Abstract
The endoplasmic reticulum (ER) has a major role in protein folding. The accumulation of unfolded proteins in the ER induces a stress, which can be resolved by the unfolded protein response (UPR). Chronicity of ER stress leads to UPR-induced apoptosis and in turn to an unbalance of tissue homeostasis. Although ER stress-dependent apoptosis is observed in a great number of devastating human diseases, how cells activate apoptosis and promote tissue homeostasis after chronic ER stress remains poorly understood. Here, using the Drosophila wing imaginal disc as a model system, we validated that Presenilin overexpression induces chronic ER stress in vivo. We observed, in this novel model of chronic ER-stress, a PERK/ATF4-dependent apoptosis requiring downregulation of the antiapoptotic diap1 gene. PERK/ATF4 also activated the JNK pathway through Rac1 and Slpr activation in apoptotic cells, leading to the expression of Dilp8. This insulin-like peptide caused a developmental delay, which partially allowed the replacement of apoptotic cells. Thanks to a novel chronic ER stress model, these results establish a new pathway that both participates in tissue homeostasis and triggers apoptosis through an original regulation.
Similar content being viewed by others
Main
Protein folding is a major role of the endoplasmic reticulum (ER) that can be challenged by modification of calcium homeostasis, elevated protein synthesis, glucose deprivation, hypoxia and altered protein glycosylation, leading to unfolded or misfolded protein accumulation in the ER. If this accumulation exceeds the folding capacity of chaperones, ER-stress is induced, triggering an adaptive response known as the unfolded protein response (UPR) to resolve the stress or eliminate the cell.1,2 Unresolved ER stress-induced apoptosis is observed in a great number of devastating diseases including neurodegenerative and renal diseases, diabetes and atherosclerosis.3 Three principal arms of the UPR have been identified and are particularly well characterized in mammals.4 Each branch is regulated by a different sensor transmembrane protein, that is, IRE1 (inositol-requiring enzyme 1), PERK (double-stranded RNA-activated Protein kinase (PKR) – like ER Kinase) or ATF6 (Activating Transcription Factor 6) – which senses unfolded protein accumulation and leads to the transcriptional activation of genes involved in the UPR. All three sensors of the UPR are conserved, though ATF6 has not yet been linked to the UPR in Drosophila.5, 6, 7 How cells activate apoptosis after a chronic ER stress remains poorly understood. This accidental programmed cell death can lead to an unbalance of tissue homeostasis. Surprisingly, the UPR has never been shown to play a role in tissue homeostasis after ER stress-mediated cell death.
Well-known strategies that allow tissue homeostasis in Drosophila wing imaginal discs are compensatory proliferation and apoptosis-induced proliferation.8 Tissue homeostasis depends on the c-Jun N-terminal kinase (JNK) pathway that is activated either in apoptotic or in proliferating cells according to the apoptotic stimuli.9, 10, 11 Both the mechanism of activation and the role of this pathway remain unclear. Indeed, the JNK signaling has been reported to be controlled by either Drosophila inhibitor of apoptosis (DIAP1) or the initiator caspase Dronc.12, 13, 14 Nevertheless, the JNK pathway components that would be targets of DIAP1 or Dronc remain unidentified. In Drosophila as in mammals, the transcription factor that represents the last component of this pathway, dAP-1, is a dimer of Jra and Kay, which are homologous to Jun and Fos.15 It is activated through a cascade of phosphorylations. Although three JNKs exist in mammals, Bsk is the only one in Drosophila. Similarly, the complexity of the pathway is lower in dipterans than in mammals with two JNKKs, six JNKKKs and many regulators of the JNKKKs described in Drosophila.16,17 The literature supports the idea that signaling specificity could be driven by the JNKKK and by the combination of activated kinases.18
The JNK pathway has also been identified as mediating developmental delay by controlling Drosophila insulin-like peptide 8 (Dilp8) expression.19 Previous studies have demonstrated that damaged imaginal discs delay the onset of metamorphosis, thus permitting tissue regeneration.20, 21, 22 This developmental delay is the result of a depletion of the Prothoracicotropic hormone (Ptth), a neuropeptide which promotes the release of the steroid hormone ecdysone that regulates developmental transitions in ecdysosoans such as Drosophila. Dilp8 and retinoids have been identified as secreted signals produced by damaged discs to promote ptth transcription inhibition.19,22,23
Although numerous studies have shown that ER stress can be induced by various stimuli, ER stress-induced cell death has only been studied once in Drosophila. This recent work showed that a strong chronic ER stress induces apoptosis in the eye imaginal discs of Drosophila thanks to a CDK5/MEKK1/JNK signaling pathway.5,24 This model of ER-induced apoptosis relies on a rhodopsin-1 mutant allele mimicking autosomal dominant retinitis pigmentosa (ADRP). The Presenilin gene encodes an eight to nine-pass transmembrane protein best described to function as the catalytic subunit of the γ-secretase multiprotein complex after an endoproteolytic processing, but also as a regulator of calcium flux in the ER. It has been previously shown that Presenilin overexpression could modify calcium homeostasis in Drosophila wing discs25 and in cultured mammalian cells.26 This expression induces ER stress in mammalian cells. Full-length Presenilin is primarily located on the ER membrane.27,28 The overexpression of mammalian Presenilin in cultured cells induced the accumulation in the ER membrane of Psn with unmodified catalytic activity.29,30 Therefore, we hypothesized that the overexpression of the Drosophila Presenilin gene (Psn) could be an appropriate model to induce ER stress in this multicellular organism.
In this study, we validate that Psn overexpression in wing imaginal discs induces an ER stress and we show that this ER stress triggers both cell death and a regulation of tissue homeostasis. Indeed, a consequence of this overexpression is the induction of a PERK-dependent apoptosis through the activation of the transcription factor ATF4 that downregulates the antiapoptotic diap1 gene. This ER stress-induced cell death does not induce apoptosis-induced proliferation. ATF4 also activates the JNK pathway through Rac1 and Slpr activation in apoptotic cells. Interestingly, the JNK pathway does not control cell death in this model, but upregulates Dilp8, which participates in tissue homeostasis maintenance by controlling the developmental clock. In summary, we present a novel in vivo model of chronic ER stress that greatly differs from the previously established ER stress model and we show that the PERK/ATF4 branch of the UPR can trigger a developmental delay favoring tissue regeneration through the Rac1/Slpr/JNK pathway in Drosophila wing imaginal discs.
