Abstract
Background:
The HMGA2 protein has experimentally been linked to EMT and cancer stemness. Recent studies imply that tumour–stroma interactions regulate these features and thereby contribute to tumour aggressiveness.
Methods:
We analysed 253 cases of pancreatic ductal adenocarcinoma (PDAC) and 155 cases of ampullary adenocarcinoma (AAC) for HMGA2 expression by IHC. The data were correlated with stroma abundance and supplemented by experimental studies.
Results:
HMGA2 acts as an independent prognostic marker associated with a significantly shorter overall survival in both tumour types. Overall, HMGA2-positivity was more frequent in patients with PDAC than with AAC. The HMGA2 status in tumour cells significantly correlated with the abundance of PDGFRβ-defined stroma cells. In vivo co-injection of Panc-1 cancer cells with pancreatic stellate cells increased tumour growth in a manner associated with increased HMGA2 expression. Furthermore, in vitro treatment of Panc-1 with conditioned media from PDGF-BB-activated stellate cells increased their ability to form tumour spheroids.
Conclusions:
This study identifies HMGA2 expression in tumour cells as an independent prognostic marker in PDAC and AAC. Correlative data analysis gives novel tissue-based evidence for a heterotypic cross-talk with stroma cells as a possible mechanism for HMGA2 induction, which is further supported by experimental models.
Similar content being viewed by others
Main
The most frequent forms of cancer in the pancreatic region are the pancreatic ductal adenocarcinoma (PDAC; 70%) followed by the ampullary adenocarcinoma (AAC; 12%). PDAC is the fourth most common cause of cancer death in Europe (Ferlay et al, 2010; Rostain et al, 2014; Ryan et al, 2014; Malvezzi et al, 2016; Siegel et al, 2016).
PDAC and AAC are very aggressive diseases with high metastatic potential and almost complete resistance to chemotherapy. Only 7% of all PDAC patients survive longer than 5 years (Lepage et al, 2015). The absence of symptoms in the early stages leads to late clinical presentation, and most patients have locally advanced or metastatic disease at time of diagnosis. As a consequence only 15–20% of PDAC patients receive curative surgery and even among those the 5-year survival rate is less than 20% (Li et al, 2004; Wagner et al, 2004). In contrast, the AAC usually causes earlier symptoms and more patients are diagnosed at earlier stages and can receive curative surgery (Howe et al, 1998). Therefore the prognosis for AAC is more favourable with ∼41% of the patients surviving 5 years after curative resection (Rostain et al, 2014).
Only few reliable prognostic or predictive biomarkers have been established for cancers in the pancreatic region, as, for example, DPC4/SMAD4 expression (Tascilar et al, 2001; Biankin et al, 2002; Iacobuzio-Donahue et al, 2009).
The high-motility group A 2 (HMGA2) protein belongs to the family of non-histone chromosomal high-mobility group proteins (reviewed in Bustin and Reeves, 1996; Bustin, 1999; Cleynen and Van de Ven, 2008; Reeves, 2015), which function as architectural factors that alter DNA structure and thereby regulate DNA-dependent activities, like transcription, replication and repair (Cleynen and Van de Ven, 2008). HMGA2 contains a structural DNA-binding motif, the AT hook, which enables binding to the minor groove of AT-rich DNA regions (Reeves, 2001). It furthermore promotes the recruitment of additional transcriptional regulators and interacts in multiple protein complexes on promoter/enhancer sites, forming the so-called enhancosome (Reeves, 2001; Watanabe et al, 2009). HMGA2 is highly expressed in the embryo, but is hardly detectable or absent in adult human tissue, suggesting a role in regulation of cell proliferation and differentiation (Abe et al, 2003; Cleynen and Van de Ven, 2008). HMGA2 expression is often re-induced in neoplastic cells and was shown to be involved in the regulation of epithelial–mesenchymal transition (EMT) and cancer stem cell properties (Thuault et al, 2006; Watanabe et al, 2009; Madison et al, 2015).
Over-expression of HMGA2 has been described to be associated with metastasis and poor prognosis, for example, in triple negative breast cancer (Wend et al, 2013), ovarian cancer (Califano et al, 2014), gastric cancer (Motoyama et al, 2008; Lee et al, 2015), colorectal cancer (Rizzi et al, 2013) and non-small cell lung cancer (Sarhadi et al, 2006). In PDAC, HMGA2 expression was found to be associated with lymph node metastases and high tumour grade (Hristov et al, 2009; Piscuoglio et al, 2012) and with loss of tumoral E-Cadherin expression (Watanabe et al, 2009). One study, restricted to 91 cases of PDAC, also detected an association with poor prognosis in evaluations limited to univariate analysis (Haselmann et al, 2014).
