Abstract
Background:
Neoadjuvant chemoradiotherapy (NCRT) for advanced rectal cancer (RC) is a well-evidenced therapy; however, some RC patients have no therapeutic response. Patient selection for NCRT so that non-responsive patients are excluded has been subjective. To date, no molecular markers indicating radiation sensitivity have been reported.
Methods:
We irradiated six colorectal cancer (CRC) cell lines and identified HCT116 cells as radiation-sensitive and HCT15 and DLD-1 cells as radiation resistant. Using a microarray, we selected candidate radiation sensitivity marker genes by choosing genes whose expression was consistent with a radiation-resistant or sensitive cell phenotype.
Results:
Among candidate genes, cellular retinol binding protein 1 (CRBP1) was of particular interest because it was not only induced in HCT116 cells by tentative 10 Gy radiation treatments, but also its expression was increased in HCT116-derived radiation-resistant cells vs parental cells. Forced expression of CRBP1 decreased the viability of both HCT15 and DLD-1 cells in response to radiation therapy. We also confirmed that CRBP1 was epigenetically silenced by hypermethylation of its promoter DNA, and that the quantitative methylation value of CRBP1 significantly correlated with histological response in RC patients with NCRT (P=0.031).
Conclusions:
Our study identified CRBP1 as a radiation-sensitive predictor in RC.
Similar content being viewed by others
Main
There is accumulated evidence to support multimodal therapy for rectal cancer (RC) for the purpose of reduction of the local recurrence rate. Radiotherapy (RT) and addition of chemotherapy to RT has been reported to decrease local recurrence (Kapiteijn et al, 2001; Gérard et al, 2006). Neoadjuvant chemoradiotherapy (NCRT) is now broadly performed as the initial treatment for locally advanced RC. Tumour response to NCRT covers a wide spectrum, ranging from no to complete response, where only 8 to 24% of the patients show a complete response (Maas et al, 2010). A molecular marker that can predict tumour response to NCRT before the therapy is eagerly anticipated because tumour response to NCRT has been shown to be associated with oncological outcomes (Park et al, 2012).
The potential of molecular biomarkers to predict tumour response to chemoradiation has been evaluated in several studies. Kuremsky et al (2009) reviewed biomarkers for tumour response to NCRT and identified TP53, epidermal growth factor (EGFR), thymidylate synthase, Ki-67, p21 and bax/bcl-2 as such biomarkers. However, none of these genes were considered appropriate for clinical use because most of the study results were controversial. In addition, in recent years, gene expression profiles associated with tumour response to chemoradiotherapy have been reported. Akiyoshi et al (2012) reviewed those studies and concluded that there were some reported gene expression signatures that were capable of high predictive accuracy, but that the compositions of these signatures differed considerably, with little gene overlap. These controversial results were probably due to intratumoral heterogeneity. Since most of those studies assessed gene expression of biopsy samples, those studies could only analyse only a part of the gene expression status of the tumour. From this point of view, the focus should be on a genetic or epigenetic change rather than on gene expression. KRAS mutation is one genetic change that has been broadly analysed. Although a correlation between KRAS mutation status and the therapeutic effect of anti-EGFR antibody was reported, only a few studies have reported its correlation with chemoradiotherapy and those results are controversial (Van Cutsem et al, 2009; Erben et al, 2011; Garcia-Aguilar et al, 2011; Russo et al, 2014; Lee et al, 2015). DNA hypermethylation in a gene promoter region is one epigenetic change that is a common event in cancer. We have found that intratumoral heterogeneity is not a limitation in analysis of DNA methylation of biopsy samples (Ooki et al, 2010). Ebert et al (2012) reported that hypermethylation of the transcription factor AP-2 epsilon gene TFAP2E was seen in 51% of the CRC cases analysed and was significantly associated with nonresponse to chemotherapy. However, only a few studies have analysed the correlation between DNA methylation and sensitivity to RT (Tsang et al, 2014; Ha et al, 2015).
We performed the present study to identify a molecular marker(s) associated with radiation sensitivity in RC. Here we planned a comprehensive exploration of genes associated with radiation sensitivity in colorectal cancer (CRC) cell lines. We analysed differences in gene expression patterns between radiation-sensitive and -resistant cell lines using an expression microarray, and subsequently identified the cellular retinol binding protein 1 (CRBP1) gene as a candidate gene whose expression represented radiation sensitivity. Furthermore, we determined the epigenetic silencing of CRBP1 through promoter hypermethylation and its clinical role in RT for RC.
Materials and methods
Cell lines
The hepatocellular carcinoma cell line HepG2 and the CRC cell lines HCT116, Colo205, Colo320, and LoVo, were purchased from the RIKEN BioResource Center (Ibaraki, Japan). DLD-1 cells were kindly provided by the Cell Response Center for Biochemical Research Institute of Development, Aging and Cancer, Tohoku University (Sendai, Japan). HCT15 cells were purchased from the American Type Culture Collection (Manassas, VA, USA). DLD-1, HCT116, HCT15, Colo205, and Colo320 cells were grown in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS). HepG2 cells were grown in Dulbecco's Modified Eagle Medium (DMEM; Gibco) supplemented with 10% FBS. LoVo cells were grown in L-15 medium (Gibco) supplemented with 10% FBS. Mycoplasma contamination was not tested because neither we, nor other researchers in our institute, have encountered mycoplasma contamination over the past 4 years.