Results
Overexpression of Presenilin leads to both ER stress and PERK/ATF4-dependent cell death
To test if the overexpression of Psn could be an appropriate model to induce ER stress in Drosophila, this gene was overexpressed in part of the vestigial (vg)-expression domain, which covers the wing-pouch dorsoventral boundary (i.e., the cells corresponding to the adult wing margin) and a part of the hinge and notum, thanks to the GAL4–UAS system (Figure 1a, top panel).31 Psn overexpression in this domain led to a notched-wing phenotype of variable expressivity (Supplementary Figure S1A). As Psn is involved in the Notch pathway, this adult phenotype could suggest that Psn overexpression inhibits the Notch pathway. However, consistent with the data indicating that Psn overexpression does not affect Psn catalytic activity,29,30 Psn overexpression and Notch depletion induced different phenotypes in third-instar wing imaginal discs. Notch depletion by using an RNAi transgene induced a complete extinction of Wg expression on the dorsoventral boundary that was not observed in discs overexpressing Psn. Conversely, Psn overexpression induced apoptosis, which was not observed upon Notch depletion (Supplementary Figure S1B).
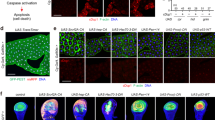
The PERK/ATF4 pathway is responsible for Psn overexpression-induced loss of DIAP1 and apoptosis. (a) Third-instar imaginal wing discs with the Psn-expressing domain detected thanks to GFP (green) and TUNEL staining (red) to detect apoptosis. Genotypes are vg-GAL4/+; UAS-EGFP/+ (top), vg-GAL4, UAS-Psn, UAS-Psn/+; UAS-EGFP/+ (middle) and UAS-p35/+; vg-GAL4, UAS-Psn, UAS-Psn/+; UAS-EGFP/+ (bottom). (b) Third-instar imaginal wing discs carrying an xbp1::EGFP reporter transgene (green) to detect ER stress and immunostained with an anti-activated Caspase 3 staining (red) to detect apoptosis. Genotypes are vg-GAL4/+; xbp1::EGFP/+ (top row) and vg-GAL4, UAS-Psn, UAS-Psn/+; xbp1::EGFP/+ (middle row). Bottom row images are enlargement of boxed regions. Colocalization between anti-activated Caspase 3 staining and Ire1 activity reporter is highlighted in white thanks to the ImageJ Colocalization plugin in the merge enlargement. Note that developmental apoptosis is only observed in the notum (top). (c) TUNEL staining of Psn-expressing third-instar imaginal wing discs in the absence or presence of dronc mutations. Genotypes are vg-GAL4, UAS-Psn, UAS-Psn/+ (left) and vg-GAL4, UAS-Psn, UAS-Psn/+; droncI24/ droncI29(right). (d, e) Third-instar imaginal wing discs with the Psn-expressing domain detected thanks to GFP (green), TUNEL stained (red) to detect apoptosis and immunostained with an anti-DIAP1 antibody (blue). (d) Genotypes are vg-GAL4/+; UAS-EGFP/+ (top), vg-GAL4, UAS-Psn, UAS-Psn/+; UAS-EGFP/+ (middle) and vg-GAL4, UAS-Psn, UAS-Psn/+; UAS-EGFP/UAS-atf4-RNAi (bottom). (e) Genotypes are vg-GAL4, UAS-Psn, UAS-Psn /UAS-nGFP; UAS-EGFP/+ (top), vg-GAL4, UAS-Psn, UAS-Psn/UAS-nGFP; UAS-p35/+ (bottom) (f) Anti-β-Galactosidase staining to detect diap1J5C8 reporter expression in vg-GAL4/+; diap1J5C8/+ (left column) vg-GAL4, UAS-Psn, UAS-Psn/+; diap1J5C8/+ (center); and vg-GAL4, UAS-Psn, UAS-Psn/+; diap1J5C8/UAS-atf4-RNAi (right column) third-instar wing imaginal discs. The black lines correspond to the projection of the corresponding enlarged transverse XZ sections shown under disk images. Enlarge areas are oriented with their basal side to the left. The asterisk indicates the diap1 expression reduction (center)
Overexpression of Psn triggered ER stress and activated the IRE1 branch of the UPR (Figure 1b), as reported by an xbp1::EGFP (enhanced green fluorescent protein) reporter, which allows EGFP protein synthesis only when ER stress stimulates IRE1-dependent XBP1 mRNA splicing.32 Cells expressing xbp1::EGFP were dying as assayed by TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling; Figure 1a) and an anti-activated caspase-3 labeling that reveals Dronc activity33 (Figure 1b). Moreover, expression of the caspase inhibitor baculovirus protein p35 and dronc loss-of-function inhibited ER stress-induced cell death (Figures 1a and c). These observations show that Psn overexpression induces both ER stress- and Dronc-dependent cell death.
To determine if this apoptosis depends on ER stress, we tested whether affecting the IRE1 and PERK branches of UPR could modulate it. Depleting components of the IRE1 branch (XBP1 or IRE1) by using efficacious RNAi transgenes did not modify TUNEL staining (data not shown and Supplementary Figure S2A), suggesting that this branch is not the main inducer of the apoptosis. Depletion of GADD34 (Growth Arrest and DNA Damage–inducible 34), the phosphatase that negatively regulates PERK activity,34 increased the cell death level specifically in Psn-overexpressing discs (Supplementary Figure S2B and data not shown). Conversely, TUNEL staining was strongly decreased upon depletion of either PERK (Supplementary Figure S2B) or its effector, ATF4 (Figure 1d). These results seemed specific and were not due to GAL4 titration as numerous transgenes inserted at the same locus revealed no effect or opposite effects to PERK and ATF4 depleting transgenes. Therefore, Psn overexpression triggers an ER stress-induced cell death that is mainly ATF4 dependent.
To better characterize the ER stress-induced cell death pathway, we focused on DIAP1, the major regulator of caspases in Drosophila. The DIAP1 protein level appeared to be decreased in the vg domain labeled by GFP (Figure 1d). Depletion of ATF4 was sufficient to re-establish a normal level of DIAP1 (Figure 1d). To rule out that this DIAP1 recovery was due to the reduction of the apoptosis observed in ATF4 depletion condition, we inhibited ER stress-induced apoptosis by expressing p35. The decrease of DIAP1 level was still observed (Figure 1e). A GAL4 titration effect could also be ruled out as the number of UAS transgenes did not affect the results. These data indicate a control of DIAP1 by the PERK/ATF4 branch of the UPR.
To test the existence of a diap1 transcriptional regulation, we used a PlacZ enhancer-trap insertion that monitors diap1 transcription.35 We observed a decrease of anti-β-Galactosidase staining in the ER-stressed domain, which was restored by ATF4 depletion (Figure 1f), demonstrating that ATF4 negatively regulates diap1 transcription after ER stress.
ER stress-induced cell death does not induce proliferation but causes a general Dilp8-dependent developmental delay
Previous studies have demonstrated that compensatory or apoptosis-induced proliferation could be activated in damaged imaginal discs to maintain tissue homeostasis.10,11,36, 37, 38, 39 We did not observe any difference in the number of mitotic cells labeled by an anti-Phospho-Histone H3 (PH3) immunostaining when compared with control discs (Supplementary Figure S3). Thus, ER stress-induced apoptosis does not seem to increase the cell proliferation rate in the surrounding tissue.