A massive desmoplastic reaction and stroma formation are characteristics for PDAC and AAC (Lunardi et al, 2014; Rucki and Zheng, 2014; Pan et al, 2015) and can also act as a physical barrier preventing proper delivery of chemotherapeutic agents (Wang et al, 2016). One major component of the reactive tumour stroma are cancer-associated fibroblasts (CAFs) and in vitro and in vivo studies have shown that CAFs have tumour-promoting functions, including support of growth and metastasis as well as suppression of immunosurveillance (Ostman and Augsten, 2009; Strell et al, 2012; Weiland et al, 2012; Lunardi et al, 2014; Pan et al, 2015). In PDAC, SDF-1α (Li et al, 2012) and Nodal/Activin (Lonardo et al, 2011) have been identified as CAF-derived factors that affect EMT and stem cell properties in cancer cells. More specifically, pancreatic tumour cells carrying oncogenic KRASG12D were identified to establish a reciprocal signalling loop with stromal cells, which regulates tumour cell proliferation and apoptosis (Tape et al, 2016). Furthermore, fibroblast activation protein-positive CAFs have been linked to tumour immune evasion (Kraman et al, 2010; Feig et al, 2013) and were found to be associated with a bad prognosis in PDAC (Kawase et al, 2015). However, two recent studies also suggested tumour-restraining functions of CAFs in pancreatic cancer. One study showed that depletion of proliferating α-smooth muscle actin (ASMA) -positive fibroblasts reduced fibrosis, but induced immunosuppression and thereby accelerated an undifferentiated tumour growth, in a mouse model of PDAC (Özdemir et al, 2014). Accordingly, a low stromal ASMA score was associated with shorter overall survival (Özdemir et al, 2014). In another study, targeted deletion of sonic hedgehog (Shh) or inhibition of canonical hedgehog signalling, an important regulator of fibroblasts, resulted in a reduced stroma cell accumulation, but faster tumour growth and a more undifferentiated phenotype (Rhim et al, 2014). Together these studies suggest that CAFs can exhibit different subset-specific tumour regulatory functions.
In the present study, we have characterised HMGA2 expression in a large collection of PDACs and AACs and related the expression to clinicopathological features and survival. In both tumour types, HMGA2-positivity was associated with a significantly shorter overall survival. Furthermore, we found that the HMGA2 status in tumour cells significantly correlated with the abundance of PDGFRβ-defined stroma cells. These findings thus present novel tissue-based evidence suggesting heterotypic stroma-epithelial cell cross-talk as a possible mechanism for HMGA2 induction. These findings are supported by experimental studies, which demonstrate fibroblast-induced regulation of HMGA2 expression, cancer stem cell properties and tumour growth in vivo and in vitro.
Materials and methods
Patients
Four hundred forty-five patients who underwent pancreatic resection at Herlev Hospital, University of Copenhagen, Denmark between 1976 and 2008 (n=277) and at Rigshospitalet, University of Copenhagen, Denmark between 2004 and 2012 (n=168) were included. Thirty-seven patients were excluded from the analyses because of an unclear cause of death. Of the included patients, 253 had PDAC and 155 had AAC. Distinction between PDAC and AAC was determined by a combination of gross pathology and microscopy. All tumours were classified and graded according to the WHO Classification of Tumours. The study was approved by the local Ethical Committee (H-KA-20060181 and VEK ref. KA-20060113) and the Danish Data Protection Agency (j.nr. 2006-41-6848).
Immunohistochemistry (IHC)
4 μm whole sections of formalin-fixed and paraffin-embedded (FFPE) tissue samples were prepared. IHC was performed on a Ventana autostainer (Ventana Benchmark Discovery, NexES V10.6; Tuscon, AZ, USA) using the rabbit monoclonal anti-HMGA2 antibody (clone D1A7, #8179 Cell Signaling, Danvers, MA, USA; 1 : 400 dilution) or the rabbit monoclonal anti-PDGFRβ antibody (clone 28E1, #3169 Cell Signaling; 1 : 100 dilution) diluted in Antibody Diluent Buffer (Antibody diluent, Ventana). The staining protocol included an extended antigen retrieval step with CC1/pH9 buffer (Discovery CC1, Ventana) for HMGA2, or with pH10 Tris buffer (Sigma-Aldrich and Merck Kgaa, Darmstadt, Germany) for PDGFRβ staining, incubation with the primary antibody for 1 h at room temperature (RT) and incubation with the secondary antibody (OmniMab anti-Rb-HRP, Ventana) for 32 min at RT.