Rectal cancer tissues
Thirty-three RC tissues and corresponding normal mucosa were obtained from patients described in our previously reported study (Nakamura et al, 2014). Briefly, a total dose of 45 Gy of RT was administered to those patients. All RC tissues were obtained from a biopsy sample of the primary cancer before therapy and the corresponding normal mucosa tissues were obtained from resected specimens after NCRT.
Genomic DNA extraction
Genomic DNA was extracted from cell lines using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). Formalin-fixed paraffin-embedded tissue was cut into six 10 μm slices before genomic DNA extraction using the QIAamp DNA FFPE Tissue Kit (Qiagen), following the manufacturer’s instructions. DNA concentration was quantified by UV spectrophotometry using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Total RNA extraction
Total RNA was extracted using an RNeasy Mini Kit (Qiagen) following the manufacturer’s protocol. RNA concentration was quantified using a NanoDrop ND-1000 spectrophotometer. The integrity of total RNA was verified by analysis of the A260/A280 ratio using NanoDrop ND-1000, and by confirming that the result of a reverse transcriptase–PCR (RT–PCR) for β-actin was clearly positive.
Total protein extraction
Cultured cells were collected using trypsin and were lysed in RIPA buffer (Thermo Scientific Wilmington, DE, USA) containing protease inhibitors. Protein concentration was measured using the Bio Photometer (Eppendorf, Hamburg, Germany).
Radiation sensitivity assay
Cells (1 × 106) of each cell line were seeded into 10-cm dishes. On the next day, each cell line was irradiated with a dose of 1 to 4 Gy per day for 5 days. Cells incubated without radiation were used as a control. On day 7, the cells were collected by trypsinisation and the cells were counted using Countess (Invitrogen, Carlsbad, CA, USA). The assay was performed once for each cell line. Radiation sensitivity was evaluated by measuring the ratio of the number of live irradiated cells compared to the number of control cells.
Microarray analysis
Total RNA was extracted from HCT15, DLD-1, and HCT116 cells. Gene profiles were compared using GeneChip 3′ IVT Express Kit microarrays (Affymetrix; Santa Clara, CA, USA) according to the manufacturer’s instructions. Genes expressed at high levels in the radiation-sensitive cells and at low levels in the radiation-resistant cells were categorised as radiation sensitivity-related genes. Genes expressed at high levels in the radiation-resistant cells and at low levels in the radiation-sensitive cells were categorised as radiation resistance-related genes.
Establishment of radiation-resistant cells
HCT116 cells (1 × 106 cells) were cultured in 10 cm dishes and irradiated with 3 Gy at 60–70% confluence. On day 7, the cells were trypsynised and 1 × 106 cells were re-seeded in 10 cm dishes and similarly irradiated. This cycle was repeated 20 times, following which the radiation dose totaled 60 Gy (I-HCT116). Concurrently, other HCT116 cells were cultured without radiation and re-seeded once a week (M-HCT116). Evaluation of resistance to radiation was performed using the following protocol. Parental HCT116 (P-HCT116), I-HCT116 and M-HCT116 cells were each seeded into two 10 cm dishes at a density of 1 × 106 cells per dish. One dish was designated as the radiation group and the other dish was used as the control. On the next day, the radiation study groups were irradiated with a dose of 1 Gy per day for 5 days. The cells were counted on day 7. The ratio of the number of cells of the radiation and control groups was calculated. Each assay was performed in triplicate.
Selection of candidate genes related to radiation sensitivity and resistance among heat map genes
To select the optimal candidate genes related to radiation sensitivity, we focused on genes that met the following criteria: (1) increased expression was seen in radiation-sensitive cells after short-term 10 Gy irradiation and expression was not seen in radiation-resistant cells; and (2) expression was further increased after long-term 60 Gy irradiation (Figure 1B). The criteria for selection of candidate genes related to radiation resistance were as follows: increased expression was seen in radiation-resistant cells after short-term 10 Gy irradiation and expression was not seen in radiation-sensitive cells.
Radiation sensitivity assay and establishment of radiation-resistant cells.(A) The protocol for selection of radiation-sensitive and resistant cells. (B) The ratio of the cell number in each cell line after to before radiation. (C) The protocol for establishment of radiation-resistant HCT116 cells. (D) Cell count after 5 Gy radiation for confirmation of the acquisition of radiation resistance following exposure of HCT116 cells to continuous RT to a total of 60 Gy.
Reverse transcriptase–PCR
First strand cDNA was synthesised from RNA using SuperScript III reverse transcriptase (Invitrogen) and Oligo (dT) primers (Invitrogen) as follows. Total RNA (2 μg) was transcribed using Oligo (dT) primers. The sample was heated to 65 °C for 5 min and then quickly chilled on ice. SuperScript III reverse transcriptase (200 units) and reaction buffer was added to the tube and incubated at 50 °C for 30 min and then 70 °C for 15 min. The obtained cDNA was used for both RT–PCR and quantitative real-time RT–PCR (qRT–PCR). RT–PCR was carried out using Platinum Taq DNA Polymerase (Invitrogen) according to the manufacturer’s protocol. The PCR conditions were: 5 min at 95 °C followed by the appropriate number of cycles of 95 °C for 1 min, the appropriate annealing temperature for 1 min, and 72 °C for 1 minute, and a subsequent final incubation at 72 °C for 10 min. Quantitative RT–PCR was performed in triplicate using TaqMan probes and iQ Supermix (Biorad, Hercules, CA, USA), and the CFX96 real-time system (Biorad). The PCR conditions were as follows: 3 min at 95 °C, followed by 40 cycles of 95 °C for 20 s, 60 °C for 30 s, and 72 °C for 30 s. The primer sequences and specific PCR conditions for each gene are shown in Supplementary Tables S1 and S2. The mRNA expression value was defined as the quantity of fluorescence intensity derived from amplification of the CRBP1 gene divided by the fluorescence intensity from amplification of β-actin, multiplied by 100.