Damages to an imaginal disc can extend the larval development to allow coordination of tissue growth and regeneration within the organism.22 Indeed, Psn overexpression in the vg domain exhibited a 20-h delay before pupariation and adult eclosion (Figure 2a). To determine if this developmental delay participates to tissue homeostasis, we shortened the third larval stage by transferring larvae at 92 h after egg deposition (AED) to food supplemented with 20-Hydroxyecdysone (20E), which is the active form of the steroid hormone that promotes the 120 h AED transition from the larval to the pupal stage.40 This treatment suppressed the ER stress-induced delay of adult eclosion (Figure 2b) and aggravated the notched-wing phenotypes that result from the ER stress (Figure 2c), showing that the developmental delay participates to tissue homeostasis. Therefore, the developmental delay favors regeneration after an ER stress in the wing imaginal disc.
Dilp8-induced developmental delay favors tissue homeostasis after ER stress. (a) Effects of Psn overexpression on pupariation and adult eclosion timing. Hatched bar (vg-GAL4/+) and open bar (vg-GAL4, UAS-Psn, UAS-Psn/+) represent mean time in hours AED of pupariation (left) or adult eclosion (right). Error bars represent the S.E. of the mean. Asterisks indicate a significant difference with the control (Left: n=4, P<10−5, ANOVA), (Right: n=6, P<10−7, ANOVA). (b) Effects of 20E food supplementation of 92 h AED old vg-GAL4, UAS-Psn, UAS-Psn/+ larvae on adult eclosion. Error bars represent the S.E.M. of three independent experiments (asterisk: P<5%, ANOVA). (c) Distribution of notched-wing phenotypes after 20E supplementation (gray bars) or not (black bars) according to their strength in a representative experiment (n=3, P<5%, ANOVA). (d, e) Third-instar wing imaginal discs carrying Dilp8::EGFP (green) and immunostained with an antiactivated caspase 3 antibody (red). Genotypes are vg-GAL4/+; dilp8MI00727/+ (d) and vg-GAL4, UAS-Psn, UAS-Psn/+; dilp8MI00727/+ (e). The enlargement of the boxed area in e, shows signal colocalization highlighted in white thanks to the ImageJ Colocalization plugin. (f, g) dilp8 RNA levels measured by quantitative reverse PCR. Data represent mean ± S.E.M. of three independent experiments. RNA was extracted from control (vg-GAL4/+, hatched bar, (f) or ER stressed (vg-GAL4, UAS-Psn, UAS-Psn/+, open bar, f and g) or ER-stressed dronc mutant (vg-GAL4, UAS-Psn, UAS-Psn/+; droncI24/ droncI29, black bar, g) wing imaginal discs. The asterisk indicates a significant difference with the control (P<10−4, Student’s t-test). (h). Effects of dilp8 homozygous mutation (dilp8MI00727/dilp8MI00727, gray bars) compared with dilp8 heterozygosity (dilp8MI00727/rheaMI00296, white bars) on adult eclosion of vg-GAL4, UAS-Psn, UAS-Psn/+ (plain bars) and vg-GAL4/+ (hatched bars) on mean time of adult eclosion. The rheaMI00296 strain is commonly used as the control for dilp8MI00727 genetic background.23 Asterisks indicate significant difference from the corresponding control that does not express Psn (P<10−4, ANOVA). Error bars represent the S.E.M. of six independent experiments. (i) Distribution of notched-wing phenotypes of vg-GAL4, UAS-Psn, UAS-Psn/+; dilp8MI00727/dilp8MI00727flies (gray bars) compared with vg-GAL4, UAS-Psn, UAS-Psn/+; dilp8MI00727/rheaMI00296 flies (open bars) (n=3, P<10−6, ANOVA)
Recently, dilp8 expression has been reported to induce developmental delay in challenged imaginal discs.19,23 Indeed, accumulation of EGFP, reporting dilp8 expression thanks to dilp8MI00727,23 was observed in cells in which caspases were activated (Figures 2d and e) and dilp8 mRNA was increased by 230-fold in ER stressed wing imaginal discs when compared with unstressed wing discs (Figure 2f). This increase was not significantly modified in dronc mutant wing imaginal discs (Figure 2g, P=27%, paired t -test), suggesting that the caspase activation is not the major control of dilp8 expression. A significant decrease of the adult eclosion delay (Figure 2h) as well as a shift toward stronger notched-wing phenotypes (Figure 2i) were observed in dilp8MI00727homozygous mutants in which dilp8 mRNA levels are strongly reduced.23 Altogether, these results indicate that Dilp8 is a major contributor to both developmental delay and tissue homeostasis after ER stress that is not directly controlled by caspase activation.
The JNK pathway activation in apoptotic cells depends on PERK/ATF4 and controls a developmental delay
The JNK signaling has been previously reported to regulate dilp8 expression but also to be activated by ER stress-induced cell death in the Drosophila model of ADRP.5,19 We also observed a JNK pathway activation after ER stress induction, thanks to the accumulation of the matrix metalloproteinase 1 (MMP1; Figures 3a and a’), as well as the expression of PlacZ enhancer traps that report the expression of downstream effectors of the JNK pathway, that is, misshapen (msn; Figures 3b and b’) and puckered (puc; Figures 3c and c’).41 puc-lacZ colocalized with TUNEL (Figure 3d) labeling but not with an anti-PH3 staining (Figure 3d'). Similar results were found with msn-lacZ (Supplementary Figure S4). Therefore, the JNK signaling is not activated in proliferating cells in response to the ER stress but in apoptotic cells.