For each sample the HMGA2 staining was scored as negative or positive. Samples were defined as positive using a cutoff of ⩾1 positive cell in 10 vision fields of 0.79 mm2. The HMGA2 scoring approach was discussed with an experienced pathologist for diseases of the pancreas, ampulla and common bile duct. The PDGFRβ staining was scored as the positive stroma fraction (negative, low, moderate or high) as previously described (Paulsson et al, 2009). IHC stainings were scored independently by two individuals blinded to survival data.
Double IHC staining was performed manually. The slides were rehydrated and antigen retrieval was performed by heating the slides in pH10 Tris buffer (Sigma Aldrich) for 5 min at 110 °C in a pressure cooker (HistoLab, Goteborg, Sweden). The slides were incubated for 30 min with serum-free protein blocking reagent (Dako, Copenhagen, Denmark) at RT. PDGFRβ antibody was diluted 1 : 100 in DAKO REAL antibody diluent (Dako) and incubated overnight at 4 °C. As a secondary antibody, the ImmPRESS-AP anti-rabbit IgG (Vector laboratories, Burlingame, CA, US) was used for 30 min at RT. Detection was performed using the Vector Blue alkaline phosphatase substrate kit (Vector laboratories) according to the manufacturer’s instructions. In order to denature primary and secondary antibodies before the second staining, a further antigen retrieval step was performed for 5 min at 95 °C with pH9 retrieval buffer (Dako). Blocking, primary and secondary antibody incubation were repeated as before but with the HMGA2 antibody at 1 : 400 dilution. Detection was performed using the Vector Red alkaline phosphatase substrate kit (Vector laboratories) according to the manufacturer’s instructions. Hematoxylin (Histolab) was shortly applied for 10 s to obtain a weak counterstain.
For IHC analysis of the Panc-1 xenograft tumours, collected tumours were inverted in cold isopentane (Sigma-Aldrich) and embedded in TissueTek (Sakura Finetek USA Inc, Torrance, CA, USA). Cryosections (4 μm) were cut and fixed with acetone (Sigma-Aldrich) for 15 min prior to staining for HMGA2 or PDGFRβ following the same protocol as for FFPE sections. The digital image analysis for HMGA2-positive areas was performed on sections before hematoxylin counterstaining. Stained sections of xenograft tumours were digitalised with the NanoZoomer digital slide scanner (Hamamatsu City, Japan) and then analysed with the ImageJ software (Karperien, A., FracLac for ImageJ. http://rsb.info.nih.gov/ij/plugins/fraclac/FLHelp/Introduction.htm. 1999–2013.) to quantify the brown-stained areas. The data was normalised to the mean value of the control group for representation.
KPC mouse model
The mouse line used was KrasLSL−G12D/+; Trp53LSL−R172H;+;Pdx-Cre (KPC) mice (Hingorani et al, 2005). The mice were maintained at a 12-h-day, 12-h-night cycle on normal chow ad libidum in line with the local ethical committee directives (Stockholm södra djurförsöksetiska nämnd). Mice were killed and pancreata were collected at different time points. Tissues were fixed in 4% para-formaldehyde at RT for 24 h and then transferred in 70% ethanol for at least another 24 h at 4 °C before further dehydration and embedding in paraffin.
Cell culture
The Panc-1 PDAC cell line was obtained from European Selection of Cell Cultures (acacc, # 87092802) and maintained in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% fetal bovine serum (FBS), 100 units per ml penicillin and streptomycin (P/S) and 2 mM glutamine (all from Hyclone, GE Healthcare, South Logan, UT, USA). Panc1-H2B-mCherry cells were a kind gift from Dr Maarten Bijlsma at the Academic Medical Center, Amsterdam, The Netherlands. The immortalised pancreatic fibroblasts (PSCs) derived from a patient with chronic pancreatitis as described in (Jesnowski et al, 2005) were cultured in Dulbecco's Modified Eagle Medium: Nutrient Mixture F-12 (DMEM:F12) supplemented with 10% FBS and 100 units per ml P/S.
Limited dilution assay (LDA)
Panc-1 cells were detached with trypsin/EDTA (0.25% trypsin and 0.2 g l−1 EDTA, Hyclone, GE Healthcare), centrifuged and incubated in dispase (Stem Cell Technologies, Grenoble, France) for 10 min at RT to generate a single-cell suspension. The dispase treatment was stopped by addition of two volumes of human NeuroCult NS-A proliferation media (#05751, Stem Cell Technologies). The cells were further filtered through a 40-μm cell strainer, counted and seeded in a descending cell number/well (16, 8, 4, 2, 1) in ultra-low attachment 96-well plates (Corning, Oneonta, New York, NY, USA) in 75 μl of fibroblast-conditioned or control medium.
