Abstract
Inhibitor of apoptosis (IAP) proteins are frequently expressed at high levels in cancer cells and represent attractive therapeutic targets. We previously reported that the Smac (second mitochondria-derived activator of caspases) mimetic BV6, which antagonizes IAP proteins, sensitizes glioblastoma cells to temozolomide (TMZ)-induced cell death in a nuclear factor-κB (NF-κB)-dependent manner. However, BV6-induced NF-κB target genes responsible for this synergistic interaction have remained elusive. Using whole-genome gene expression profiling, we here identify BV6-stimulated, NF-κB-dependent transcriptional upregulation of interferon-β (IFNβ) and IFN-mediated proapoptotic signaling as critical events that mediate BV6/TMZ-induced apoptosis. Knockdown of IFNβ significantly rescues cells from BV6/TMZ-induced cell death. Similarly, silencing of the corresponding receptor IFNα/β receptor (IFNAR) confers a significant protection against apoptosis, demonstrating that IFNβ and IFN signaling are required for BV6/TMZ-mediated cell death. Moreover, BV6 and TMZ cooperate to transcriptionally upregulate the proapoptotic B-cell lymphoma 2 family proteins Bax (Bcl-2-associated X protein) or Puma (p53-upregulated modulator of apoptosis). Knockdown of Bax or Puma significantly decreases BV6/TMZ-induced apoptosis, showing that both proteins are necessary for apoptosis. By identifying IFNβ as a key mediator of BV6/TMZ-induced apoptosis, our study provides novel insights into the underlying molecular mechanisms of Smac mimetic-mediated chemosensitization with important implications for the development of novel treatment strategies for glioblastoma.
Similar content being viewed by others
Main
Glioblastoma is the most common primary malignant brain tumor and current treatment options include surgical resection, radiation and chemotherapy with the alkylating agent temozolomide (TMZ).1 However, despite aggressive treatment regimens, the prognosis of patients suffering from glioblastoma is still very poor,2 highlighting the high medical need for novel treatment strategies.
Evasion of programmed cell death is one of the hallmarks of human cancers3 and promotes tumorigenesis as well as treatment resistance.4 Apoptosis is a common form of programmed cell death that can be engaged via the intrinsic (mitochondrial) or extrinsic (death receptor) pathway.5 Activation of the intrinsic pathway is controlled by pro- and antiapoptotic B-cell lymphoma 2 (Bcl-2) family protein, including Bcl-2 family proteins such as p53-upregulated modulator of apoptosis (Puma) or Bcl-2-associated X protein (Bax). Following engagement of the mitochondrial pathway, mitochondrial intermembrane space proteins are released into the cytosol, including second mitochondria-derived activator of caspases (Smac).6 Smac binds to and neutralizes Inhibitor of Apoptosis (IAP) proteins, a family of antiapoptotic proteins, thereby promoting activation of caspases and apoptosis.7 In addition, binding of Smac to IAP proteins that harbor a Really Interesting New Gene (RING) domain with E3 ligase activity triggers their autoubiquitination and proteasomal degradation, which in turn leads to stabilization of nuclear factor-κB (NF-κB)-inducing kinase (NIK) and activation of noncanonical NF-κB signaling.8, 9
IAP proteins are expressed at high levels in various cancers and represent attractive targets for therapeutic intervention.7 BV6 is a synthetically designed Smac mimetic that mimics the N-terminal part of endogenous Smac protein.8 We previously reported that Smac mimetics such as BV6 sensitize glioblastoma cells to chemotherapy- or γ-irradiation-induced apoptosis in an NF-κB-dependent manner.10, 11, 12 Although BV6-stimulated NF-κB activation was demonstrated to be critically required for Smac mimetic-mediated sensitization of glioblastoma cells towards TMZ, the proapoptotic NF-κB-regulated target genes that mediate this chemosensitization have so far remained elusive.12 While tumor necrosis factor α (TNFα), a prototypic NF-κB target gene, was shown to mediate apoptosis via an autocrine/paracrine loop upon treatment with Smac mimetic alone8, 9, 13 or in combination with anticancer drugs in different carcinoma cell lines,14 TNFα was found to be largely dispensable for BV6/TMZ-induced apoptosis in glioblastoma cells.12 In the present study, we therefore aimed at discovering novel NF-κB-dependent factors that are required for the cooperative anticancer activity of BV6 and TMZ, the prototypic chemotherapeutic agent used for the treatment of glioblastoma.