Psn overexpression induces a JNK pathway-independent apoptosis. (a, a') Anti-MMP1 staining in vg-GAL4/+ (a) and vg-GAL4, UAS-Psn, UAS-Psn/+ (a') third-instar wing imaginal discs. (b–c') Anti-β-Galactosidase staining to detect the expression of msn- (b) or puc-lacZ (c, d) reporters in vg-GAL4/+; msn06946/+ (b), vg-GAL4, UAS-Psn, UAS-Psn/+; msn06946/+ (b'), vg-GAL4/+; pucE69/+ (c) and vg-GAL4, UAS-Psn, UAS-Psn/+; pucE69/+ (c') third-instar wing imaginal discs. Note MMP1 staining (a) and msn-lacZ expression (b) in the notum (white arrowheads) are constitutive and independent from Psn overexpression. Note that puc-lacZ expression at the tip of the notum reflects the JNK pathway activation required for thorax closure (c, white arrow). (d, d') Colocalization (white) between anti-β-Galactosidase staining (green) to detect puc expression and either TUNEL labeling (d, red) to detect apoptosis or anti-PH3 (d', red) to detect mitotic cells in vg-GAL4, UAS-Psn, UAS-Psn/+; pucE69/+ third-instar larvae wing imaginal discs. (e–e''') TUNEL staining in vg-GAL4, UAS-Psn, UAS-Psn/+ (e), UAS-bskDN/+ vg-GAL4, UAS-Psn, UAS-Psn/+ (e'), vg-GAL4, UAS-Psn, UAS-Psn/+; UAS-hep-RNAi/+ (e') and in vg-GAL4, UAS-Psn, UAS-Psn/UAS-cdk5-RNAi (e''') third-instar wing imaginal discs
We thus asked whether the ER stress-induced apoptosis was JNK dependent. To address this question, we checked whether the inhibition of the JNK signaling was able to modify ER stress-induced cell death. Neither the expression of a dominant negative form of the JNK Basket (BskDN) nor the RNAi-mediated depletion of the Drosophila JNKK MMK7 homolog, Hemipterous (Hep), had any obvious effect on apoptosis (Figures 3e and e’’). The interfering RNA was verified for its efficacy on Eiger-induced cell death and was validated by Marchal et al.42 This could suggest that our model of ER stress differs from the Retinitis Pigmentosa model, in which apoptosis induction involves a CDK5-dependent activation of JNK signaling.5 Confirming our hypothesis, CDK5 depletion did not modify the ER stress-induced cell death triggered by Psn overexpression (Figure 3e’’’). These results indicate that, although activated, the JNK pathway does not regulate ER stress-induced apoptosis in our model of ER stress.
Next, we tested whether the JNK pathway could modulate ER stress-induced dilp8 expression and therefore developmental timing. Reducing the JNK activity by overexpressing puc, which encodes a phosphatase negatively regulating JNK signaling,43,44 significantly rescued the developmental delay (Figure 4a), led to an aggravation of the wing phenotype (Figure 4b) and suppressed dilp8 expression (Figures 4c and d). Opposite effects on developmental delay, wing phenotype and dilp8 expression were observed in flies heterozygous for the pucE69 loss-of-function mutant allele (Figures 4a–d). Altogether, these results show that JNK signaling is involved in maintaining tissue homeostasis after ER-stress, by stimulating dilp8 expression.
The JNK pathway regulates a Dilp8-dependent developmental delay. (a) Effects of puc overexpression (UAS-puc) (gray) and puc mutant heterozygosity (pucE69/+) (black) compared with control (white) in a vg-GAL4, UAS-Psn, UAS-Psn/+ (plain bars) or vg-GAL4/+ (hatched bars) background on the mean time of adult eclosion. Error bars represent the S.E.M. (n=4). Asterisks indicate significant difference (P<10−4, ANOVA). (b) Distribution of notched-wing phenotypes of vg-GAL4, UAS-Psn, UAS-Psn/+; UAS-puc/+ (gray bars) and vg-GAL4, UAS-Psn, UAS-Psn/+; pucE69/+ (black bars) flies compared with vg-GAL4, UAS-Psn, UAS-Psn/+ flies (open bars). (c, d) Intensity of GFP reflecting dilp8 expression in vg-GAL4, UAS-Psn, UAS-Psn/+; dilp8MI00727/+ (c left, d white bars), vg-GAL4, UAS-Psn, UAS-Psn/+; dilp8MI00727/UAS-puc (c center, d gray bar), and vg-GAL4, UAS-Psn, UAS-Psn/+; dilp8MI00727/pucE69(c right, d black bar) third-instar wing imaginal discs. Error bars represent the S.E.M. of at least eight independent experiments. Asterisks indicate significant difference (P<10−3, ANOVA)
To address the question of the mechanism of activation of the JNK pathway, we tested whether the JNK signaling depends on DIAP1/Dronc. Given that the ER stress involved Dronc activation (Figures 1a and c) and DIAP1 downregulation (Figures 1d and f), we overexpressed both Psn and diap1 to counteract DIAP1 decrease and block Dronc activation. Under these conditions, caspase activation was strongly suppressed but no significant modification of the JNK activation was revealed by anti-MMP1 staining or by detecting dilp8 expression (Figures 5a and b). Altogether, these results suggest that JNK signaling is not mainly controlled by DIAP1/Dronc.
ATF4 activates the JNK pathway. (a, left) Anti-activated caspase 3 (green) and anti-MMP1 (red) staining in vg-GAL4, UAS-Psn, UAS-Psn/+ (top) and vg-GAL4, UAS-Psn, UAS-Psn/+; UAS-diap1/+ (bottom) third-instar wing imaginal discs. (a, right) Intensity of GFP in vg-GAL4, UAS-Psn, UAS-Psn/+; dilp8MI00727/+ (top) and vg-GAL4, UAS-Psn, UAS-Psn/+; dilp8MI00727/UAS-diap1 (bottom) third-instar wing imaginal discs. (b) Quantification of relative dilp8::EGFP expression. Error bars represent the S.E.M. of at least nine independent experiments. The asterisk indicates significant difference between the indicated genotype and the control (P<10−5, ANOVA). (c) Anti-MMP1 staining (left) and intensity of Dilp8::EGFP detection (right) in vg-GAL4, UAS-Psn, UAS-Psn/+ (top) and vg-GAL4, UAS-Psn, UAS-Psn/+; UAS-atf4-RNAi (bottom) third-instar wing imaginal discs. (d) Quantification of relative dilp8 expression. Error bars represent the S.E.M. of at least four independent experiments. The asterisk indicates significant difference between the indicated genotype and the control (P<10−4, ANOVA). (e) Effects of ATF4 depletion (gray bars) compared with control (white bars) on vg-GAL4, UAS-Psn, UAS-Psn/+ (solid bars) and vg-GAL4/+ (streaked bars) on the mean time of adult eclosion. Error bars represent the S.E.M. (n=3). The asterisk corresponds to a statistical difference (P<10−3, ANOVA)
We next checked whether the components of the PERK branch of the UPR could modulate the JNK pathway activation. Depletion of ATF4 strongly reduced the JNK activation that was directly assayed by an anti-MMP1 staining or indirectly by detecting dilp8 expression (Figures 5c and d). Similar results were observed by reducing the dosage of the ATF4-encoding gene, crc, thanks to crc1 and crcBG00047 hypomorphic mutant alleles (data not shown). In agreement with dilp8 expression reduction, ATF4 depletion also suppressed the ER stress-induced developmental delay (Figure 5e). Similar results were found by depleting PERK and converse effects were observed when GADD34 was depleted (Supplementary Figure S5), confirming that the PERK/ATF4 branch of the UPR activates JNK signaling. Therefore, tissue homeostasis is maintained when ER stress induces cell death thanks to a developmental delay controlled by the JNK pathway in an ATF4-dependent manner.