Plasmid construction for transfection into cell lines
Full-length CRBP1 cDNA was synthesised from total RNA extracted from HCT116 cells by PCR using Platinum Taq Polymerase and Oligo (dT) primers, and was cloned into the pcDNA3.1 myc-His C expression vector (Invitrogen) using the restriction enzymes EcoRI and XhoI, and the T4 Ligase. The plasmid sequence was directly sequenced to confirm the fidelity of the CRBP1 insert. Cells were transfected with this plasmid using Lipofectamine 2000 (Invitrogen) in Opti-MEM (Invitrogen) as per the manufacturer’s instructions.
Confirmation of forced expression of CRBP1 after transfection
The mRNA and protein expression of CRBP1 after transfection was confirmed using RT–PCR and western blotting, respectively. Total RNA and total protein was extracted 48 h after transfection for further RT–PCR and western blotting.
Western blotting analysis
Total cellular protein (60 μg) was loaded onto a NuPAGE 4–12% Bis-Tris gel (Invitrogen) and electrophoresis was performed followed by electroblotting to a PVDF membrane (Invitrogen). The blots were incubated with anti-myc (Invitrogen) and anti β-actin (Invitrogen) antibodies that were used as primary antibodies. The blots were developed using Western Breeze (Invitrogen), which contains alkaline phosphatase-conjugated anti-mouse immunoglobulin and a chemiluminescent substrate for alkaline phosphatase. Signals were detected using the luminescent image analyzer ImageQuant LAS 4000 (GE Healthcare, CT, USA).
Cell proliferation assay
Cell proliferation was assayed using the CytoSelect water-soluble tetrazolium salt (WST-1) Cell Proliferation Assay Reagent (Cell Biolabs, San Diego, CA, USA). On day 1, the cells were cultured in a 96-well plate at a density of 1 × 104 cells per plate. On day 2, the cells were transiently transfected with CRBP1. On day 3, cell proliferation was evaluated by measuring the optical density (OD) at 450 nm.
Radiation sensitivity assay after cell transfection
We analysed increased cellular radiation sensitivity by the WST-1 assay and cell counting. In the WST-1 assay, on day 1, the cells were seeded in a 96-well plate (1 × 104 cells per plate). On day 2, the cells were transiently transfected with CRBP1. The cells were irradiated with a dose of 5 Gy per day on days 3 and 4, and were analysed with the WST-1 assay on day 6. For cell counting, 2 × 105 cells were seeded in a 6-well plate on day 1. On day 2, the cells were transiently transfected with CRBP1. The cells were irradiated with a dose of 5 Gy per day on days 3 and 4, and were counted on day 6. Non-radiated but transfected cells were incubated concurrently as controls for both the WST-1 and the cell count assays. The ratio of the absorption of irradiated and non-irradiated cells (control) was calculated.
Bisulfite treatment of genomic DNA
Genomic DNA (2 μg) was bisulfite converted using the EZ DNA Methylation-Gold Kit (Zimo Research, Irvine, CA, USA) following the manufacturer’s instructions.
Bisulfite sequence analysis
Bisulfite treated DNA from cell lines was amplified using PCR. Primer sequences for amplification of the CRBP1 promoter region were designed to recognise DNA alterations caused by bisulfite treatment (Supplementary Table S1). The PCR products were purified using QIAquick Spin (Qiagen) and were either directly sequenced or were inserted into the pCR4-TOPO vector using the TOPO TA Cloning Kit for Sequence (Invitrogen). Ten clones were analysed for each cell line. We performed direct sequence analysis to screen for the existence of methylation and we used the cloned sequence to confirm the exact status of methylation.
Quantitative methylation-specific PCR (qMSP)
For qMSP analysis of CRBP1, we performed real-time PCR using iQ Supermix and CFX96 real-time systems. Two micrograms of bisulfite treated samples were loaded into each well. The PCR conditions were as follows: 3 min at 95 °C, followed by 40 cycles of 95 °C for 20 s, 62 °C for 30 s, and 72 °C for 30 s. Primer and hybridisation probe sequences are shown in Supplementary Table S1. Bisulfite treated DNA from DLD-1 cells was used as the methylation positive control for construction of a calibration curve on each plate, because cloned sequence analysis showed the cytosines in all of the clones of DLD-1 were almost completely methylated. Bisulfite treated DNA from HepG2 cells was used as the negative control because almost all of the cytosines were unmethylated. All reactions were performed in triplicate. The methylation value TaqMeth V) was defined as the quantity of fluorescence intensity derived from amplification of the CRBP1 gene divided by the fluorescence intensity derived from amplification of β-actin, multiplied by 100. This ratio was used as a measure for the relative level of methylated DNA in samples.
5-Aza-dC and TSA treatment
Cells were seeded in a 10 cm dish on day 0. The cells were then treated every 24 h for 4 days with either 1 or 5 μM 5-Aza-dC (5-aza-2′-deoxycytidine) dissolved in 50% acetic acid or were mock treated with PBS including the same amount of acetic acid. Trichostatin A (TSA; 300 nM; Sigma Aldrich, Inc, St Louis, MO, USA) was added to the medium for the final 24 h. On day 5, the cells were harvested and mRNA was extracted. RT–PCR was performed to confirm the re-expression of silenced genes.