Results
BV6/TMZ cotreatment upregulates IFN-responsive genes before cell death induction
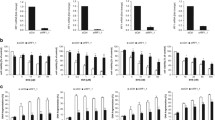
Initially, we assessed cell death upon treatment with the chemotherapeutic agent TMZ and the Smac mimetic BV6 using the glioblastoma cell lines A172 and T98G to confirm that Smac mimetic enhances TMZ-induced apoptosis, as we reported previously.12 Indeed, determination of DNA fragmentation as a marker of apoptosis demonstrated that BV6 sensitizes glioblastoma cells to TMZ-induced apoptotic cell death (Figure 1a and Supplementary Figure 1). As Smac mimetics have been described to activate NF-κB signaling, we stably overexpressed dominant-negative IκBα-superrepressor (IκBα-SR) to block NF-κB signaling15 (Figure 1c). Remarkably, inhibition of NF-κB by IκBα-SR almost completely rescued cells from BV6/TMZ-induced apoptosis (Figure 1b), underlining the proapoptotic role of NF-κB signaling in this context. To further investigate which BV6-induced NF-κB target genes are responsible for proapoptotic signaling upon BV6/TMZ cotreatment, we performed whole-genome expression profiling using an expression bead chip hybridization assay. Whole-genome expression data were ranked according to fold upregulation comparing A172 glioblastoma cells expressing empty vector (EV) with and without BV6/TMZ treatment. Expression data showing upregulation in A172 glioblastoma cells expressing IκBα-SR served as control to identify background expression of non-NF-κB-regulated genes. BV6-treated cells showed a similar expression pattern as BV6/TMZ cotreated cells (data not shown). Interestingly, gene set enrichment analysis (GSEA) revealed upregulation of interferon (IFN)-responsive genes after BV6/TMZ treatment (Table 1 and Supplementary Figure 2). These results demonstrate that BV6/TMZ treatment upregulates IFN-responsive genes in an NF-κB-dependent manner.
BV6/TMZ cotreatment upregulates IFN-responsive genes. (a) A172 cells (left) or T98G cells (right) were treated for indicated times with 100 μM TMZ and/or 2 μM BV6 (A172) or 4 μM BV6 (T98G) or dimethyl sulfoxide (DMSO). Apoptosis was determined by fluorescence-activated cell sorting (FACS) analysis of DNA fragmentation of PI-stained nuclei. Mean values +S.D. of three to four independent experiments performed in triplicate are shown; *P<0.05; **P<0.01 compared with all other settings. (b) A172 cells (left) or T98G cells (right) stably expressing IκBα-SR or EV were treated for 120 h with 100 μM TMZ and/or 2 μM BV6 (A172) or 4 μM BV6 (T98G) or DMSO. Apoptosis was determined by FACS analysis of DNA fragmentation of PI-stained nuclei. Mean values +S.D. of three independent experiments performed in triplicate are shown; *P<0.05; **P<0.01 compared with all other settings. (c) A172 cells (left) or T98G cells (right) stably expressing IκBα-SR or EV were analyzed for IκBα expression levels by western blotting. Expression of β-actin served as a loading control. A representative experiment of two independent experiments is shown
BV6-mediated upregulation of IFNβ sensitizes glioblastoma cells to TMZ-induced apoptosis
Next, we investigated whether IFNs are involved in BV6/TMZ-induced cell death. Type I IFNs such as IFNα and IFNβ have been reported to synergize with TMZ in cell death induction in glioblastoma cells.16, 17 Therefore, we analyzed mRNA expression levels of IFNα and IFNβ upon treatment with BV6 and/or TMZ using quantitative real-time-PCR (qRT-PCR) analysis, as IFNs were not represented on the expression bead chip hybridization assay. Interestingly, IFNβ was upregulated upon BV6 single treatment, as well as upon BV6/TMZ cotreatment (Figure 2a). In addition, BV6-stimulated transcriptional upregulation of IFNβ was inhibited in IκBα-SR-overexpressing cells (Figure 2a), demonstrating that it occurs in an NF-κB-dependent manner. In contrast to IFNβ, IFNα mRNA expression levels remained largely unchanged upon treatment with BV6 and/or TMZ (Figure 2b).