A Rac1/Slpr-dependent JNK pathway regulates developmental delay after ER-stress
The core of the JNK pathway is composed by Jra and Kay, which dimerize to form the dAP-1 transcription factor, and the unique JNK, Bsk. We observed that Jra or kay depletion, which had no effect in the absence of ER stress, aggravated the wing phenotype (Supplementary Table S1), showing that the JNK pathway core is involved in tissue homeostasis after an ER stress.
We decided to identify the upstream JNK pathway components that promote ER stress-induced developmental delay. A shift toward stronger wing phenotypes was observed when the JNKKs dMKK4 and Hep (Supplementary Table S1) or the JNKKK Slpr were depleted by RNAi in the presence of ER stress-induced cell death (Figure 6a,Supplementary Table S1). These depletions did not have any detectable effect by themselves (data not shown). Slpr depletion also decreased dilp8 expression and the ER stress-induced developmental delay (Figures 6d, g and j). Slpr can be activated by direct interaction with either Msn or the Rac1 GTPase.45 We thus examined if these interactors participate to ER stress-associated JNK signaling. Surprisingly, msn depletion induced a distribution shift toward weaker phenotypes (Figure 6b). It also increased both dilp8 expression (Figures 6e and h) and developmental delay (Figure 6k) in response to ER stress. These results suggest that Msn may antagonize JNK signaling, which, to our knowledge, has only been observed once.46 As ER stress induces msn expression (Figure 3b), we thus asked whether this was in a JNK-dependent manner. Indeed, the expression of a Bsk dominant negative form strongly reduced msn expression (data not shown), suggesting that Msn could belong to a JNK pathway negative feedback induced after ER stress.
The Rac1/Slpr JNK pathway triggers an ER stress-induced developmental delay negatively controlled by Msn. (a–c) Distribution of notched-wing phenotypes of vg-GAL4, UAS-Psn, UAS-Psn/+; UAS-slpr-RNAi/+ (a, red), vg-GAL4, UAS-Psn, UAS-Psn/+; UAS-msn-RNAi/+ (b, blue) and vg-GAL4, UAS-Psn, UAS-Psn/+; UAS-Rac1-RNAi/+ (c, yellow) flies compared with vg-GAL4, UAS-Psn, UAS-Psn/+ flies (a–c, white). (d–i) Intensity of GFP emitted by the dilp8::EGFP reporter in vg-GAL4, UAS-Psn, UAS-Psn/+ (d–i), vg-GAL4, UAS-Psn, UAS-Psn/+; UAS-slpr-RNAi/+ (d, g), vg-GAL4, UAS-Psn, UAS-Psn/+; UAS-msn-RNAi/+ (e, h) and vg-GAL4, UAS-Psn, UAS-Psn/+; UAS-Rac1-RNAi/+ (f, i) third-instar wing imaginal discs. Error bars represent the S.E.M. of at least six independent experiments. Asterisks indicate significant difference between the indicated genotype and its control (P<10−4). (j–l) Effects of Slpr (j, red), Msn (k, blue) and Rac1 (l, yellow) depletion compared with the control (white) in a vg-GAL4, UAS-Psn, UAS-Psn/+ (plain bars) or vg-GAL4/+ (hatched bars) background on mean time of adult eclosion. Error bars represent the S.E.M. of seven independent experiments. Asterisks correspond to significant difference between controls without Psn (hatched bars) and the indicated genotype (P<10−4, ANOVA)
Finally, depletion of rac1 induced a shift toward stronger wing phenotypes (Figure 6c) and a decrease of dilp8 expression (Figures 6f and i) resulting in a developmental delay suppression (Figure 6l). Therefore, Rac1 seems to be the JNK pathway activator that triggers ER stress-induced developmental delay. In conclusion, the developmental delay seems to be dependent of a Rac1/Slpr/JNK pathway.
Discussion
As previously reported in mammalian cells, we have validated that Psn overexpression can provoke chronic ER stress in Drosophila.26 In mammalian models, the UPR branches can display opposite roles depending on the model. For example, Perk can be either anti or proapoptotic.47, 48, 49 Thanks to a new model of chronic ER stress, we have demonstrated in this study that the PERK/ATF4 pathway has a fundamental role in Drosophila tissue homeostasis (Figure 7). So far, the ADRP model was the only model of strong chronic ER stress reported in Drosophila.5,24 We have validated that Psn overexpression can also provoke a chronic ER stress in Drosophila, as previously reported in mammalian cells.26 In both Drosophila models, apoptosis is induced by UPR in response to ER stress. Nevertheless, this induction involves totally different pathways. In our chronic ER stress model, cell death induction is PERK/ATF4 dependent and JNK independent, contrarily to the ADRP model in which CDK5 activates JNK signaling that triggers apoptosis.5 These differences show that the complexity of ER stress-induced signaling found in mammals is conserved in Drosophila, thus highlighting the usefulness of ER stress models plurality.
Model of tissue homeostasis maintenance after an ER stress. Chronic ER stress activates the PERK pathway, which results in ATF4 expression. ATF4 has two antagonistic functions. On one hand, ATF4 induces a caspase-dependent apoptosis by repressing diap1 expression. On the other hand, it favors tissue homeostasis through the induction of dilp8 expression by Rac1/Slpr/JNK pathway activation. The role of Msn in the control of the observed developmental delay remains to be elucidated. It could either negatively regulate the JNK pathway or have a JNK pathway-independent mechanism of action
We have shown that the PERK/ATF4 pathway induces a caspase-dependent apoptosis by repressing diap1 transcription. However, PERK has been described to exert some antiapoptotic activity by inducing IAP gene expression in mammals.47 This effect does not seem to rely on direct targets of PERK, ATF4 and CHOP.50 Similarly, we did not find any ATF4 consensus binding sequence (5'-RTTRCRTCA-3') in the diap1 promoter region and no CHOP homolog has been found in Drosophila. Therefore, the mechanisms involved in PERK regulation of IAPs remain to be clarified.
In our chronic ER stress model, the JNK pathway is activated in apoptotic cells to favor tissue homeostasis without stimulating cell proliferation. This is in contrast to a JNK activation in cells neighboring apoptotic cells, which results in an increase of the proliferation rate.11 Similar to our observation, JNK activation in apoptotic cells has been observed in ‘undead cell’ models.9,12, 13, 14 In these models, the JNK pathway could be activated by DIAP1 or DRONC, whereas JNK signaling seems to be primarily independent from DIAP1/DRONC in our model. In a mammalian model of chronic ER stress, the IRE1 branch of the UPR activated the JNK pathway to trigger apoptosis thanks to TRAF2/ASK1.51 In our ER stress model, depletion of traf2 or ask1 had no effect (Supplementary Table S1 and data not shown). Instead, we have shown for the first time that JNK pathway activation mainly depends on the PERK/ATF4 pathway. Interestingly, this particular JNK pathway is not mainly activated by apoptosis and does not modulate cell death or proliferation.