Evaluation of pathological specimens
Tumour responses to NCRT were evaluated by histopathological examination of serial sections of resected specimens after surgery. Responses were evaluated according to the General Rules for Clinical and Pathological Studies on Cancer of the Colon, Rectum and Anus (Japanese Society for Cancer of the Colon and Rectum, 2009). This study was conducted in accordance with the Declaration of Helsinki, and all patients signed a consent form approved by the Research Ethics Committee of Kitasato University School of Medicine.
Statistical analysis
Either χ2 or Fisher exact tests were used for categorical variables. Student’s t-test was used for continuous variables (cell count and cell proliferation assays). P-values <0.05 were considered to represent statistical significance.
Results
Identification of radiation-sensitive and -resistant CRC cell lines
Initially, we irradiated six CRC cell lines to determine their radiation sensitivity. DLD-1, HCT116, HCT15, LoVo, Colo320 and Colo205 cells were irradiated with various doses (from 1–4 Gy per day) and the cell count was performed after the radiation treatments (Figure 1A). With the dose of 1 Gy per day, the highest percentage of live cells was found in HCT15 and DLD-1 cells (86% and 73%, respectively), while the lowest percentage was found in HCT116 cells (27%; Figure 1B). This trend in the percentage of live cells per cell line was preserved with radiation doses of 2, 3 and 4 Gy per day although the difference in the number of live cells between the cell lines became smaller than that at 1 Gy per day presumably due to the toxic effect of the radiation treatments. Based on these results, we designated HCT15 and DLD-1 cells as radiation-resistant and HCT116 cells as radiation-sensitive for the purposes of this study.
Establishing radiation-resistant cells by repeated irradiation
We investigated if radiation-sensitive cells could be converted into radiation-resistant cells by continuous irradiation. HCT116 cells were therefore repeatedly irradiated with a dose of 3 Gy per week, to a final total dose of 60 Gy as shown in Figure 1C. Subsequently, we evaluated if these I-HCT116 cells had indeed became resistant to radiation treatment by comparison of the cell number of I-HCT116, P-HCT116 and M-HCT116 after 5 Gy radiation treatments (Figure 1D). The survival of I-HCT116 cells was significantly higher than that of either P-HCT116 (P=0.005) or M-HCT 116 (P=0.003) cells. We, therefore, concluded that the I-HCT116 cells had become resistant to radiation treatment over time.
Comprehensive exploration of genes related to radiation sensitivity
To identify candidate genes underlying radiation sensitivity, gene expression of both radiation-sensitive and radiation-resistant cells was analysed using expression microarrays. Candidate genes were initially restricted to those abundantly expressed in each cell line (defined as ‘present expression’ in the microarray), resulting in 14 235 candidate genes in the radiation-sensitive cell line HCT116 and 13 288 candidate genes in the radiation-resistant cell lines HCT15 and DLD-1. Genes that showed a high sensitive cells/resistant cells expression level ratio in HCT116 cells were then chosen, of which we selected the top 40 genes. Additionally, genes that showed a high resistant cells/sensitive cells expression level ratio were also chosen, of which we chose 26 genes that were common to both resistant cell lines (Figure 2). Heat maps of the differentially expressed genes between radiation-resistant and radiation-sensitive cell lines are shown in Figure 3A. The microarray results for all candidate genes were confirmed using RT–PCR (Figure 3B).
Selection of candidate genes related to radiation sensitivity among the heat map genes.(A) Heat map of Affymetrix GeneChip gene expression microarray in radiation-resistant and -sensitive CRC cells. Red coloured bars indicate upregulated genes and green coloured bars indicate downregulated genes. (B) Relative results of RT–PCR analysis, which was performed to confirm microarray analysis results. (C) The selection criteria for sensitivity-related and resistance-related genes. (D) The results of RT–PCR for the selection of sensitivity-related genes. P, Parental cell; 10 Gy, after 10 Gy radiation; I, I-HCT116; M, M-HCT116 (Figure 1C) (E) The results of quantitative RT–PCR according to CRBP1 expression.
Selection of candidate genes related to radiation sensitivity among heat map genes
Three genes (CRBP1, STC2 and SLCO3A1) matched our criteria for involvement in cellular radiation sensitivity (Figure 3C and D). Of these genes, we focused on CRBP1 because several other studies have reported CRBP1 as a tumour suppressor gene (TSG) candidate. Increased expression of CRBP1 in cell lines was validated using qRT–PCR (Figure 3E). CRBP1 expression increased in HCT116 cells after tentative 10 Gy irradiation, but was counterintuitively also increased in the I-HCT116 cells. CRBP1 expression was not detected in HCT15 or DLD-1 cells, even after the tentative irradiation. We also tried to identify genes related to radiation-resistance by using the converse criteria to those used for the radiation-sensitive genes, but none of the genes matched these criteria. Therefore, we only focused on the radiation-sensitive candidate genes in the present study.
Forced expression of CRBP1 increased radiation sensitivity in the radiation-resistant cell lines
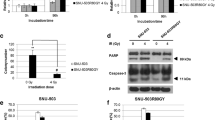
To determine the functional relevance of CRBP1 for radiation sensitivity, we first analysed CRBP1 expression in the cell lines before and after short-term radiation using qRT–PCR. The basal expression of CRBP1 tended to increase as cellular radiation sensitivity increased. Radiation-induced increased expression of CRBP1 was detected in Colo205 and HCT116 cells (Figure 4A).