BV6-mediated upregulation of IFNβ sensitizes glioblastoma cells to TMZ-induced apoptosis. A172 cells (left) or T98G cells (right) stably expressing IκBα-SR or EV were treated for 6 h with 100 μM TMZ and/or 2 μM BV6 (A172) or 4 μM BV6 (T98G) or dimethyl sulfoxide (DMSO). IFNβ (a) or IFNα (b) mRNA levels were analyzed by qRT-PCR, normalized to 28S rRNA expression and fold increase in mRNA levels are shown. Mean values+S.D. of three to four independent experiments performed in duplicate are shown. *P<0.05; **P<0.01. (c) A172 cells (left) or T98G cells (right) were treated for 120 h with 100 μM TMZ and/or 1 ng/ml IFNβ or DMSO. Apoptosis was determined by fluorescence-activated cell sorting (FACS) analysis of DNA fragmentation of PI-stained nuclei. Mean values+S.D. of three independent experiments performed in triplicate are shown; *P<0.05; **P<0.01
To explore whether IFNβ acts in concert with TMZ to cause cell death, we treated glioblastoma cells with IFNβ alone and in combination with TMZ. Intriguingly, IFNβ significantly increased TMZ-induced cell death in A172 and T98G cells compared to treatment with either agent alone (Figure 2c). In addition to IFNβ, IFNα significantly enhanced TMZ-induced cell death in glioblastoma cells (Supplementary Figure 3). This set of experiments demonstrates that BV6/TMZ induces upregulation of IFNβ in an NF-κB-dependent manner and that IFNβ and TMZ cooperate to induce apoptosis in glioblastoma cells.
IFNβ is required for BV6/TMZ-induced apoptosis
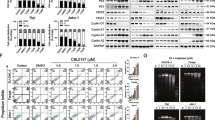
To examine whether IFNβ is required for BV6/TMZ-induced cell death, we created IFNβ-knockdown cells (Figure 3a). Remarkably, silencing of IFNβ significantly inhibited BV6/TMZ-mediated cell death (Figure 3b). Type I IFNs such as IFNα and IFNβ bind to a transmembrane receptor termed IFNα/β receptor (IFNAR) and thereby induce transcriptional activation of IFN-stimulated genes (ISGs).18 To examine whether IFNAR signaling is involved in BV6/TMZ-induced cell death, we generated IFNAR1-knockdown cells (Figure 3c). Silencing of IFNAR1 significantly reduced BV6/TMZ-mediated cell death (Figure 3d). In addition, knockdown of IFNAR1 rescued cells from IFNβ/TMZ-induced cell death (Figure 3e). Taken together, this set of experiments demonstrates that IFNβ has an important role in mediating BV6/TMZ-induced cell death.
IFNβ is a crucial mediator of BV6/TMZ-induced cell death. (a) A172 cells (left) or T98G cells (right) were transiently transfected with small interfering RNA (siRNA) against IFNβ. IFNβ mRNA levels were analyzed after 120 h by qRT-PCR, normalized to 28S rRNA expression and fold increase in mRNA levels are shown. Mean values+S.D. of three independent experiments performed in duplicate are shown. *P<0.05; **P<0.01. (b) A172 cells (left) or T98G cells (right) were transiently transfected with siRNA against IFNβ. Cells were treated for 120 h with 100 μM TMZ and/or 2 μM BV6 (A172) or 4 μM BV6 (T98G) or dimethyl sulfoxide (DMSO). Apoptosis was determined by fluorescence-activated cell sorting (FACS) analysis of DNA fragmentation of PI-stained nuclei. Mean values+S.D. of three to four independent experiments performed in triplicate are shown; *P<0.05; **P<0.01. (c) A172 cells (left) or T98G cells (right) were transiently transfected with siRNA against IFNAR1. IFNAR1 mRNA levels were analyzed after 120 h by qRT-PCR, normalized to 28S rRNA expression and fold increase in mRNA levels are shown. Mean values+S.D. of three to four independent experiments performed in duplicate are shown. *P<0.05; **P<0.01. (d) A172 cells (left) or T98G cells (right) were transiently transfected with siRNA against IFNAR1. Cells were treated for 120 h with 100 μM TMZ and/or 2 μM BV6 (A172) or 4 μM BV6 (T98G) or DMSO. Apoptosis was determined by FACS analysis of DNA fragmentation of PI-stained nuclei. Mean values+S.D. of three independent experiments performed in triplicate are shown; *P<0.05; **P<0.01. (e) A172 cells (left) or T98G cells (right) were transiently transfected with siRNA against IFNAR1. Cells were treated for 120 h with 100 μM TMZ and/or 1 ng/ml IFNβ or DMSO. Apoptosis was determined by FACS analysis of DNA fragmentation of PI-stained nuclei. Mean values+S.D. of four independent experiments performed in triplicate are shown; *P<0.05; **P<0.01
BV6/TMZ-induced apoptosis is mediated by cooperative upregulation of Puma and Bax
ISGs have been described to mediate IFN-induced apoptosis via upregulation of proapoptotic proteins, including proteins of the mitochondrial-dependent cell death pathway.19 To link BV6/TMZ-mediated activation of IFN signaling to activation of the mitochondrial apoptotic pathway, we analyzed the expression levels of various proapoptotic Bcl-2 family members (Supplementary Figure 4a). BV6/TMZ cotreatment significantly upregulated mRNA levels of Puma and Bax, whereas no consistent upregulation was observed for Bak (Bcl-2 homologous antagonist/killer), Noxa, Bim (Bcl-2-interacting mediator of cell death), Bid (BH3-interacting domain death agonist) and Bmf (Bcl-2-modifying factor; Figures 4a and b and Supplementary Figure 4a). Also, Puma and Bax were upregulated on the protein level upon BV6/TMZ treatment (Supplementary Figure 4b). To determine whether Puma and Bax are required of BV6/TMZ-induced cell death, we created Puma- or Bax-knockdown cells (Figures 4c and e). Interestingly, knockdown of either Puma or Bax significantly reduced BV6/TMZ-mediated cell death (Figures 4d and f). These results demonstrate that Puma and Bax contribute to BV6/TMZ-induced cell death.