Fibroblast-conditioned medium was generated by incubation of the cells at 75% confluence for 16 h with NeuroCult media without growth factors. Medium was collected, centrifuged at 4000 r.p.m. for 10 min at 4 °C to remove any cell debris and diluted 1 : 2 with complete NeuroCult media including 20 ng ml−1 EGF and 10 ng ml−1 bFGF (both Life Technologies, Carlsbad, CA, USA) as well as 4 μg ml−1 heparin (Stem Cell Technologies). Control medium was prepared accordingly. Activation of fibroblasts with 50 ng ml−1 PDGF-BB (RnD Systems, Minneapolis, MN, USA) was performed for 6 h and washed away before the collection of conditioned media.
Fresh conditioned/control medium was added every fifth day. After 2 weeks, sphere-containing wells were counted and collected for subsequent RNA isolation.
The data obtained by LDA were analysed with extreme limiting dilution analysis software (ELDA) (Hu and Smyth, 2009) to estimate the frequency of sphere forming cells. The data is presented as the estimated frequency of sphere forming cells±upper/lower confidence interval.
Direct co-culture spheroid assay
Panc1-H2B-mCherry cells were seeded alone or in co-culture with PSCs (1 : 1), at a total concentration of 2500 cells/well. Non-cell culture treated round bottom 96-well plates (Falcon, BD NJ, USA) were used, and cells were seeded in culture media with a final concentration of 0.24% methylcellulose, as previously described (Longati et al, 2013). After 5 days, the formed spheroids were collected. For dissociation spheroids were treated twice as follows, 5 min incubation in trypsin/EDTA at 37 °C followed by trituration. The trypsin was inactivated by the addition of culture media containing 10% FBS. The cells were collected by centrifugation, washed once with cold PBS and then re-suspended in cold sorting buffer containing 25 mM HEPES, 2 mM EDTA and 10 units per ml DNAse II (Sigma Aldrich) in PBS. Cells were filtered through 50 μm cell strainers and kept on ice until sorting for mCherry-positive Panc1 cells by fluorescence activated cell sorting (FACSAria, BD Biosciences, Franklin Lakes, NJ, USA). Cells from mono-spheroids were also sorted with the same gating. Sorted cells were collected by centrifugation for subsequent RNA isolation.
RNA isolation and quantitative reverse transcriptase PCR
RNA was isolated with GenElute Mammalian Total RNA Miniprep Kit (Sigma-Aldrich) according to manufacturer’s instructions, including DNaseI treatment. RNA concentration was measured with a NanoDrop ND-1000 Spectrophotometer (Thermo Scientific, Wilmington, USA). CDNA synthesis from 500 ng RNA was performed with the SuperScript III Reverse Transcriptase (LifeTechnologies) kit using random hexamer primer according to manufacturer’s instructions.
For RNA isolation from frozen tumour tissue, ∼20 mg of tumour tissue was collected from frozen xenograft tumours and homogenised using a T18 basic Ultra Turrax (IKA, Stauffen, Germany). Total RNA was isolated using the SV Total RNA Isolation System (Promega, Madison, WI, USA).
Quantitative real-time PCR was performed using the 2 × Power SYBR Green PCR Master Mix (LifeTechnologies), 200 nM as final primer concentration and 10 ng of cDNA per 10 μl reaction. The real-time PCR primers for human HMGA2, PDGFRβ, CK18, CK19, RPL13a and RPS18 were purchased from Sigma-Aldrich, the RPL13a primers from Life Technologies and the GAPDH Quantitec primer from Qiagen (Hilden, Germany). The expression values were calculated by the comparative ΔΔCT-method for the gene-of- interest relative to the expression level of the house-keeping gene. For the HMGA2 determination in xenograft tumours, results were also corrected for differences in epithelial fraction by dividing the HMGA2 expression with the expression average of Cytokeratin (CK)18 and CK19 of the corresponding samples. Thereafter, the expression-values were normalised to the mean value of the control group for representation.
Primer Sequences: HMGA2 Fwd 5′-CGGCCAAGAGGCAGACCTAGG-3′ and Rev 5′-GTTGGCGCCCCCTAGTCCTCT-3′; PDGFRβ Fwd 5′-AGGCAAGCTGGTCAAGATCT-3′ and Rev 5′-GCTGTTGAAGATGCTCTCCG-3′; CK18 Fwd 5′-TCAGCAGATTGAGGAGAGCAC-3′ and Rev 5′-CGTCTCAGCTCTGTGAGCGTCG-3′; CK19 Fwd 5′-CATCCAGGACCTGCGGGACA-3′ and Rev 5′-GCGCAGAGCCTGTTCCGTCTC-3′; RPS18 Fwd 5′-TGAGGTGGAACGTGTGATCA-3′ and Rev 5′-CCTCTATGGGCCCGAATCTT-3′; RPL13a Fwd 5′ AGATGGCGGAGGTGCAG-3′ and Rev 5′-GGCCCAGCAGTACCTGTTTA-3′.
