimulated with Pam3CSK4 (200 ng/ml) and poly I:C (10 μg/ml) for 30 min and 1 h. Cytoplasmic and nuclear fractions were probed as indicated. (d) WT and Sam68 KO primary BMDMs (1 × 106) were stimulated with Pam3CSK4 (200 ng/ml) and FSL-1 (300 ng/ml) for the indicated time points. Cytoplasmic and nuclear fractions were probed for NF-κB p65 and c-Rel to determine their nuclear translocation. Actin, lamin and hnRNPA1 were used as loading controls and PLCγ1 was used as nuclear fractionation purity control. BMDMs, bone marrow-derived macrophages; KO, knockout; LPS, lipopolysaccharide; NF-κB, nuclear factor kappaB; WT, wild type.
To demonstrate that Sam68 is indeed required for P3C-induced NF-κB c-Rel activation and the observed defective c-Rel nuclear translocation in Sam68 KO cells does not occur as an artifact in Sam68 KO cells, we reconstituted Sam68 KO macrophages with HA-tagged Sam68 cDNA. We stimulated WT, Sam68 KO and HA-Sam68 (Sam68+) macrophages with P3C and poly I:C for 30 min and 1 h. This gain-of-function experiment using HA-Sam68 reconstituted Sam68 KO cells rescued the defective NF-κB c-Rel nuclear translocation (Figure 2c), confirming that loss of P3C and poly (I:C)-induced nuclear translocation of NF-κB c-Rel in Sam68 KO cells is directly related to the absence of Sam68.
To further confirm the role of Sam68 in regulation of NF-κB using a primary cellular system, we generated bone marrow-derived macrophages (BMDMs) from WT and Sam68 KO mice. Both WT and Sam68 KO cells were similar in terms of cellular morphology and cellular yields during the differentiation process. Similar to RAW cells, Sam68 KO BMDMs also showed almost complete ablation of NF-κB c-Rel nuclear translocation following stimulation with two TLR2 ligands, P3C that uses TLR2/1 and FSL-1 that uses TLR2/6 receptors (Figure 2d). Interestingly, FSL-1-induced p65 activation was compromised in Sam68 KO cells, although it retained residual p65 activation following P3C treatment as observed in RAW cells and macrophages. Similar results were observed in bone marrow-derived dendritic cells following P3C stimulation (Supplementary Figure 4). Taken together, these data indicate that Sam68 has a critical role in specifically mediating NF-κB nuclear translocation, following TLR signaling in innate immune cells.
We next tested whether diminished nuclear NF-κB translocation in the absence of Sam68 is a global occurrence or is limited to immune receptor-induced signaling. To this end, we studied NF-κB activation induced by a completely unrelated mechanism, that is, UV radiation.34 We exposed WT and Sam68 KO RAW cells to UVC radiation (254 nm at 100 mJ/cm2) and monitored NF-κB activation at 1, 2, 4 and 8 h post UVC exposure. We found no significant differences in nuclear NF-κB translocation of either p65 or c-Rel between the WT and Sam68 KO cells following UV treatment, suggesting that the NF-κB activation pathway is intact in Sam68 KO cells (Supplementary Figure 5). This result suggests that the role of Sam68 in mediating NF-κB activation is, at least in part, limited to traditional immune receptor signaling.