BV6/TMZ-induced apoptosis is mediated by the upregulation of Puma and Bax. A172 cells (left) or T98G cells (right) were treated for indicated times with 100 μM TMZ and/or 2 μM BV6 (A172) or 4 μM BV6 (T98G) or dimethyl sulfoxide (DMSO). Puma (a) or Bax (b) mRNA levels were analyzed by qRT-PCR, normalized to 28S rRNA expression and fold increase in mRNA levels are shown. Mean values+S.D. of three independent experiments performed in duplicate are shown. *P<0.05; **P<0.01 compared with DMSO control. (c) A172 cells (left) or T98G cells (right) were transiently transfected with small interfering RNA (siRNA) against Puma. Puma mRNA levels were analyzed after 120 h by qRT-PCR, normalized to 28S rRNA expression and fold increase in mRNA levels are shown. Mean values+S.D. of three independent experiments performed in duplicate are shown. *P<0.05; **P<0.01 compared with all other settings, if not indicated otherwise. (d) A172 cells (left) or T98G cells (right) were transiently transfected with siRNA against Puma. Cells were treated for 120 h with 100 μM TMZ and/or 2 μM BV6 (A172) or 4 μM BV6 (T98G) or DMSO. Apoptosis was determined by fluorescence-activated cell sorting (FACS) analysis of DNA fragmentation of PI-stained nuclei. Mean values+S.D. of three to four independent experiments performed in triplicate are shown; *P<0.05; **P<0.01. (e) A172 cells (left) or T98G cells (right) were transiently transfected with siRNA against Bax. Bax mRNA levels were analyzed after 120 h by qRT-PCR, normalized to 28 S rRNA expression and fold increase in mRNA levels are shown. Mean values+S.D. of three independent experiments performed in duplicate are shown. *P<0.05; **P<0.01. (f) A172 cells (left) or T98G cells (right) were transiently transfected with siRNA against Bax. Cells were treated for 120 h with 100 μM TMZ and/or 2 μM BV6 (A172) or 4 μM BV6 (T98G) or DMSO. Apoptosis was determined by FACS analysis of DNA fragmentation of PI-stained nuclei. Mean values+S.D. of three independent experiments performed in triplicate are shown; *P<0.05; **P<0.01
Discussion
We previously reported NF-κB-dependent sensitization of glioblastoma cells to TMZ-induced apoptosis by the Smac mimetic BV6 as a novel approach to enhance the efficacy of conventional chemotherapy in glioblastoma.12 However, the proapoptotic NF-κB target genes mediating this synergistic interaction have so far remained elusive, as autocrine/paracrine TNFα/TNF receptor 1 (TNFR1) signaling turned out to be largely dispensable.12 In the present study, we identify Smac mimetic-stimulated, NF-κB-dependent upregulation of IFNβ and IFN-mediated proapoptotic signaling as critical events that mediate BV6/TMZ-induced apoptosis (Supplementary Figure 5). This conclusion is based on the following lines of evidence:
First, treatment with BV6 alone or in combination with TMZ triggers transcriptional upregulation of IFNβ in an NF-κB-dependent manner, as this increase in IFNβ mRNA levels is blocked by IκBα-SR-mediated inhibition of NF-κB. In addition, gene expression profiling shows an NF-κB-dependent upregulation of ISGs upon BV6/TMZ treatment. Second, BV6-induced upregulation of IFNβ- and IFN-mediated signaling are required for the induction of apoptosis, as genetic silencing of either IFNβ or its corresponding receptor IFNAR significantly reduces BV6/TMZ-induced apoptosis. The notion that BV6-stimulated upregulation of IFNβ promotes TMZ-induced apoptosis is further underscored by data showing that exogenous supply of IFNβ cooperates with TMZ to trigger apoptosis in glioblastoma cells. Third, we show that IFNβ and TMZ cooperate to upregulate the proapoptotic Bcl-2 family proteins Puma and Bax, which both contribute to BV6/TMZ-induced apoptosis, as knockdown of Bax or Puma significantly rescues cells from BV6/TMZ-induced apoptosis. Taken together, this identification of Smac mimetic-stimulated, NF-κB-dependent upregulation of IFNβ and engagement of proapoptotic IFN signaling pathways provides novel insights into the molecular mechanisms that are responsible for Smac mimetic-mediated sensitization of glioblastoma cells to TMZ-induced cell death.