Xenograft model
Six- to seven-week-old male CB17 SCID mice (Janvier Labs, Le Genest-Saint-Isle, France) were injected subcutaneously with either 4 × 106 Panc-1 cells alone or Panc-1 together with PSCs at a 1 : 1 ratio. Each group consisted of five mice. Tumour growth was monitored two times per week. Tumour volumes (V) were calculated from the formula (V=length × height × width/2). The mice were killed under anaesthesia when V gained approximately 500 mm3. Tumours were collected and immediately snap frozen in liquid nitrogen. The xenograft experiments were performed in agreement with the local ethical committee (Utrecht University animal ethical board; ethical permit number is 2014.III.02.022).
Statistical analysis
Associations between HMGA2 expression and clinicopathological parameters and PDGFRβ expression were analysed with contingency tables and Fishers’ exact test, two-sided. The Kaplan–Meier and log rank test were used to compare cancer-specific survival. A Cox proportional hazards model was used for estimation of hazard ratios (HR) in both uni- and multivariate analyses including relevant risk factors as age, sex, tumour localisation, treatment, stage group and grade of differentiation in the model. The P-value for cox regression is based on Wald test. Calculations were performed with the statistical package IBM SPSS Statistics Version 22 (SPSS Inc., Chicago, IL, USA).
The TCGA data on PDAC were used for Kaplan–Meier analysis and multivariate analysis of overall survival. Cases with histological subtype other than PDAC were excluded (43 cases excluded out of 197 cases total). Dichotomisation of this data set was performed by splitting the gene-expression data at the median. The results shown in Figure 2B are in whole based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
The GSE21501 data set was only used for Kaplan–Meier analysis of overall survival. Hundred and two PDAC patients for which clinical follow-up was available were included. Since only T and N-stage as additional clinical data was provided no further multivariate analysis was performed. Dichotomisation of this data set was performed by splitting the gene-expression data at the median.
The results from the experimental studies were evaluated by group comparison using Student’s t-test, two-sided, unpaired.
Two-way ANOVA with Bonferroni post hoc test for comparison of means was used to evaluate differences in the tumour growth curves between the two groups of the xenograft model.
Differences with P-values <0.05 were considered as significant.
Results
HMGA2 expression in cancers of the pancreas and ampulla
The expression pattern of HMGA2 was analysed by IHC on tumour sections from 408 patients diagnosed with pancreatic adenocarcinoma (PDAC; n=253) or AAC (n=155). For each sample, the staining was scored as negative or positive using a cutoff of ⩾1 positive cell in 10 vision fields (see Material and Methods for details).
HMGA2 expression was found in tumour cells, in nuclear localisation, while it was absent in stroma cells, including fibroblasts, immune cells and vascular cells. However, the amount of HMGA2-positive tumour cells varied between cases, and positive and negative tumours were found both in the PDAC and AAC group (Figure 1).
Together, these initial analyses thus confirm earlier findings of variable HMGA2 expression by malignant cells in PDAC (Hristov et al, 2009; Piscuoglio et al, 2012; Haselmann et al, 2014), and extend these studies by including AAC in the investigations.
Correlations between HMGA2 expression and clinicopathological parameters
A set of analyses was performed to investigate potential associations between HMGA2 expression and clinicopathological features.
A significant association was observed between HMGA2-positivity and tumour localisation (Table 1). While 56.6% of the PDAC tumours were positive for HMGA2, only 32.7% of the AACs were positive (P<0.001; Fishers Exact, two-sided). In addition, we found overall significant correlations between HMGA2-positivity and lower tumour differentiation as well as higher stage group (P<0.001 for differentiation; P<0.028 for stage group).
The correlation between differentiation and HMGA2-positivity remained strongly significant only in the group of patients with PDAC (P<0.001). In the smaller AAC patient group, HMGA2-positivity was correlated only with older age (P<0.020) (Table 1).