We have also shown that PERK/ATF4 regulates an ER stress-induced developmental delay. As previously reported, we observed that Dilp8 is a major contributor to developmental delay.19,23 An obvious candidate for a Dilp8-independent developmental delay regulation was the retinoic acid signaling that has been reported to modulate an irradiation-induced developmental delay.22 We have tested if this pathway could also regulate the developmental delay caused by Psn overexpression. No wing phenotype modification was detected upon the downregulation of this pathway (data not shown).
We have characterized the components of the JNK signaling that is activated in response to chronic ER stress in Drosophila wing imaginal discs (Figure 7). The small GTPase Rac1 would activate the JNKKK Slpr, which in turn would activate JNK signaling core to regulate dilp8 expression and ultimately favor development delay and tissue homeostasis maintenance. How the ATF4/PERK branch activates Rac1 remains to be elucidated. Our results also suggest the existence of a negative feedback loop regulating the JNK pathway, which would involve the JNKKKK, Msn. This is in agreement with a genetic and phosphoproteomic study showing that Msn is able to inhibit the phosphorylation of Jun.46 Considering that the JNK pathway induces dilp8 expression in abnormally growing imaginal discs in other stress models,19 one may wonder whether the same JNK pathway is implicated in these models. Moreover, one may wonder whether dilp8 control during tissue homeostasis-associated developmental delay is always JNK-dependent and relies on a Rac1/Slpr pathway.
To summarize, we have shown in this study that in response to an ER stress induced by Psn overexpression, the PERK pathway is activated resulting in a Janus-faced ATF4 role. On one hand, ATF4 induces caspase-dependent apoptosis by repressing diap1 expression and on the other hand, it favors tissue homeostasis maintenance through the induction of a Rac1/Slpr/JNK pathway and the resulting dilp8 expression. More investigations on this new Drosophila chronic ER-stress model should allow the identification of novel regulators of UPR-dependent tissue and organism homeostasis that may be conserved in mammals.
Materials and Methods
Drosophila crosses and strains
Flies were raised on standard corn-agar medium. All crosses were performed at 25 °C. The Drosophila strains used carried vg-GAL4,52 pucE69,41 UAS-puc,43 UAS-Psn (Psn+14),53 UAS-xbp1::EGFP reporter,32 UAS-diap1,54 UAS-EGFP9.2,55 UAS-p35 (Bl#6298), msn06946(Bl#11707), thj5C8 (Bl#12093),35 UAS-bskDN (Bl#108773),56 dilp8MI00727 (Bl#33079). The UAS-cdk5-RNAi (Bl#35287), UAS-atf4-RNAi (Bl#25985), UAS-Rac1-RNAi (Bl#28985) strains from the Transgenic RNAi Project (TRIP) were provided by the Bloomington Drosophila Stock Center (BDRC, Bloomington, IN, USA). The UAS-slpr-RNAi (ID 106449) and UAS-msn-RNAi (ID 101517) strains were obtained from the Vienna Drosophila RNAi Center (VDRC, Vienna, Austria), while the UAS-hep-RNAi (4353R2) strain was provided by the National Institute of Genetics stock center (NIG-Fly, Kyoto, Japan). Genetic background control strains were adapted to the different transgenic lines we used. The rheaMI00296 (Bl#30955) strain was used as the control for dilp8MI00727 (Bl#33079) genetic background, the y,w[1118];P{attP,y[+],w[3]} (ID 60100) line was the control of the KK library hosted by the VDRC, the y1 v1; P{UAS-GFP.VALIUM10}attP2 (Bl#35786) was used as a control for the TRIP V10 library and y1 sc1 v1 P{nos-phiC31\int.NLS}X; P{CaryP}attP2 (Bl#25710) for the TRIP V20 library. Canton S flies were used as reference for all the other strains.
Immunostaining, TUNEL assay and microscopy
Wing discs of third-instar male larvae were dissected in PBS and fixed in 3.7% formaldehyde for 20 min at room temperature. Samples were then washed thrice in PBT 0.3%. TUNEL staining was performed following manufacturer’s instructions (ApopTag Red in situ apoptosis detection kit, Millipore, Temecula, CA, USA). Immunostaining was always performed, according to standard protocols, before the TUNEL assay. The following primary antibodies were used: rabbit anti-Caspase-3 (Asp175, Cell Signaling, Danvers, MA, USA, 1 : 20), mouse anti-β-galactosidase (40-1a, DSHB, University of Iowa, IA, USA, 1 : 200), mouse anti-MMP1 (5H7B11, DSHB, University of Iowa, IA, USA, 1 : 30), mouse anti-DIAP1 (kind gift from B Hay, 1 : 200). Secondary antibodies conjugated to Alexa Fluor 488, 568 or 647 were purchased from Molecular Probes (Eugen, OR, USA 1 : 400). Discs were mounted in Citifluor (Biovalley, Marne-La-Vallée, France). Images were captured using a Leica SPE confocal laser-scanning microscope (Leica, Wetzlar, Germany). Images were processed and treated with ImageJ and Adobe Photoshop 8.0. We used the Colocalization plugin (Pierre Bourdoncle, 2003), which considers that two signals colocalize if their respective intensities are strictly higher than the threshold of their channels and if their ratio of intensity is strictly higher than the ratio setting value (here, 75%). Transverse sections were computationally generated after reslicing confocal stacks using ImageJ. GFP mean intensity of the Dilp8MI00727-encoded protein trap was evaluated using the 'measure' function of ImageJ.
Developmental timing and 20E treatment
Eighty females were crossed to 40 males and transferred to fresh food after 2 days for 1 h to deposit eggs on culture medium. Flies reaching adulthood or pupariating were then counted every 3 h. Addition of ecdysteroid (Sigma Aldrich, Saint Louis, MO, USA) was performed accordingly to standard protocols.22 Each experiment was realized at least thrice independently and data were statistically analyzed by ANOVA.
Genetic interaction test
Genetic interactions were tested based on the modulation of the severity of Psn overexpression-dependent notched-wing phenotype. Wings were categorized according to the number and size of notches observed in their margin (Supplementary Figure S1A). To control the genetic background, mutant strains and control strains were first crossed with males containing reference chromosomes carrying markers, that is, BI for chromosome II and Dr for chromosome III. Male progeny carrying Bl, Dr and the mutation to be tested were crossed with vg-GAL4, UAS-Psn, UAS-Psn flies. To rule out any effect by itself of the tested strains, control crosses with vg-GAL4 females were performed for each strain. Only the results obtained with males are depicted in figures although all data were statistically analyzed by ANOVA, which allows to determine the effect of the chromosome of interest compared with the same chromosome in the control genetic background.