In the present study, we identify IFNβ as a key mediator of BV6/TMZ-induced cell death that is transcriptionally upregulated in an NF-κB-dependent manner upon treatment with the Smac mimetic BV6. Whether or not this increase in IFNβ is directly mediated via NF-κB transcription factors20 or indirectly mediated via NF-κB-dependent upregulation or activation of transcription factors regulating IFNβ21 remains to be investigated in future studies. It is interesting to note that type I IFNs such as IFNβ or IFNα have recently been reported to act in concert with TMZ to trigger cell death in glioblastoma cells, although the mechanisms responsible for this cooperative effect have so far remained elusive.16, 17 In contrast to the critical role of IFNβ for BV6/TMZ-induced apoptosis that we discovered in the current study, we previously reported that TNFα, another prototypic NF-κB target gene, is largely dispensable for BV6/TMZ-induced apoptosis, as addition of the TNFα-blocking antibody Enbrel or TNFR1 knockdown failed to rescue apoptosis upon combination treatment.12
Although our study demonstrates for the first time that the Smac mimetic BV6 can transcriptionally induce IFNβ as an important mediator of Smac mimetic-conferred chemosensitization in glioblastoma cells, IFN signaling has been implicated in the past to foster cell death by Smac mimetics. For example, we recently reported that BV6 synergizes with IFNα to trigger apoptosis in acute myeloid leukemia cells.22 Of note, BV6 was found in the present study to transcriptionally upregulate IFNβ, but not IFNα in glioblastoma cells, pointing to distinct roles of type I IFNs in this context. Moreover, we identified IFN regulatory factor 1 (IRF1) as a novel dual regulator of Smac mimetic BV6-induced apoptosis and proinflammatory cytokine secretion with impact on the immune response.23, 24 Furthermore, Smac mimetics have been described to act in concert with innate immune stimuli such as oncolytic viruses and adjuvants, which stimulate a cytokine storm of TNFα, TNF-related apoptosis-inducing ligand and IFNβ, to trigger cancer cell death.25
Induction of cell death by IFNβ has been described to involve ISGs.19 However, little is yet known about which ISGs mediate these apoptotic functions. Transcription factors regulated via IFNs such as IRF1 and IRF3 have been reported to promote upregulation or activation of Puma and Bax.26, 27 Puma and Bax were also described as DNA damage-induced target genes that are upregulated by TMZ treatment.28 Consistently, we show in the present study that IFNβ and TMZ cooperate to upregulate Puma and Bax, which both contribute to BV6/TMZ-induced apoptosis, as genetic silencing of either Bax or Puma, two Bcl-2 family proteins known to promote mitochondrial apoptosis, rescues glioblastoma cells from cell death. In line with these findings, we previously reported that cotreatment with BV6/TMZ activates the mitochondrial pathway of apoptosis as demonstrated by the loss of mitochondrial membrane potential and cytochrome C release.12
Furthermore, context-specific settings have an impact on the regulation of signaling pathways and cellular functions by Smac mimetics, depending, for example, on additional cytotoxic stimuli and/or cell types. We demonstrated that Smac mimetics can exert non-apoptotic functions and can stimulate migration and invasion of glioblastoma cells via activation of NF-κB and TNFα/TNFR1 autocrine/paracrine signaling.23, 24 In glioblastoma cancer stem-like cells, Smac mimetics at non-toxic concentrations can promote astrocytic differentiation by activating NF-κB.29
Smac mimetics are currently evaluated in early clinical trials.30 By identifying IFNβ as a novel Smac mimetic-induced and NF-κB-mediated target gene that has an important role in mediating chemosensitization by Smac mimetic, our findings provide novel mechanistic insights into this combination regimen. Additionally, our present study emphasizes that Smac mimetics are effective sensitizers for TMZ-induced apoptosis in glioblastoma cells with implications for the development of experimental treatment approaches.