The finding that HMGA2-positivity correlated with lower tumour cell differentiation prompted analyses on potential associations between HMGA2 and EMT markers. IHC analyses combining HMGA2 and cytokeratin antibodies indicated that HMGA2 expression was not restricted to cells with low cytokeratin expression (Supplementary Figure 2A). Furthermore, HMGA2 expression was not strongly linked to EMT-associated miRNAs in the present cohort (Supplementary Figure 2B). Finally, analyses of the TCGA data set did not detect strong associations between HMGA2 and selected EMT-related genes (data not shown) or an established EMT core signature (Taube et al, 2010; Supplementary Figure 2C).
Together, these analyses demonstrate a relation between HMGA2 expression and clinicopathological parameters known to be associated with worse prognosis, such as poor tumour differentiation and advanced stage group.
HMGA2 expression and overall survival
We used the HMGA2 IHC data together with clinical data on overall survival (OS) with the aim to investigate a possible prognostic significance of HMGA2 expression in PDAC and AAC.
The presence of HMGA2-positive cells was associated with a significantly shorter OS in the whole cohort (log Rank P<0.001), as well as in the patient groups with PDAC (log Rank P<0.001) or AAC (log Rank P<0.001) (Figure 2A;) separately. In a Cox Proportional Hazards model, HMGA2-positivity increased the hazard ratio (HR) for death in the whole cohort (HR 2.29 (1.83–2.86 95% confidence interval); P<0.001). This association was also detected in separate analysis of patients with PDAC (HR 1.74 (1.33–2.29 95% confidence interval); P<0.001) and AAC (HR 3.12 (2.07–4.70 95% confidence interval); P<0.001) (Figure 2A).
HMGA2 expression in PDAC and AAC is associated with a shorter cancer-specific survival. (A) Kaplan–Meier graphs of the cancer-specific survival in HMGA2-negative (black line) and -positive (red line) patients. HMGA2 expression was defined by IHC staining. Survival analyses were performed on the entire cancer cohort (upper panel) or restricted to the subsets of patients with PDAC (middle panel) and AAC (lower panel). (B) Kaplan–Meier analyses of overall survival of PDAC patients of the TCGA and GSE21501 cohorts. HMGA2 status is defined as low (black line) or high (red line) based on gene expression level (see Materials and Methods for cutoff definition). The P values from log-rank tests comparing the two Kaplan–Meier curves are indicated. The hazard ratio (HR) was calculated with a Cox Proportional Hazards model and the corresponding P-value is based on Wald test statistic.
The associations between poor prognosis and HMGA2-positivity within the whole cohort remained highly significant in multivariate analysis, including age, sex, gemcitabine treatment, grade of differentiation and clinical stage (P<0.001) (Table 2). Similar results were obtained in analyses in which PDAC and AAC were evaluated separately (P<0.001 for both PDAC and AAC) (Table 2).
To obtain independent support of the findings from the IHC analysis, we used the TCGA PDAC gene-expression data set with corresponding OS information. Patients with high HMGA2 gene-expression were found to have a significant shorter OS (log Rank P<0.009). Also in this cohort, the Cox Proportional Hazards model showed a significant increased hazard ratio (HR 1.78 (1.15–2.76 95% confidence interval); P<0.010) in patients with high HMGA2 gene-expression (Figure 2B), which remained significant in multivariate analysis (P<0.010) (Supplementary Table 1).
Furthermore, in the GSE21501 cohort, including 102 PDAC patients, we observed a strong trend towards shorter OS (log Rank P<0.094) for HMGA2-high patients, which remained in the Cox Proportional Hazards model (HR 1.50 (0.92–2.45 95% confidence interval); P<0.103) (Figure 2B).
The analyses thus identify HMGA2-positivity as an independent prognostic marker for patients with PDAC or AAC.
HMGA2 expression is induced at early stages of tumour development in a genetic mouse model for PDAC
To the best of our knowledge, there is no report in the literature at which phase of tumour progression HMGA2 expression is induced. A set of studies, using both the clinical cohort and the KPC-model of PDAC, was therefore performed to address this issue.
The human samples contained predominantly carcinoma and only small amount of surrounding tissue, limiting the ability to identify PanIN lesions. However, the detected PanIN-1 and PanIN-2 lesions were all negative for HMGA2 (n=18; data not shown), while no PanIN-3 lesions were identified.
In the KPC-model we detected very weak positive staining for Hmga2-positive tumour cells already in pancreatic intraepithelial neoplasia (PanIN)-1 lesions (Figure 3), with an increasing frequency and intensity in PanIN-2 and PanIN-3 lesions. In the carcinomas we observed heterogeneous staining, with positivity predominantly in the more poorly differentiated parts (Figure 3).