Changes of radiation sensitivity by the forced expression of CRBP1 gene.(A) The result of quantitative RT–PCR analysis of CRBP1 expression in CRC cell lines. P, Parental; R, after 10 Gy radiation. (B) RT–PCR and Western blotting to confirm CRBP1 expression in CRBP1-transfected HCT15 and DLD-1 cells. (C) Cell proliferation assay after CRBP1 transfection. (D) Cell proliferation assay after radiation of transfected CRC cell lines. (E) Cell counts after radiation of transfected CRC cell lines. (F) Scheme of CRBP1 function.
We then transiently transfected a plasmid vector encoding full-length CRBP1 under the control of the cytomegalovirus promoter region into the radiation-resistant CRC cell lines (HCT15 and DLD-1; Figure 4B). We first assessed the effect of CRBP1 transfection on cell viability using the WST-1 assay. A significant decrease in DLD-1 and HCT15 cell viability was observed after CRBP1 transfection (Figure 4C; P<0.0001 and P=0.0004, respectively, vs vector-transfected cells). Next, we similarly assayed the viability of CRBP1-transfected DLD-1 and HCT15 cells after radiation. A significant decrease in cell viability (radiation vs control) was observed in the two cell types (Figure 4D; P=0.0291 and P=0.0312, respectively). A significant decrease in cell number ratio (radiation vs control) was observed only in DLD-1 cells (P=0.0071; Figure 4E). On the basis of these results, we considered that CRBP1 acted as a TSG, and that its forced expression resulted in the acquisition of a phenotype of RT sensitivity by DLD-1 and HCT15 cells (Figure 4F).
DNA methylation of the CRBP1 promoter in the CRC cell lines
We next analysed the methylation pattern of the CRBP1 gene in the CRC cell lines. For this purpose, we designed a unique primer for bisulfite sequencing and qMSP using ‘Meth primer’ software (Li and Dahiya, 2002). Two regions within the CRBP1 promoter region were indicated as CpG islands. One region is situated upstream of the transcription start site (TSS), and the other region is situated from upstream of the TSS to the end of exon 1 (Figure 5A). We focused on the former region in the present study. First, we performed direct bisulfite sequencing of this CRBP1 promoter region in HCT15, DLD-1, and HCT116 cells (Figure 5B). HCT15 and DLD-1 cells harboured densely methylated CpG islands whereas CRBP1 genomic DNA in HCT116 cells was not methylated. The methylation patterns of the CRBP1 promoter region in the 6 CRC cell lines were then analysed by bisulfite sequencing of cloned PCR products (Figure 5C). Finally, we quantified the CRBP1 gene methylation level of each CRC cell line using qMSP. Radiation-resistant cells displayed a significantly higher TaqMeth V than radiation-sensitive cells (Figure 5E). Furthermore, re-expression of CRBP1 was observed in HCT15 and DLD-1 cells following their treatment with demethylating agents (Figure 5D). A close association between the TaqMeth V and suppressed CRBP1 mRNA expression was found in CRC cell lines (Figure 5E).
Analysis of promoter hypermethylation of CRBP1 gene in CRC cell lines and clinical samples.(A) Schematic diagram of CpG islands within the CRBP1 promoter region. TSS; translation start site. (B) Representative results of bisulfite sequencing of HCT15, DLD-1, and HCT116. The boxed arrow indicates methylated cytosine. (C) Methylation status of 26 individual CpG sites (columns) of 10 cloned PCR products (rows) within the CRBP1 promoter region by bisulfite sequencing. Open and filled circles, unmethylated and methylated CpG sites, respectively. Numbers in parenthesis denote TaqMeth V. (D) The result of RT–PCR analysis of CRBP1 after demethylation treatments in HCT15 and DLD1 cells. TSA, trichostatin A; Aza, 5-Aza-dC. (E) Correlation between expression and methylation status in CRC cell lines. (F) Distribution of the CRBP1 TaqMeth V in tumour tissue and corresponding normal mucosa. (G) Correlation between therapeutic response of 33 RC tissues and the TaqMeth V.
CRBP1 promoter DNA methylation in preoperative biopsy samples and corresponding normal mucosa of RC patients with NCRT
We considered that qMSP was the optimal method for high throughput analysis to quantify the CRBP1 promoter DNA methylation level in patient samples. We therefore analysed 33 biopsy samples of RC patients before NCRT and the 33 corresponding normal mucosa using qMSP. The clinicopathological parameters of all patients analysed are shown in Table 1. Tumour samples showed significantly higher TaqMeth Vs than the corresponding normal mucosa (P=0.0122; Figure 5F). Tumour specific CRBP1 promoter hypermethylation was seen in RC patients. Grade 3 was seen in 8 cases, among which 7 cases showed a low CRBP1 TaqMeth V, and surprisingly, only one grade 3 patient showed a high TaqMeth V. Of 25 cases with Grade 1–2, 13 patients had a low CRBP1 TaqMeth V (Figure 5G). There was a significant correlation between the CRBP1 TaqMeth V and histological response (P=0.031). Taking into consideration the basic experiments reported herein, the patients with a low TaqMeth V were regarded as radiation-sensitive.