Materials and Methods
Cell culture and chemicals
The human glioblastoma cell lines A172 and T98G were obtained from the American Type Culture Collection (Manassas, VA, USA) and maintained in DMEM medium (Invitrogen, Karlsruhe, Germany) supplemented with 1% penicillin/streptomycin, 1% sodium pyruvate and 10% fetal calf serum (Invitrogen). For experiments, cells were seeded at 5 × 103 cells/cm2. Smac mimetic BV6, which neutralizes XIAP, cIAP1 and cIAP2,8 was kindly provided by Genentech Inc. (South San Francisco, CA, USA), TMZ was purchased from Sigma (Taufkirchen, Germany) and recombinant human IFNα and IFNβ from Biochrom (Berlin, Germany). All chemicals were obtained from Sigma, unless indicated otherwise.
Determination of apoptosis
Apoptosis was assessed by flow cytometric analysis (FACSCanto II; BD Biosciences, Heidelberg, Germany) of DNA fragmentation of propidium iodide (PI)-stained nuclei as described previously.31
Western blotting
Western blot analysis was performed as described previously12 using the following antibodies: anti-IκBα (Cell Signaling, Beverly, MA, USA), anti-Bax (BD Biosciences), anti-Puma (Cell Signaling) and anti-β-actin (Sigma). Donkey anti-mouse IgG or donkey anti-rabbit IgG labeled with IRDye infrared dyes were used for fluorescence detection at 680–800 nm (LI-COR Biotechnology, Bad Homburg, Germany).
Whole-genome gene expression array and GSEA
Gene expression profiling was performed as described previously32 using Illumina Whole-Genome Expression Beadchips Human HT12v4 (Illumina, San Diego, CA, USA). Expression data were ranked according to fold upregulation comparing A172 glioblastoma cells expressing EV with and without BV6/TMZ treatment. Expression data showing upregulation with and without BV6/TMZ treatment in A172 glioblastoma cells expressing IκBα-SR served as control to identify background expression of non-NF-κB-regulated genes. GSEA was performed using software provided by the Broad Institute website (http://www.broadinstitute.org/gsea/index.jsp).33
Transduction and siRNA transfection
Overexpression of the dominant-negative IκBα-SR was performed by retroviral transduction as described previously.15 For transient knockdown by siRNA, cells were transfected with 20 nM Silencer Select siRNA (Invitrogen) control siRNA (no. 4390844) or targeting siRNAs (s7188 and s7189 for IFNβ, s782 and s784 for IFNAR1, s1888 and s1890 for Bax, s25840 and s25842 for Puma) using Neon Transfection System (Invitrogen) according to the manufacturer's instructions.
qRT-PCR analysis
Total RNA extraction and qRT-PCR analysis was performed as described previously32 using 7900HT Fast Real-Time PCR System (Applied Biosystems, Darmstadt, Germany). The following primers were used: 28 S (forward, 5′-TTGAAAATCCGGGGGAGAG-3′; reverse, 5′-ACATTGTTCCAACATGCCAG-3′), IFNAR1 (forward, 5′-TCCAGTACATTGTATAAAGACCACAGT-3′; reverse, 5′-GTTCTGATTTTGGACACTGACTTC-3′), Puma (forward, 5′-GACCTCAACGCACAGTACGA-3′; reverse, 5′-GAGATTGTACAGGACCCTCCA-3′), Bax (forward, 5′-AGCAAACTGGTGCTCAAGG-3′; reverse, 5′-TCTTGGATCCAGCCCAAC-3′), Bak (forward, 5′-CCTGCCCTCTGCTTCTGA-3′; reverse, 5′-CTGCTGATGGCGGTAAAAA-3′), Noxa (forward, 5′-GGAGATGCCTGGGAAGAAG-3′; reverse, 5′-CCTGAGTTGAGTAGCACACTCG-3′), Bid (forward, 5′-TGCAGCTCAGGAACACCA-3′; reverse, 5′-TCTCCATGTCTCTAGGGTAGGC-3′), Bim (forward, 5′-CATCGCGGTATTCGGTTC-3′; reverse, 5′-GCTTTGCCATTTGGTCTTTTT-3′), Bmf (forward, 5′-GAGACTCTCTCCTGGAGTCACC-3′; reverse, 5′-CTGGTTGGAACACATCATCCT-3′). Melting curves were plotted to verify the specificity of the amplified products. IFNα and IFNβ mRNA levels were assessed by TaqMan Gene Expression Assay (Life Technologies, Darmstadt, Germany; IFNαHs01077958_s1, IFNβHs00855471_g1) according to the manufacturer’s protocol. The relative expression of the target gene transcript and reference gene transcript was calculated as ΔΔCt.