Hmga2 expression during different stages of PDAC development in the KPC mouse model. Representative microphotographs of Hmga2 expression in the KPC mouse model of pancreatic cancer. KPC PanIN-1 (upper left): minimal positivity of weak intensity; KPC PanIN-1/−2 (upper right): partial positive staining with weak to moderate intensity mostly in PanIN-2; KPC PanIN-2 (middle left): partial positive staining of weak to strong intensity; KPC PanIN-3 (middle right): diffuse positive staining of moderate to strong intensity; KPC PDAC (lower left): heterogeneous positive staining, predominantly in the poorer differentiated and sarcomatoid differentiated component; KPC PDAC (lower right): heterogeneous positive staining, predominantly in the poorer differentiated areas. Scale bar, 100 μm.
Therefore, HMGA2 seems to be differentially expressed within precursor lesions in mice and humans.
HMGA2-positive cells are located in close proximity to cancer-associated fibroblasts
PDGFRβ is an important regulator of mesenchymal cells, and is highly expressed on tumour stroma cells such as cancer-associated fibroblasts (CAFs) and pericytes (Pietras et al, 2002; Paulsson et al, 2009).
In a case-based analysis, we found a positive association between the expression of HMGA2 and the abundance of PDGFRβ-defined fibroblasts (P<0.008, Fishers exact test) (Figure 4A).
HMGA2-positive tumour cells are located in close proximity to PDGFR β expressing cancer-associated fibroblasts. (A) Correlation analysis of the abundance of PDGFRβ-positive stroma cells and HMGA2-positivity of tumour cells in patients with PDAC or AAC (P-value based on Fisher’s Exact test). (B) Double IHC staining for PDGFRβ (blue) and HMGA2 (red) on samples selected to represent human PDAC cases with low (upper picture) and high (lower picture) HMGA2 expression. Scale bar, 100 μm. **P<0.01.
To investigate the spatial localisation of the two cell types in more detail, we performed double IHC staining on four different specimens and found that HMGA2 positive tumour cells appeared in higher numbers in tumour areas with a strong abundance of PDGFRβ-positive fibroblasts in human PDAC (Figure 4B). However, one could also detect PDGFRβ-positive stromal areas which were not linked to high expression in the epithelium and vice versa but these were less frequent (Supplementary Figure 3A).
These observations suggest a role of fibroblasts in the induction of HMGA2 in tumour cells.
Pancreatic stellate cells support HMGA2 expression and tumorigenic properties in pancreatic cancer cells
With the aim to test whether HMGA2 expression and tumourigenic properties of pancreatic cancer cells are regulated by paracrine interaction, we performed ‘subcutaneous’ injections of Panc-1 cells either alone or together with pancreatic stellate cells (PSCs) in SCID mice. Notably, tumours formed following co-injection grew faster than tumours derived from Panc-1 mono-injections (310 mm3 vs 159 mm3 at collection on day 16 after injection; P<0.05 two-way ANOVA with Bonferroni post hoc test) (Figure 5A).
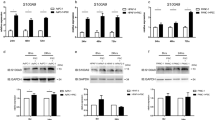
Pancreatic fibroblasts support HMGA2 expression and tumourigenic properties in pancreatic cancer cells. (A) Tumour growth following ‘subcutaneous’ injections of Panc-1 cells alone or co-injections together with PSCs in SCID mice. Each injection group consisted of 5 animals and tumour formation was observed in all animals. Two-way ANOVA with Bonferroni post hoc was used for statistical group comparison. (B) Gene expression of HMGA2 and the stromal marker PDGFRβ in tumours formed after mono-injection of cancer cells or co-injection with PSCs. Data is presented as average ± s.e.m. (P-value based on student’s t-test). (C) IHC analyses of control tumours and tumours formed after co-injection. Data is presented as average ± s.e.m. (P-value based on student’s t-test). Scale bar=100 μm. (D) Limited dilution assay (analysed with ELDA software) of Panc-1 cells in the presence of conditioned media from PSCs or PSCs pre-stimulated with PDGF-BB. The frequency of sphere forming cells is given as 1/n cells±upper/lower 95% confidence interval based on three independent experiments. (E) Gene expression levels of HMGA2 in Panc-1 spheroids grown in conditioned media of control-PSCs or PDGF-BB-stimulated PSCs. Expression values were normalised to the mean of the control group (set to 1.0 arbitrary units). Data is presented as average ± s.e.m. from three independent experiments. (F) Gene expression levels of HMGA2 in Panc-1 cells grown in mono-culture spheroids (control) or co-culture heterospheroids with direct cell–cell contact to PSCs. Expression values were normalised to the mean of the control group (set to 1.0 arbitrary units). Data are presented as average ±s.e.m. from two independent experiments. *P<0.05, **P<0.01, ***P<0.001.