Loss of Sam68 enhances ERK and JNK activation following TLR engagement
Cellular responses to TLR stimulation activate MAP kinase pathways, including ERK and JNK that control cell survival, proliferation, differentiation and apoptosis.27, 28 We examined the role of Sam68 in P3C, poly (I:C) and LPS-induced MAPK activation, using WT and Sam68 KO RAW cells. We found that P3C treatment resulted in enhanced phosphorylation of JNKp46 (bottom band), as well as ERK1 (top band) and ERK2 (bottom band) in Sam68 KO RAW cells compared with WT cells (Figure 3a). Of note, the concentrations of TLR ligands in our experiments were carefully chosen as the minimal dose at which we saw robust NF-κB activation and all fall below 150 nM. Interestingly, this low-grade activation of TLR2 using P3C showed only modest phosphorylation of JNK in WT RAW cells. However, it has been previously shown that P3C induces appreciable phosphorylation of JNK when used at 1 μg/ml, which is over 5 × the level used in our study.35 To confirm our cellular system is capable of robust phosphorylation of JNKp54 (top) and JNKp46 (bottom), we stimulated our WT RAW cells using 1 μg/ml P3C and observed substantial phosphorylation of JNK (Supplementary Figure 6). We observed similar enhancements in basal and stimulation-induced phosphorylation of JNKp46 and ERK1/2 in Sam68 KO RAWs treated with poly (I:C) (Figure 3b) and LPS (Figure 3c). Given a recent report, showing that low-dose LPS activates, whereas super-low-dose LPS antagonizes ERK activation,36 we studied whether there is a dose-dependent modulation of ERK signaling in Sam68 KO cells. We stimulated WT and Sam68 KO RAWs with titrating doses of LPS from 1000 to 0.01 ng/ml. We observed that for high doses of LPS, 500–1000 ng/ml, there was enhanced ERK phosphorylation in Sam68 KO cells compared with WT cells. Interestingly, enhanced ERK1/2 phosphorylation was also observed in Sam68 KO cells following 100 and 1 ng/ml of LPS treatment. These results indicate that both high and low doses of LPS activate ERK signaling in Sam68 KO cells and the role of Sam68 in LPS-induced ERK activation is not highly dependent on the dose of LPS (Supplementary Figure 7). In combination with our MEF data from Figure 1, these results demonstrate that Sam68 deficiency results in enhanced MAPK activation and decreased NF-κB activation.
Loss of Sam68 enhances basal and TLR-induced MAPK activation. WT and Sam68 KO RAWs (2.5 × 105) were plated overnight and switched to serum-free DMEM media for an additional day. Cells were then stimulated with (a) Pam3CSK4 (200 ng/ml), (b) poly (I:C) (10 μg/ml) and (c) LPS (1 μg/ml) for up to 30 min. Cells were collected and lysed in Triton lysis buffer containing protease and phosphatase inhibitors. Total lysates were probed for MAPK activation using phospho-JNK and phospho-ERK antibodies. Samples were probed for loading controls using total ERK and actin. a–c are representative of 3 independent experiments. ERK, extracellular signal-regulated kinases; JNK, c-Jun N-terminal kinasesKO, knockout; LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinases; TLR, Toll-like receptor; WT, wild type.
Sam68 is required for P3C-induced gene expression
Our studies on the role of Sam68 in TLR-induced NF-κB activation identify a marked defect in nuclear translocation of NF-κB in the absence of Sam68. In order to confirm this defect results in altered NF-κB-dependent gene expression, we performed quantitative real-time PCR on WT and Sam68 KO RAWs stimulated with P3C and poly (I:C). We have shown previously that NF-κB target genes such as TNF, CXCL10 (IP10), VCAM1 and CCL5 are dependent on Sam68 in TNF signaling.23 Stimulation of WT RAWs with P3C resulted in rapid induction of transcripts for TNF, VCAM1, IL-10, CXCL10 (IP10) and CCL5 (Figures 4a–c). Absence of Sam68 substantially decreased P3C-induced expression of TNF, VCAM1 and IL10 (Figure 4a). Expression of CXCL10 was also impaired in Sam68 KO cells (Figure 4b). Interestingly, unlike TNF signaling, we observed comparable expression of CCL5 in both WT and Sam68 KO cells following P3C stimulation (Figure 4c). Consistent to this, it has been previously shown that CCL5 expression could be under the regulation of JNK pathway activation, which is not compromised in Sam68 KO cells.37 We next examined the impact of Sam68 deficiency on poly (I:C)-induced gene expression. We found that loss of Sam68 did not block the expression of TNF and IL10, and only had a modest effect in decreasing the expression of VCAM1 (Figure 4d), suggesting that the residual NF-κB p65 activation observed in these cells (Figure 2b) likely sufficed to cause induction of these genes. Early induction of CXCL10 by poly (I:C) was dependent on Sam68. However, unlike P3C stimulation, prolonged poly (I:C) treatment for 3 h showed elevation in the expression of CXCL10 in Sam68 KO cells (Figure 4e). Expression of CCL5 was found enhanced at both short and long poly (I:C) stimulations in Sam68 KO cells (Figure 4f), suggesting a suppressive role of Sam68 in poly (I:C)-induced CCL5 expression. This enhancement in CCL5 expression might also be attributed to the enhanced MAPK activation we observed in these cells (Figure 3b).