Discussion and conclusion
In our screen, we identified three potential radiation sensitivity-related genes using our criteria: CRBP1, STC2 and SLCO3A1. The functional role of SLCO3A1 has not been studied extensively, although transcripts of SLCO3A1 were upregulated in pancreatic cancer (Hays et al, 2013). Stanniocalcins (STCs) are glycoprotein hormones, and several investigators have reported their role in calcium and phosphate homeostasis (Ishibashi and Imai, 2002; Chang et al, 2003). Upregulation of STC2 has been reported in several cancers (Ieta et al, 2009; Kita et al, 2011; Arigami et al, 2013; Fang et al, 2014; Hashemzadeh et al, 2014). High levels of STC2 expression were correlated with worse prognoses of cervical cancer patients after RT (Shen et al, 2014). Since both STC2 and SLCO3A1 are thought to have oncogenic roles in cancer they were considered inappropriate for further investigation in our study. On the other hand, decreased expression of CRBP1 or hypermethylation of the CRBP1 promoter have been reported in multiple cancers in several reports (Esteller et al, 2002; Yamashita et al, 2002; Jerónimo et al, 2004; Kwong et al, 2005; Toki et al, 2010; Colvin et al, 2011; Peralta et al, 2012; Mendoza-Rodriguez et al, 2013). Analysis of the expression of CRBP1 in CRC cell lines suggested that it may have a functional role in radiation sensitivity in at least four cell lines. Cell lines with higher basal expression of CRBP1 (HCT116 and LoVo) tended to be radiation-sensitive, and HCT116 cells could express higher CRBP1 by short-term radiation. Conversely, cell lines with lower basal CRBP1 expression (HCT15 and DLD-1) were radiation-resistant and neither of these cell lines could induce CRBP1 expression after short-term radiation. Moreover, enforced CRBP1 expression reduced DLD-1 and HCT15 cell viability. A reduction in cell viability was also seen even after RT of CRBP1-expressing DLD-1 and HCT15 cells. Based on these results, we concluded that CRBP1 acts as a TSG and strongly contributes to cellular radiation sensitivity (Figure 5F). CRBP1 contribution to radiation sensitivity might be higher in DLD-1 cells than in HCT15 cells, because the cell number of CRBP1-transfected HCT15 cells was not decreased by radiation, while that of CRBP1-transfected DLD1 cells was decreased. On the basis of these results it might be expected that the established radiation-resistant cell line (I-HCT116) would express less CRBP1 than the parental cell, but in fact its CRBP1 expression was somewhat higher. This finding suggested that there may be a critical pathway related to radiation sensitivity that is regulated by a gene downstream of CRBP1, and that this downstream gene was inactivated by a mechanism such as DNA methylation or gene mutation. Connected with this finding, there was one clinical sample of a rectal cancer case that showed grade 3 after NCRT, but in which there was high CRBP1 promoter DNA methylation. Qualitative or semi-quantitative analyses of CRBP1 promoter methylation have been reported in several studies (Jerónimo et al, 2004; Kwong et al, 2005; Toki et al, 2010; Peralta et al, 2012; Mendoza-Rodriguez et al, 2013). In this study, we performed quantitative analysis by using qMSP. There was a clear correlation between the degree of CRBP1 methylation and CRBP1 expression in DLD-1 and HCT116 cells, which had distinguishable radiation-sensitivity phenotypes. However, in HCT15 cells, while their CRBP1 expression was relatively low and their phenotype was radiation-resistant, CRBP1 methylation was low. We speculate that, in HCT15 cells, silencing of CRBP1 expression was regulated not by its promoter hypermethylation but by its upstream genes. Although Colo205 cells showed a high CRBP1 expression level, the CRBP1 promoter region that we analysed was hypermethylated. This result suggested the existence of other CpG islands in the CRBP1 promoter whose hypermethylation should be analysed. It was a limitation of our study.
A significant correlation between the TaqMeth V and the therapeutic response of RC was seen in the present study, in which hypermethylation of the CRBP1 promoter was correlated with a poor therapeutic effect of RT. Of the 14 cases with a high TaqMeth V, 13 cases (93%) were grade 1–2 cases. These 13 cases constituted 52% (13/25) of the grade 1–2 cases. These data indicate that CRBP1 promoter methylation status could be a good indicator to preoperatively identify radiation-resistant patients. However, we could not predict non-responders to NCRT among these 13 patients. A biomarker that predicts non-responders to NCRT is required since non-responders should not be given NCRT and must be operated on immediately. Another marker for detecting such patients should be explored. It is a limitation of our study.
In conclusion, to date, there is no good predictor of tumour response to RT. However, our comprehensive molecular exploration identified CRBP1 as a candidate mediator of cellular radiation sensitivity, and our functional studies of CRBP1 confirmed that it plays an important role in radiation sensitivity, and that hypermethylation of its promoter region was correlated with resistance to RT. However, CRBP1 was not completely consistent with radiation sensitivity in all tested cell lines or clinical samples. This present investigation may have suggested many other potential candidate genes apart from CRBP1. Therefore, future rigorous investigation of these genes are warranted to elucidate radio-sensitive predictors in combination with CRBP1, which would be beneficial for selection of the most appropriate therapy for RC patients.
Change history
11 April 2017
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Akiyoshi T, Kobunai T, Watanabe T (2012) Predicting the response to preoperative radiation or chemoradiation by a microarray analysis of the gene expression profiles in rectal cancer. Surg Today 42: 713–719.
Arigami T, Uenosono Y, Ishigami S, Yanagita S, Hagihara T, Haraguchi N, Matsushita D, Hirahara T, Okumura H, Uchikado Y, Nakajo A, Hokita S, Natsugoe S (2013) Clinical significance of stanniocalcin 2 expression as a predictor of tumor progression in gastric cancer. Oncol Rep 30: 2838–2844.
Chang AC-M, Jellinek DA, Reddel RR (2003) Mammalian stanniocalcins and cancer. Endocr Relat Cancer 10: 359–373.