Statistical analysis
Statistical significance was assessed by Student's t-test (two-tailed distribution, two-sample unequal variance).
Abbreviations
- Bak:
-
Bcl-2 homologous antagonist/killer
- Bax:
-
Bcl-2-associated X protein
- Bcl-2:
-
B-cell lymphoma 2
- Bid:
-
BH3-interacting domain death agonist
- Bim:
-
Bcl-2-interacting mediator of cell death
- Bmf:
-
Bcl-2-modifying factor
- IAP:
-
inhibitor of apoptosis
- IFN:
-
interferon
- IFNAR:
-
IFNα/β receptor
- IκBα-SR:
-
IκBα superrepressor
- IRF:
-
interferon regulatory factor
- ISG:
-
IFN-stimulated gene
- NF-κB:
-
nuclear factor-κB
- NIK:
-
nuclear factor-κB-inducing kinase
- Noxa/PMAIP1:
-
phorbol-12-myristate-13-acetate-induced protein 1
- Puma:
-
p53-upregulated modulator of apoptosis
- RING:
-
Really Interesting New Gene
- Smac:
-
second mitochondria-derived activator of caspases
- TMZ:
-
temozolomide
- TNFα:
-
tumor necrosis factor-α
- TNFR1:
-
tumor necrosis factor-α receptor 1
References
Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 2009; 10: 459–466.
Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ . Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin 2010; 60: 166–193.
Hanahan D, Weinberg RA . Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674.
Fulda S . Tumor resistance to apoptosis. Int J Cancer 2009; 124: 511–515.
Fulda S, Debatin KM . Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene 2006; 25: 4798–4811.
Fulda S, Galluzzi L, Kroemer G . Targeting mitochondria for cancer therapy. Nat Rev Drug Discov 2010; 9: 447–464.
Fulda S, Vucic D . Targeting IAP proteins for therapeutic intervention in cancer. Nat Rev Drug Discov 2012; 11: 109–124.
Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell 2007; 131: 669–681.
Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell 2007; 131: 682–693.
Vellanki SH, Grabrucker A, Liebau S, Proepper C, Eramo A, Braun V et al. Small-molecule XIAP inhibitors enhance gamma-irradiation-induced apoptosis in glioblastoma. Neoplasia 2009; 11: 743–752.
Berger R, Jennewein C, Marschall V, Karl S, Cristofanon S, Wagner L et al. NF-{kappa}B is required for Smac mimetic-mediated sensitization of glioblastoma cells for {gamma}-irradiation-induced apoptosis. Mol Cancer Ther 2011; 10: 1867–1875.
Wagner L, Marschall V, Karl S, Cristofanon S, Zobel K, Deshayes K et al. Smac mimetic sensitizes glioblastoma cells to temozolomide-induced apoptosis in a RIP1- and NF-kappaB-dependent manner. Oncogene 2013; 32: 988–997.
Petersen SL, Wang L, Yalcin-Chin A, Li L, Peyton M, Minna J et al. Autocrine TNFalpha signaling renders human cancer cells susceptible to Smac-mimetic-induced apoptosis. Cancer Cell 2007; 12: 445–456.
Probst BL, Liu L, Ramesh V, Li L, Sun H, Minna JD et al. Smac mimetics increase cancer cell response to chemotherapeutics in a TNF-alpha-dependent manner. Cell Death Differ 2010; 17: 1645–1654.
Karl S, Pritschow Y, Volcic M, Hacker S, Baumann B, Wiesmuller L et al. Identification of a novel pro-apopotic function of NF-kappaB in the DNA damage response. J Cell Mol Med 2009; 13: 4239–4256.
Park JA, Joe YA, Kim TG, Hong YK . Potentiation of antiglioma effect with combined temozolomide and interferon-beta. Oncol Rep 2006; 16: 1253–1260.
Yoshino A, Tashiro S, Ogino A, Yachi K, Ohta T, Fukushima T et al. Gene expression profiles predicting the response to IFN-beta and a combination of temozolomide and IFN-beta in malignant gliomas. Int J Oncol 2011; 39: 529–542.