TLR2- but not TLR3-induced gene expression is ablated in Sam68-deficient macrophages. Gene expression of key NF-κB target genes was determined by quantitative real-time PCR. WT and Sam68 KO RAW macrophages (2 × 105) were stimulated with Pam3CSK4 (200 ng/ml) or poly (I:C; 10 μg/ml) for 1 and 3 h. Total cellular RNA was isolated and 1 μg was converted to cDNA. Expression of the genes (a, d) TNF, VCAM1, IL10, (b, e) CXCL10 and (c, f) CCL5 was determined. Values are expressed as fold induction over the mock treatment using the ΔΔCt method. All values are normalized to the housekeeping gene L32. All samples were run in duplicate and ligand stimulation conditions represent n⩾2 independent samples. Statistical analysis was performed in GraphPad (La Jolla, CA, USA) for Student’s t-test *P<0.05. cDNA, complementary DNA; KO, knockout; NF-κB, nuclear factor kappaB; TLR, Toll-like receptor; WT, wild type.
On the basis of our data, we propose a model whereby cytoplasmic Sam68 serves as a cellular rheostat, which functions to balance NF-κB and MAPK activation following TLR stimulation (Figure 5).
Schematic model of the role of Sam68 in TLR2 and TLR3 signaling pathways. In normal cells, Sam68 is required for TLR2 and TLR3-induced NF-κB activation and inflammatory gene expression. In the absence of Sam68, NF-κB nuclear translocation and subsequent inflammatory gene induction are substantially reduced in response to TLR2 activation that signals through MyD88. Although NF-κB nuclear translocation is substantially reduced following TLR3 stimulation, TLR3-induced gene expression is largely unaffected in Sam68-deficient cells. Both TLR2 and TLR3-induced MAPK activation is enhanced in Sam68-deficient cells. NF-κB independent pathways, including MAPK and IRFs, may have a role in gene expression in Sam68-deficient cells in response to TLR3 activation. Black arrow—normal signaling, bold black arrow—enhanced signaling and gray arrow—diminished signaling. IRFs, interferon regulatory factors; MAPK, mitogen-activated protein kinases; MyD88, myeloid differentiation primary response gene 88; NF-κB, nuclear factor kappaB; TLR, Toll-like receptor.
DISCUSSION
One of the key roles of the immune system is to drive inflammation and cytotoxic responses following microbial infection. The immune system recognizes pathogens via PRRs in cells, which bind to conserved microbial structures and endogenous danger signals indicative of cellular insult.1 Despite the growing literature on PRR structure and function over ‘the past 20 years, our knowledge on how these variant receptors activate differential signaling cascades within a cell to coordinate an appropriate immune response remains poorly understood. In this study, we identified that Sam68 (a) keeps TLR-induced MAPK activation in check, and (b) is a key regulator of TLR-induced NF-κB activation. We demonstrate that the absence of Sam68 decreases TLR-induced NF-κB nuclear translocation in both MEFs and RAW macrophages, as well as in primary macrophages and dendritic cells. It appears that Sam68 is an essential component of signaling by the members of TLR family. We saw NF-κB activation defects in Sam68-deficient cells following activation by multiple TLR ligands, including MyD88-dependent (P3C and FSL-1) and TRIF-dependent poly (I:C) ligands, as well as LPS that utilizes both MyD88 and TRIF pathways. Nuclear level of NF-κB p65 is reduced, whereas nuclear levels of c-Rel are ablated in Sam68 KO cells. This loss of TLR-induced NF-κB activation in Sam68 KO cells is not simply a temporal or cell-type-specific phenomenon. First, we observed only minimal nuclear translocation of NF-κB c-Rel even after prolonged TLR2 activation of 6 h. Second, this phenomenon is not limited to immortalized MEF and RAW cells, as the same defect is observed in primary BMDMs and dendritic cells isolated from Sam68 KO mice. In order to eliminate the possibility that this was a global defect in NF-κB response, we tested nuclear NF-κB translocation following UV stimulation, to mimic stress induced NF-κB activation. We found that both WT and Sam68 KO RAW macrophages responded to UV treatment in a similar manner and had comparable nuclear NF-κB p65 and c-Rel levels. This data indicates that the role of Sam68 is specific to immune receptor-induced NF-κB signaling and not the result of a global defect in NF-κB activity. This is consistent with Sam68 KO mice being viable,38 whereas we would expect a global NF-κB defect to result in embryonic or early life lethality.