Colvin EK, Susanto JM, Kench JG, Ong VN, Mawson A, Pinese M, Chang DK, Rooman I, O’Toole SA, Segara D, Musgrove EA, Sutherland RL, Apte MV, Scarlett CJ, Biankin AV (2011) Retinoid signaling in pancreatic cancer, injury and regeneration. PLoS One 6: e29075.
Ebert MPA, Tänzer M, Balluff B, Burgermeister E, Kretzschmar AK, Hughes DJ, Tetzner R, Lofton-Day C, Rosenberg R, Reinacher-Schick AC, Schulmann K, Tannapfel A, Hofheinz R, Röcken C, Keller G, Langer R, Specht K, Porschen R, Stöhlmacher-Williams J, Schuster T, Ströbel P, Schmid RM (2012) TFAP2E-DKK4 and chemoresistance in colorectal cancer. N Engl J Med 366: 44–53.
Erben P, Ströbel P, Horisberger K, Popa J, Bohn B, Hanfstein B, Kähler G, Kienle P, Post S, Wenz F, Hochhaus A, Hofheinz R-D (2011) KRAS and BRAF mutations and PTEN expression do not predict efficacy of cetuximab-based chemoradiotherapy in locally advanced rectal cancer. Int J Radiat Oncol Biol Phys 81: 1032–1038.
Esteller M, Guo M, Moreno V, Peinado MA, Capella G, Galm O, Baylin SB, Herman JG (2002) Hypermethylation-associated Inactivation of the Cellular Retinol-Binding-Protein 1 Gene in Human Cancer. Cancer Res 62: 5902–5905.
Fang Z, Tian Z, Luo K, Song H, Yi J (2014) Clinical significance of stanniocalcin expression in tissue and serum of gastric cancer patients. Chin J Cancer Res 26: 602–610.
Garcia-Aguilar J, Chen Z, Smith DD, Li W, Madoff RD, Cataldo P, Marcet J, Pastor C (2011) Identification of a biomarker profile associated with resistance to neoadjuvant chemoradiation therapy in rectal cancer. Ann Surg 254: 486–92-3.
Gérard J-P, Conroy T, Bonnetain F, Bouché O, Chapet O, Closon-Dejardin M-T, Untereiner M, Leduc B, Francois E, Maurel J, Seitz J-F, Buecher B, Mackiewicz R, Ducreux M, Bedenne L (2006) Preoperative radiotherapy with or without concurrent fluorouracil and leucovorin in T3-4 rectal cancers: results of FFCD 9203. J Clin Oncol 24: 4620–4625.
Ha YJ, Kim CW, Roh SA, Cho DH, Park JL, Kim SY, Kim JH, Choi EK, Kim YS, Kim JC (2015) Epigenetic regulation of KLHL34 predictive of pathologic response to preoperative chemoradiation therapy in rectal cancer patients. Int J Radiat Oncol Biol Phys 91: 650–658.
Hashemzadeh S, Arabzadeh AA, Estiar MA, Sakhinia M, Mesbahi N, Emrahi L, Ghojazadeh M, Sakhinia E (2014) Clinical utility of measuring expression levels of Stanniocalcin 2 in patients with colorectal cancer. Med Oncol 31: 237.
Hays A, Apte U, Hagenbuch B (2013) Organic anion transporting polypeptides expressed in pancreatic cancer may serve as potential diagnostic markers and therapeutic targets for early stage adenocarcinomas. Pharm Res 30: 2260–2269.
Ieta K, Tanaka F, Yokobori T, Kita Y, Haraguchi N, Mimori K, Kato H, Asao T, Inoue H, Kuwano H, Mori M (2009) Clinicopathological significance of stanniocalcin 2 gene expression in colorectal cancer. Int J cancer 125: 926–931.
Ishibashi K, Imai M (2002) Prospect of a stanniocalcin endocrine/paracrine system in mammals. Am J Physiol Renal Physiol 282: F367–F375.
Japanese Society for Cancer of the Colon and Rectum (2009) General rules for clinical and pathological studies on cancer of the colon, rectum, and anus 7th edn, (Kanehara), p30.
Jerónimo C, Henrique R, Oliveira J, Lobo F, Pais I, Teixeira MR, Lopes C (2004) Aberrant cellular retinol binding protein 1 (CRBP1) gene expression and promoter methylation in prostate cancer. J Clin Pathol 57: 872–876.
Kapiteijn E, Marijnen CA, Nagtegaal ID, Putter H, Steup WH, Wiggers T, Rutten HJ, Pahlman L, Glimelius B, van Krieken JH, Leer JW, van de Velde CJ Dutch Colorectal Cancer Group (2001) Preoperative radiotherapy combined with total mesorectal excision for resectable rectal cancer. N Engl J Med 345: 638–646.
Kita Y, Mimori K, Iwatsuki M, Yokobori T, Ieta K, Tanaka F, Ishii H, Okumura H, Natsugoe S, Mori M (2011) STC2: a predictive marker for lymph node metastasis in esophageal squamous-cell carcinoma. Ann Surg Oncol 18: 261–272.
Kuremsky JG, Tepper JE, McLeod HL (2009) Biomarkers for response to neoadjuvant chemoradiation for rectal cancer. Int J Radiat Oncol Biol Phys 74: 673–688.
Kwong J, Lo K-W, Chow LS-N, To K-F, Choy K-W, Chan FL, Mok SC, Huang DP (2005) Epigenetic silencing of cellular retinol-binding proteins in nasopharyngeal carcinoma. Neoplasia 7: 67–74.