Stark GR, Darnell JE Jr . The JAK-STAT pathway at twenty. Immunity 2012; 36: 503–514.
Kotredes KP, Gamero AM . Interferons as inducers of apoptosis in malignant cells. J Interferon Cytokine Res 2013; 33: 162–170.
Lenardo MJ, Fan CM, Maniatis T, Baltimore D . The involvement of NF-kappa B in beta-interferon gene regulation reveals its role as widely inducible mediator of signal transduction. Cell 1989; 57: 287–294.
Ivashkiv LB, Donlin LT . Regulation of type I interferon responses. Nat Rev Immunol 2014; 14: 36–49.
Bake V, Roesler S, Eckhardt I, Belz K, Fulda S . Synergistic interaction of Smac mimetic and IFNalpha to trigger apoptosis in acute myeloid leukemia cells. Cancer Lett 2014; 355: 224–231.
Tchoghandjian A, Jennewein C, Eckhardt I, Rajalingam K, Fulda S . Identification of non-canonical NF-kappaB signaling as a critical mediator of Smac mimetic-stimulated migration and invasion of glioblastoma cells. Cell Death Dis 2013; 4: e564.
Eckhardt I, Weigert A, Fulda S . Identification of IRF1 as critical dual regulator of Smac mimetic-induced apoptosis and inflammatory cytokine response. Cell Death Dis 2014; 5: e1562.
Beug ST, Tang VA, LaCasse EC, Cheung HH, Beauregard CE, Brun J et al. Smac mimetics and innate immune stimuli synergize to promote tumor death. Nat Biotechnol 2014; 32: 182–190.
Chattopadhyay S, Marques JT, Yamashita M, Peters KL, Smith K, Desai et al. Viral apoptosis is induced by IRF-3-mediated activation of Bax. EMBO J 2010; 29: 1762–1773.
Gao J, Senthil M, Ren B, Yan J, Xing Q, Yu J et al. IRF-1 transcriptionally upregulates PUMA, which mediates the mitochondrial apoptotic pathway in IRF-1-induced apoptosis in cancer cells. Cell Death Differ 2010; 17: 699–709.
Borges HL, Linden R, Wang JY . DNA damage-induced cell death: lessons from the central nervous system. Cell Res 2008; 18: 17–26.
Tchoghandjian A, Jennewein C, Eckhardt I, Momma S, Figarella-Branger D, Fulda S . Smac mimetic promotes glioblastoma cancer stem-like cell differentiation by activating NF-kappaB. Cell Death Differ 2014; 21: 735–747.
Fulda S . Molecular pathways: targeting inhibitor of apoptosis proteins in cancer – from molecular mechanism to therapeutic application. Clin Cancer Res 2014; 20: 289–295.
Fulda S, Sieverts H, Friesen C, Herr I, Debatin KM . The CD95 (APO-1/Fas) system mediates drug-induced apoptosis in neuroblastoma cells. Cancer Res 1997; 57: 3823–3829.
Eckhardt I, Roesler S, Fulda S . Identification of DR5 as a critical, NF-kappaB-regulated mediator of Smac-induced apoptosis. Cell Death Dis 2013; 4: e936.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Acknowledgements
We thank D Vucic (Genentech Inc.) for providing BV6, the microarray unit of the DKFZ Genomics and Proteomics Core Facility (Heidelberg, Germany) for providing the Illumina Whole-Genome Expression Beadchips and related services, Ronald Sauter for expert technical assistance and C Hugenberg for expert secretarial assistance. This work has been partially supported by grants from the Deutsche Forschungsgemeinschaft, the BMBF and IUAPVII (to SF).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Dewson
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Marschall, V., Fulda, S. Smac mimetic-induced upregulation of interferon-β sensitizes glioblastoma to temozolomide-induced cell death. Cell Death Dis 6, e1888 (2015). https://doi.org/10.1038/cddis.2015.235
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2015.235
- Springer Nature Limited
This article is cited by
-
BH3-mimetics: recent developments in cancer therapy
Journal of Experimental & Clinical Cancer Research (2021)
-
The Smac mimetic BV6 cooperates with STING to induce necroptosis in apoptosis-resistant pancreatic carcinoma cells
Cell Death & Disease (2021)
-
Cell cycle arrest in mitosis promotes interferon-induced necroptosis
Cell Death & Differentiation (2019)
-
Cell death-based treatment of glioblastoma
Cell Death & Disease (2018)
-
PD-L1 and IAPs co-operate to protect tumors from cytotoxic lymphocyte-derived TNF
Cell Death & Differentiation (2017)