Activation of TLRs not only result in potent NF-κB activation, but can also lead to the activation of MAPK pathways, including ERK and JNK.26 The balance between these signaling pathways directs the functioning of innate immune cells and their capacity to activate other immune cells during an infection. Our results show a bimodal regulatory role of Sam68 in controlling the activation of NF-κB and MAPK pathways. Although the deficiency of Sam68 decreases TLR-induced NF-κB activation, it increased the activation of ERK and JNK pathways. It appears that Sam68 acts as a factor that keeps MAPK activation in check during TLR activation, although allowing NF-κB activation. A thorough study of gene expression patterns in WT and Sam68 KO macrophages, following TLR activation will decipher the biological significance of this differential regulatory role of Sam68. Interestingly, we also found that loss of Sam68 also elevates the basal MAPK activation, suggesting a role for Sam68 in cellular homeostasis.
The MAPK activation we observed in this study is at a diminished level compared with several other previous studies.35, 39 This is likely due to the lower doses of TLR ligands we are using in our experiments. We found that the commonly used concentrations of TLR ligands are extremely high and may not accurately correspond to the stimulation conditions of live infection. For instance, we and others commonly use LPS at a concentration of 1 μg/ml; however, a study of ~250 septic patients found median levels in the blood of nonsurvivors was 515 pg/ml, almost 2000 × lower than used in vitro.40 Therefore, first we determined the minimal concentration of TLR ligands required to activate NF-κB in our cellular system and used these concentrations in an attempt to stay as close to the in vivo pathological conditions as possible. During a natural course of infection, innate immune cells will initially come into contact with limited amounts of pathogenic stimuli during the first few days as the pathogen begins to reproduce. By using the minimal dose required for activation, we are able to more accurately model how these pathogenic molecules activate innate immunity early during infection, providing better insight into the role of Sam68 in live infections. Our data on decreased NF-κB and enhanced MAPK activation represents the first evidence that Sam68 differentially regulates NF-κB and MAPK signaling downstream of TLR activation. Intriguingly, our results also highlight a critical role of Sam68 in differentially activating two subunits of NF-κB, p65 and c-Rel, which suggest a highly selective role of Sam68 within NF-κB pathway activation.
Given our prior findings that Sam68 is rapidly and robustly recruited to the TNFR complex following activation,23 it is possible that pro-inflammatory TNF receptor signaling may also alter the cellular response to TLR ligands. Further understanding how immune cells balance NF-κB and MAPK signaling cascades will not only enhance our basal knowledge on how TLR signaling is regulated during an immune response, but also provide potential therapeutic targets for altering the balance of these signaling cascades in patients.
Our gene expression studies yielded some interesting insights into the role of Sam68 in regulating the expression of pro-inflammatory genes. The most profound effect of Sam68 deficiency was observed in P3C-induced TLR2/1-dependent pathway where the expression of TNF, VCAM1, IL-10 and CXCL10 were compromised. But the expression of CCL5, a gene with NF-κB signatures in its promoter,41 was comparable between WT and Sam68 KO cells. Although this shows Sam68 KO cells are responsive to P3C stimulation and validates our gene expression data, it also raises some interesting questions. Does TLR2/1 pathway induce CCL5 independent of NF-κB or dependent on NF-κB, but independent of Sam68? Alternatively, it has also been shown that CCL5 is a target of MAPK signaling and the active MAPK pathways in Sam68 KO cells may induce CCL5.37
Alluding to the complex inter-pathway variations of signaling from receptors belonging to the same family, unlike P3C treatment, we found that Sam68 deficiency had only minor effects on the expression of NF-κB-targeted genes TNF, VCAM1 and IL10 following poly (I:C) treatment. Expression of CCL5 was substantially enhanced in Sam68 KO cells, and CXCL10 showed only a transient defect in expression. One possibility is that these genes are partially controlled by other signaling pathways such as the IRFs, which are induced by TLR3 but not TLR2 signaling.42 This also shows a role of Sam68 in temporal regulation of IP10 expression and a negative regulatory role in TLR3-induced CCL5 expression.