Lee JW, Lee JH, Shim BY, Kim SH, Chung M-J, Kye B-H, Kim HJ, Cho HM, Jang HS (2015) KRAS mutation status is not a predictor for tumor response and survival in rectal cancer patients who received preoperative radiotherapy with 5-fluoropyrimidine followed by curative surgery. Medicine (Baltimore) 94: e1284.
Li L-C, Dahiya R (2002) MethPrimer: designing primers for methylation PCRs. Bioinformatics 18: 1427–1431.
Maas M, Nelemans PJ, Valentini V, Das P, Rödel C, Kuo L-J, Calvo FA, García-Aguilar J, Glynne-Jones R, Haustermans K, Mohiuddin M, Pucciarelli S, Small W, Suárez J, Theodoropoulos G, Biondo S, Beets-Tan RGH, Beets GL (2010) Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: a pooled analysis of individual patient data. Lancet Oncol 11: 835–844.
Mendoza-Rodriguez M, Arreola H, Valdivia A, Peralta R, Serna H, Villegas V, Romero P, Alvarado-Hernández B, Paniagua L, Marrero-Rodríguez D, Meraz MA, Salcedo M (2013) Cellular retinol binding protein 1 could be a tumor suppressor gene in cervical cancer. Int J Clin Exp Pathol 6: 1817–1825.
Nakamura T, Yamashita K, Sato T, Ema A, Naito M, Watanabe M (2014) Neoadjuvant chemoradiation therapy using concurrent S-1 and irinotecan in rectal cancer: impact on long-term clinical outcomes and prognostic factors. Int J Radiat Oncol Biol Phys 89: 547–555.
Ooki A, Yamashita K, Kikuchi S, Sakuramoto S, Katada N, Kokubo K, Kobayashi H, Kim MS, Sidransky D, Watanabe M (2010) Potential utility of HOP homeobox gene promoter methylation as a marker of tumor aggressiveness in gastric cancer. Oncogene 29: 3263–3275.
Park IJ, You YN, Agarwal A, Skibber JM, Rodriguez-Bigas MA, Eng C, Feig BW, Das P, Krishnan S, Crane CH, Hu C-Y, Chang GJ (2012) Neoadjuvant treatment response as an early response indicator for patients with rectal cancer. J Clin Oncol 30: 1770–1776.
Peralta R, Valdivia A, Alvarado-Cabrero I, Gallegos F, Apresa T, Hernández D, Mendoza M, Romero P, Paniagua L, Ibáñez M, Cabrera L, Salcedo M (2012) Correlation between expression of cellular retinol-binding protein 1 and its methylation status in larynx cancer. J Clin Pathol 65: 46–50.
Russo AL, Ryan DP, Borger DR, Wo JY, Szymonifka J, Liang W-Y, Kwak EL, Blaszkowsky LS, Clark JW, Allen JN, Zhu AX, Berger DL, Cusack JC, Mamon HJ, Haigis KM, Hong TS (2014) Mutational and clinical predictors of pathologic complete response in the treatment of locally advanced rectal cancer. J Gastrointest Cancer 45: 34–39.
Shen X-J, Gu K, Shi J-P, Yao J-Q, Wu J-C (2014) Increased expression of stanniocalcin 2 is associated with tumor progression after radiotherapy in patients with cervical carcinoma. Int J Clin Exp Pathol 7: 8770–8776.
Toki K, Enokida H, Kawakami K, Chiyomaru T, Tatarano S, Yoshino H, Uchida Y, Kawahara K, Nishiyama K, Seki N, Nakagawa M (2010) CpG hypermethylation of cellular retinol-binding protein 1 contributes to cell proliferation and migration in bladder cancer. Int J Oncol 37: 1379–1388.
Tsang JS, Vencken S, Sharaf O, Leen E, Kay EW, McNamara DA, Deasy J, Mulligan ED (2014) Global DNA methylation is altered by neoadjuvant chemoradiotherapy in rectal cancer and may predict response to treatment—A pilot study. Eur J Surg Oncol 40: 1459–1466.
Van Cutsem E, Köhne C-H, Hitre E, Zaluski J, Chang Chien C-R, Makhson A, D’Haens G, Pintér T, Lim R, Bodoky G, Roh JK, Folprecht G, Ruff P, Stroh C, Tejpar S, Schlichting M, Nippgen J, Rougier P (2009) Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med 360: 1408–1417.
Yamashita K, Upadhyay S, Osada M, Hoque MO, Xiao Y, Mori M, Sato F, Meltzer SJ, Sidransky D (2002) Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell 2: 485–495.
Acknowledgements
We thank Mrs Kazuko Fujitani for her technical assistance. This work was supported by grants from Chugai Pharmacological, Co, Ltd (MW) and YASUDA Medical Foundation (HK). The authors received a grant from Chugai Pharmaceutical, Co., Ltd and YASUDA Medical Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Yokoi, K., Yamashita, K., Ishii, S. et al. Comprehensive molecular exploration identified promoter DNA methylation of the CRBP1 gene as a determinant of radiation sensitivity in rectal cancer. Br J Cancer 116, 1046–1056 (2017). https://doi.org/10.1038/bjc.2017.65
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2017.65
- Springer Nature Limited
Keywords
This article is cited by
-
Cellular retinol-binding protein 1: a therapeutic and diagnostic tumor marker
Molecular Biology Reports (2023)
-
Downregulation of KRAB zinc finger proteins in 5-fluorouracil resistant colorectal cancer cells
BMC Cancer (2022)
-
The RBP1–CKAP4 axis activates oncogenic autophagy and promotes cancer progression in oral squamous cell carcinoma
Cell Death & Disease (2020)
-
Epigenetic heterogeneity in cancer
Biomarker Research (2019)