RNA extraction and RT-qPCR
Fifty wing imaginal discs were dissected in the RNA XS NucleoSpin kit’s lysis buffer on ice for each genotype. Total RNAs were extracted using this kit (Macherey-Nagel, Düren, Germany). RT was performed using 4.8 μg of RNA incubated with random primer oligonucleotides and Recombinant Taq DNA Polymerase (Invitrogen, Life Technologies, Carlsbad, USA). Real-time PCR was performed using the ABI Prism 7700 HT (Applied Biosystems, Life Technologies, Carlsbad, USA) with SYBRGreen mastermix (Abgene, Thermo Fisher Scientific, Waltham, MA, USA) and 11 ng of cDNA. The real-time PCR primers (Invitrogen, Life Technologies) were designed as in Garelli et al.23 Three independent RT experiments were performed and data were normalized against rp49 mRNA levels.
Abbreviations
- 20E:
-
20-Hydroxyecdysone
- ADRP:
-
autosomal dominant retinitis pigmentosa
- AED:
-
after egg deposition
- ATF6:
-
activating transcription factor 6
- Bsk:
-
Basket
- DIAP1:
-
Drosophila inhibitor of apoptosis 1
- Dilp8:
-
Drosophila insulin-like peptide 8
- EGFP:
-
enhanced green fluorescent protein
- GADD34:
-
growth arrest and DNA damage-inducible 34
- ER:
-
endoplasmic reticulum
- Hep:
-
Hemipterous
- IRE1:
-
inositol-requiring enzyme 1
- JNKKK:
-
c-Jun N-terminal kinase kinase kinase
- MMP1:
-
matrix metalloproteinase
- Msn:
-
Misshapen
- PERK:
-
double-stranded RNA-activated protein kinase (PKR)-like ER kinase
- PH3:
-
phospho-histone H3
- PSN:
-
presenilin
- Puc:
-
Puckered
- TUNEL:
-
terminal deoxynucleotidyl transferase dUTP nick end labeling
- UPR:
-
unfolded protein response
- vg:
-
vestigial
- XBP1:
-
X-box binding protein 1
References
Patil C, Walter P . Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr Opin Cell Biol 2001; 13: 349–355.
Harding HP, Calfon M, Urano F, Novoa I, Ron D . Transcriptional and translational control in the Mammalian unfolded protein response. Annu Rev Cell Dev Biol 2002; 18: 575–599.
Kaufman RJ . Orchestrating the unfolded protein response in health and disease. J Clin Invest 2002; 110: 1389–1398.
Walter P, Ron D . The unfolded protein response: from stress pathway to homeostatic regulation. Science 2011; 334: 1081–1086.
Kang MJ, Chung J, Ryoo HD . CDK5 and MEKK1 mediate pro-apoptotic signalling following endoplasmic reticulum stress in an autosomal dominant retinitis pigmentosa model. Nat Cell Biol 2012; 14: 409–415.
Rasheva VI, Domingos PM . Cellular responses to endoplasmic reticulum stress and apoptosis. Apoptosis 2009; 14: 996–1007.
Ryoo HD, Steller H . Unfolded protein response in Drosophila: why another model can make it fly. Cell Cycle 2007; 6: 830–835.
Mollereau B, Perez-Garijo A, Bergmann A, Miura M, Gerlitz O et al. Compensatory proliferation and apoptosis-induced proliferation: a need for clarification. Cell Death Differ 2013; 20: 181.
Ryoo HD, Gorenc T, Steller H . Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev Cell 2004; 7: 491–501.
Perez-Garijo A, Shlevkov E, Morata G . The role of Dpp and Wg in compensatory proliferation and in the formation of hyperplastic overgrowths caused by apoptotic cells in the Drosophila wing disc. Development 2009; 136: 1169–1177.
Bergantinos C, Corominas M, Serras F . Cell death-induced regeneration in wing imaginal discs requires JNK signalling. Development 2010; 137: 1169–1179.
Huh JR, Guo M, Hay BA . Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase Dronc in a nonapoptotic role. Curr Biol 2004; 14: 1262–1266.
Wells BS, Yoshida E, Johnston A . Compensatory proliferation in Drosophila imaginal discs requires Dronc-dependent p53 activity. Curr Biol 2006; 16: 1606–1615.
Kondo S, Senoo-Matsuda N, Hiromi Y, Miura M . DRONC coordinates cell death and compensatory proliferation. Mol Cell Biol 2006; 26: 7258–7268.
Perkins KK, Dailey GM, Tjian R . Novel Jun- and Fos-related proteins in Drosophila are functionally homologous to enhancer factor AP-1. EMBO J 1988; 7: 4265–4273.
Chen W, White MA, Cobb MH . Stimulus-specific requirements for MAP3 kinases in activating the JNK pathway. J Biol Chem 2002; 277: 49105–49110.
Stronach B, Perrimon N . Activation of the JNK pathway during dorsal closure in Drosophila requires the mixed lineage kinase, slipper. Genes Dev 2002; 16: 377–387.
Stronach B . Dissecting JNK signaling, one KKKinase at a time. Dev Dyn 2005; 232: 575–584.
Colombani J, Andersen DS, Leopold P . Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science 2012; 336: 582–585.
Hussey RG, Thompson WR, Calhoun ET . The Influence of X-Rays on the Development of DrosophilaLarvae. Science 1927; 66: 65–66.
Simpson P, Berreur P, Berreur-Bonnenfant J . The initiation of pupariation in Drosophila: dependence on growth of the imaginal discs. J Embryol Exp Morphol 1980; 57: 155–165.
Halme A, Cheng M, Hariharan IK . Retinoids regulate a developmental checkpoint for tissue regeneration in Drosophila. Curr Biol 2010; 20: 458–463.
Garelli A, Gontijo AM, Miguela V, Caparros E, Dominguez M . Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science 2012; 336: 579–582.
Ryoo HD, Domingos PM, Kang MJ, Steller H . Unfolded protein response in a Drosophila model for retinal degeneration. EMBO J 2007; 26: 242–252.
Michno K, Knight D, Campusano JM, van de Hoef D, Boulianne GL . Intracellular calcium deficits in Drosophila cholinergic neurons expressing wild type or FAD-mutant presenilin. PLoS One 2009; 4: e6904.
Honarnejad K, Jung CK, Lammich S, Arzberger T, Kretzschmar H, Herms J . Involvement of presenilin holoprotein upregulation in calcium dyshomeostasis of Alzheimer's disease. J Cell Mol Med 2013; 17: 293–302.
Cook DG, Sung JC, Golde TE, Felsenstein KM, Wojczyk BS, Tanzi RE et al. Expression and analysis of presenilin 1 in a human neuronal system: localization in cell bodies and dendrites. Proc Natl Acad Sci USA 1996; 93: 9223–9228.