These gene expression data are in agreement with the already established critical role of the TLR-induced inflammation and role of adapter proteins, MyD88 and TRIF, in determining the pathway specificity. Here we show Sam68 as a new player in TLR signaling with apparently more crucial role in inducing gene expression downstream of MyD88-dependent receptors. It will be interesting to explore further and dissect the discrete regulatory role of Sam68 in evolutionarily similar, but functionally different signaling cascades initiated by the members of TLR family.
References
Di Gioia M, Zanoni I . Toll-like receptor co-receptors as master regulators of the immune response. Mol Immunol 2015; 63: 143–152.
Takeda K, Akira S . Toll-like receptors in innate immunity. Int Immunol 2005; 17: 1–14.
Huysamen C, Brown GD . The fungal pattern recognition receptor, Dectin-1, and the associated cluster of C-type lectin-like receptors. FEMS Microbiol Lett 2009; 290: 121–128.
Kanneganti TD, Lamkanfi M, Nunez G . Intracellular NOD-like receptors in host defense and disease. Immunity 2007; 27: 549–559.
Franchi L, Munoz-Planillo R, Nunez G . Sensing and reacting to microbes through the inflammasomes. Nat Immunol 2012; 13: 325–332.
Yoneyama M, Fujita T . RIG-I family RNA helicases: cytoplasmic sensor for antiviral innate immunity. Cytokine Growth Factor Rev 2007; 18: 545–551.
Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T et al. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 1999; 11: 443–451.
Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 1998; 282: 2085–2088.
Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL . Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol 2001; 167: 1882–1885.
Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol 2006; 7: 569–575.
Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Ozoren N, Jagirdar R et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol 2006; 7: 576–582.
Bauer S, Kirschning CJ, Hacker H, Redecke V, Hausmann S, Akira S et al. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc Natl Acad Sci USA 2001; 98: 9237–9242.
Alexopoulou L, Holt AC, Medzhitov R, Flavell RA . Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 2001; 413: 732–738.
Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 2004; 303: 1526–1529.
Brown GD, Gordon S . Immune recognition. A new receptor for beta-glucans. Nature 2001; 413: 36–37.
Tomalka J, Ganesan S, Azodi E, Patel K, Majmudar P, Hall BA et al. A novel role for the NLRC4 inflammasome in mucosal defenses against the fungal pathogen Candida albicans. PLoS Pathog 2011; 7: e1002379.
Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J . Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 2006; 440: 237–241.
Dostert C, Petrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J . Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science 2008; 320: 674–677.
Medzhitov R, Preston-Hurlburt P, Janeway CA Jr . A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 1997; 388: 394–397.
Dunne A, O'Neill LA . The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci STKE 2003; 2003: re3.
Colonna M . TLR pathways and IFN-regulatory factors: to each its own. Eur J Immunol 2007; 37: 306–309.
Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R . TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat Immunol 2008; 9: 361–368.
Ramakrishnan P, Baltimore D . Sam68 is required for both NF-kappaB activation and apoptosis signaling by the TNF receptor. Molecular Cell 2011; 43: 167–179.
Lukong KE, Richard S . Sam68, the KH domain-containing superSTAR. Biochimic Biophys Acta 2003; 1653: 73–86.
Hayden MS, Ghosh S . NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev 2012; 26: 203–234.
Dong C, Davis RJ, Flavell RA . MAP kinases in the immune response. Ann Rev Immunol 2002; 20: 55–72.
Seger R, Krebs EG . The MAPK signaling cascade. FASEB J 1995; 9: 726–735.
Leppa S, Bohmann D . Diverse functions of JNK signaling and c-Jun in stress response and apoptosis. Oncogene 1999; 18: 6158–6162.
Sanjana NE, Shalem O, Zhang F . Improved vectors and genome-wide libraries for CRISPR screening. Nat Meth 2014; 11: 783–784.
Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014; 343: 84–87.
Fujita M, Into T, Yasuda M, Okusawa T, Hamahira S, Kuroki Y et al. Involvement of leucine residues at positions 107, 112, and 115 in a leucine-rich repeat motif of human Toll-like receptor 2 in the recognition of diacylated lipoproteins and lipopeptides and Staphylococcus aureus peptidoglycans. J Immunol 2003; 171: 3675–3683.
Takeuchi O, Sato S, Horiuchi T, Hoshino K, Takeda K, Dong Z et al. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J Immunol 2002; 169: 10–14.
Tseng PH, Matsuzawa A, Zhang W, Mino T, Vignali DA, Karin M . Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat Immunol 2010; 11: 70–75.
Li N, Karin M . Ionizing radiation and short wavelength UV activate NF-kappaB through two distinct mechanisms. Proc Natl Acad Sci USA 1998; 95: 13012–13017.
Goh ET, Arthur JS, Cheung PC, Akira S, Toth R, Cohen P . Identification of the protein kinases that activate the E3 ubiquitin ligase Pellino 1 in the innate immune system. Biochem J 2012; 441: 339–346.
Chen K, Geng S, Yuan R, Diao N, Upchurch Z, Li L . Super-low dose endotoxin pre-conditioning exacerbates sepsis mortality. EBioMedicine 2015; 2: 324–333.
Kumar D, Hosse J, von Toerne C, Noessner E, Nelson PJ . JNK MAPK pathway regulates constitutive transcription of CCL5 by human NK cells through SP1. J Immunol 2009; 182: 1011–1020.
Richard S, Torabi N, Franco GV, Tremblay GA, Chen T, Vogel G et al. Ablation of the Sam68 RNA binding protein protects mice from age-related bone loss. PLoS Genet 2005; 1: e74.
Jiang Z, Zamanian-Daryoush M, Nie H, Silva AM, Williams BR, Li X . Poly(I-C)-induced Toll-like receptor 3 (TLR3)-mediated activation of NFkappa B and MAP kinase is through an interleukin-1 receptor-associated kinase (IRAK)-independent pathway employing the signaling components TLR3-TRAF6-TAK1-TAB2-PKR. J Biol Chem 2003; 278: 16713–16719.
Opal SM, Scannon PJ, Vincent JL, White M, Carroll SF, Palardy JE et al. Relationship between plasma levels of lipopolysaccharide (LPS) and LPS-binding protein in patients with severe sepsis and septic shock. J Infect Dis 1999; 180: 1584–1589.
Moriuchi H, Moriuchi M, Fauci AS . Nuclear factor-kappa B potently up-regulates the promoter activity of RANTES, a chemokine that blocks HIV infection. J Immunol 1997; 158: 3483–3491.
O'Neill LA, Fitzgerald KA, Bowie AG . The Toll-IL-1 receptor adaptor family grows to five members. Trends Immunol 2003; 24: 286–290.
Acknowledgements
We acknowledge the Visual Sciences Training Program training grant (EY007157) for providing salary and reagent support to JAT. We thank American Association of Immunologists for Careers in Immunology Fellowship support to PR and JAT. We thank American Cancer Society Institutional Research grant (IRG-91-022-18) and National Institutes of Health grant 1R01AI116730-01A1 to PR. We thank Dr Stephane Richard, McGill University for providing the Sam68 KO mice and Case Western Reserve University Animal Resource Center for support with housing of mice. We thank Elizabeth Akinbiyi for technical guidance, Dr Clifford Harding for providing qPCR primers, Steven Chirieleison and Dr Derek Abbott for the guidance in the generation of CRISPR guide RNAs and optimization of viral infection.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information for this article can be found on the Cellular & Molecular Immunology website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Tomalka, J., de Jesus, T. & Ramakrishnan, P. Sam68 is a regulator of Toll-like receptor signaling. Cell Mol Immunol 14, 107–117 (2017). https://doi.org/10.1038/cmi.2016.32
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cmi.2016.32
- Springer Nature Limited
Keywords
This article is cited by
-
Role of Sam68 as an adaptor protein in inflammatory signaling
Cellular and Molecular Life Sciences (2024)
-
LncRNA Gm15834 Aggravates Cardiac Hypertrophy by Interacting with Sam68 and Activating NF-κB Mediated Inflammation
Cardiovascular Drugs and Therapy (2024)
-
Sam68 promotes osteogenic differentiation of aortic valvular interstitial cells by TNF-α/STAT3/autophagy axis
Journal of Cell Communication and Signaling (2023)
-
Sam68 contributes to intestinal inflammation in experimental and human colitis
Cellular and Molecular Life Sciences (2021)
-
Differential regulation of basal expression of inflammatory genes by NF-κB family subunits
Cellular & Molecular Immunology (2019)