Annaert WG, Levesque L, Craessaerts K, Dierinck I, Snellings G, Westaway D et al. Presenilin 1 controls gamma-secretase processing of amyloid precursor protein in pre-golgi compartments of hippocampal neurons. J Cell Biol 1999; 147: 277–294.
Levitan D, Lee J, Song L, Manning R, Wong G, Parker E et al. PS1 N- and C-terminal fragments form a complex that functions in APP processing and Notch signaling. Proc Natl Acad Sci USA 2001; 98: 12186–12190.
Kim J, Kleizen B, Choy R, Thinakaran G, Sisodia SS, Schekman RW . Biogenesis of gamma-secretase early in the secretory pathway. J Cell Biol 2007; 179: 951–963.
Brand AH, Perrimon N . Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993; 118: 401–415.
Souid S, Lepesant JA, Yanicostas C . The xbp-1 gene is essential for development in Drosophila. Dev Genes Evol 2007; 217: 159–167.
Fan Y, Bergmann A . The cleaved-caspase-3 antibody is a marker of caspase-9-like DRONC activity in Drosophila. Cell Death Differ 2010; 17: 534–539.
Novoa I, Zeng H, Harding HP, Ron D . Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol 2001; 153: 1011–1022.
Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H . Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat Cell Biol 2002; 4: 432–438.
Smith-Bolton RK, Worley MI, Kanda H, Hariharan IK . Regenerative growth in Drosophila imaginal discs is regulated by wingless and Myc. Dev Cell 2009; 16: 797–809.
Herrera SC, Martin R, Morata G . Tissue homeostasis in the wing disc of Drosophila melanogaster: immediate response to massive damage during development. PLoS Genet 2013; 9: e1003446.
Bryant PJ, Simpson P . Intrinsic and extrinsic control of growth in developing organs. Q Rev Biol 1984; 59: 387–415.
Haynie JL, Bryant PJ . Intercalary regeneration in imaginal wing disk of Drosophila melanogaster. Nature 1976; 259: 659–662.
Colombani J, Bianchini L, Layalle S, Pondeville E, Dauphin-Villemant C, Antoniewski C et al. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science 2005; 310: 667–670.
Ring JM, Martinez Arias A . Puckered, a gene involved in position-specific cell differentiation in the dorsal epidermis of the Drosophila larva. Dev Suppl 1993; 121: 251–259.
Marchal C, Vinatier G, Sanial M, Plessis A, Pret AM, Limbourg-Bouchon B et al. The HIV-1 Vpu protein induces apoptosis in Drosophila via activation of JNK signaling. PLoS One 2012; 7: e34310.
Martin-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM et al. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev 1998; 12: 557–570.
McEwen DG, Peifer M . Puckered, a Drosophila MAPK phosphatase, ensures cell viability by antagonizing JNK-induced apoptosis. Development 2005; 132: 3935–3946.
Garlena RA, Gonda RL, Green AB, Pileggi RM, Stronach B . Regulation of mixed-lineage kinase activation in JNK-dependent morphogenesis. J Cell Sci 2010; 123: 3177–3188.
Bakal C, Linding R, Llense F, Heffern E, Martin-Blanco E, Pawson T et al. Phosphorylation networks regulating JNK activity in diverse genetic backgrounds. Science 2008; 322: 453–456.
Hamanaka RB, Bobrovnikova-Marjon E, Ji X, Liebhaber SA, Diehl JA . PERK-dependent regulation of IAP translation during ER stress. Oncogene 2009; 28: 910–920.
Verfaillie T, van Vliet A, Garg AD, Dewaele M, Rubio N, Gupta S et al. Pro-apoptotic signaling induced by photo-oxidative ER stress is amplified by Noxa, not Bim. Biochem Biophys Res Commun 2013; 438: 500–506.
Oommen D, Prise KM . Down-regulation of PERK enhances resistance to ionizing radiation. Biochem Biophys Res Commun 2013; 441: 31–35.
Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol 2013; 15: 481–490.
Nishitoh H, Matsuzawa A, Tobiume K, Saegusa K, Takeda K, Inoue K et al. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev 2002; 16: 1345–1355.
Simmonds AJ, Brook WJ, Cohen SM, Bell JB . Distinguishable functions for engrailed and invected in anterior-posterior patterning in the Drosophila wing. Nature 1995; 376: 424–427.
Ye Y, Fortini ME . Characterization of Drosophila Presenilin and its colocalization with Notch during development. Mech Dev 1998; 79: 199–211.
Hay BA, Wassarman DA, Rubin GM . Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell 1995; 83: 1253–1262.
Netter S, Faucheux M, Theodore L . Developmental dynamics of a polyhomeotic-EGFP fusion in vivo. DNA Cell Biol 2001; 20: 483–492.
Adachi-Yamada T, Nakamura M, Irie K, Tomoyasu Y, Sano Y, Mori E et al. p38 mitogen-activated protein kinase can be involved in transforming growth factor beta superfamily signal transduction in Drosophila wing morphogenesis. Mol Cell Biol 1999; 19: 2322–2329.
Acknowledgements
Confocal image acquisition and analysis were performed on CYMAGES imaging facility. We thank S Netter, B Limbourg-Bouchon, B Hay, J-A Lepesant and ME Fortini for providing fly stocks and antibodies. We are very grateful to S Netter and I Guenal for suggestions on the manuscript and helpful discussions throughout this work. We thank C Pirou, P Gandille, C Wintz and our team for their contribution to this project. This article is dedicated to the memory of Didier Contamine who succumbed to a long illness after initiating this project. We acknowledge the NIG (National Institute of Genetics, Kyoto, Japan), Vienna Drosophila RNAi Center (VDRC, Vienna, Austria) and Bloomington Stock center (Bloomington, IN, USA) for providing fly stocks.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by E Baehrecke
Supplementary Information accompanies this paper on Cell Death and Disease website
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Demay, Y., Perochon, J., Szuplewski, S. et al. The PERK pathway independently triggers apoptosis and a Rac1/Slpr/JNK/Dilp8 signaling favoring tissue homeostasis in a chronic ER stress Drosophila model. Cell Death Dis 5, e1452 (2014). https://doi.org/10.1038/cddis.2014.403
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2014.403
- Springer Nature Limited
This article is cited by
-
Slik maintains tissue homeostasis by preventing JNK-mediated apoptosis
Cell Division (2023)
-
Nacα protects the larval fat body from cell death by maintaining cellular proteostasis in Drosophila
Nature Communications (2023)
-
The steroid-hormone ecdysone coordinates parallel pupariation neuromotor and morphogenetic subprograms via epidermis-to-neuron Dilp8-Lgr3 signal induction
Nature Communications (2021)
-
Upregulated TNF/Eiger signaling mediates stem cell recovery and tissue homeostasis during nutrient resupply in Drosophila testis
Scientific Reports (2020)
-
An ADAMTS Sol narae is required for cell survival in Drosophila
Scientific Reports (2019)