Gene expression and protein analysis confirmed a higher abundance of PDGFRβ-positive stroma cells in tumours derived from co-injection (mRNA level: 4.12-fold increase, P<0.004; protein based on IHC: 2.1-fold increase, P<0.003; P-values calculated with Students’ t-Test). Furthermore, the co-injection-derived tumours showed an increased HMGA2 expression on gene as well as on protein level (mRNA level: 2.6-fold increase after correction for epithelial fraction, P<0.012; protein based on IHC: 1.4-fold increase, P<0.019; P-values calculated with Student’s t-Test) as compared to tumours derived from mono-injection (Figure 5B and C).
To further analyse a potential impact of PSCs on HMGA2 expression in vitro, we seeded Panc-1 cells under anchorage-independent conditions in control medium, conditioned medium from PSCs or conditioned medium from PDGF-BB pre-stimulated fibroblasts. Panc-1 cells seeded in PSC-conditioned media displayed an increased ability to form tumour cell spheroids under anchorage-independent conditions (1/13 cells (11.4–15.7) 95% CI; P<0.003) than control Panc-1 cells (1/19 cells (16.0–23.0) 95% CI). This effect was further enhanced with conditioned medium derived from PSCs pre-stimulated with PDGF-BB (1/10 cells (8.8–11.8) 95% CI; P<0.001 vs control and P<0.014 vs PSCs) (Figure 5D). The spheroids grown in conditioned medium of PSCs and pre-stimulated PSCs also showed a slightly increased expression of HMGA2 mRNA when compared to the control spheroids (1.4-fold increase for PSC-conditioned medium and 1.6-fold increase for conditioned medium from PDGF-BB pre-stimulated PSCs; not significant) (Figure 5E).
We obtained similar data in a 3D spheroid co-culture model allowing direct cell–cell contact between Panc-1 cells and PSCs. The HMGA2 mRNA level was increased 1.7-fold in Panc-1 cells grown in co-culture heterospheroids together with PSCs as compared to Panc-1 control spheroids (Figure 5F).
Together these experiments demonstrate that fibroblasts can support HMGA2 expression in a manner that is associated with increased tumourigenic properties.
Discussion
In the present study, we show by IHC analysis that HMGA2-positivity is a strong prognostic factor for shorter overall survival in PDAC and AAC (Tables 1 and 2). Notably, prognostic association was also detected in multivariate analyses where HMGA2 expression and tumour staging were the only significant features (Table 1). Additional evidence for a role of HMGA2 expression as an independent marker of poor prognosis was obtained by analyses of gene expression data from an independent cohort of PDAC (Table 2). These findings, including multivariable analyses and separate analyses of PDAC and AAC, thus confirm and go well beyond findings of one earlier study, which was limited to PDAC and included only univariate survival analysis (Haselmann et al, 2014). Collectively the findings suggest that HMGA2 IHC can identify patients with a worse prognosis.
The long duration of sample collection, along with changes in the clinical management of PDAC, make this tumour collection unsuitable for analyses of possible relevance of HMGA2 as a response-predictive marker. Such studies are warranted using other tumour collections. Explorative analysis of the present collection indicated that the survival association was equally strong in cases that received or did not receive gemcitabine (Supplementary Figure 1).
Among clinicopathological parameters a significant correlation between HMGA2-positivity and lower tumour differentiation was observed. However, within the clinical samples we did not find a clear correlation between the HMGA2-protein and the expression of EMT-specific miRNAs. Likewise, the HMGA2 gene expression level showed no strong association with the expression of EMT associated genes. HMGA2 could be detected in tumour cells independent of their epithelial cell differentiation status, although it appears enriched in tumour cells with low cytokeratin level (Supplementary Figure 2B). Taken together, this data could suggest that HMGA2 may be already induced at very early steps of tumour dedifferentiation before the onset of EMT marker expression. Thus far, it is unknown, when exactly HMGA2, normally expressed during embryonic development, is re-induced during malignant transformation. We and others (Abe et al, 2003; Piscuoglio et al, 2012), found HMGA2 to be absent in ductal epithelial cells of normal pancreas tissue. Pancreatic ductal cells in patients with chronic pancreatitis were also reported to be negative (Abe et al, 2003). Nevertheless, two studies observed an incipient positivity for HMGA2 in non-neoplastic ductal cells from peritumoural tissues of PDAC patients (Haselmann et al, 2014) as well as within PanIN lesions (Piscuoglio et al, 2012). In the KPC mouse model for PDAC, we detected weak Hmga2-positive cells already within PanIN1 lesions (Figure 3). In contrast, we did not detect any HMGA2-positive PanIN lesions within the human samples.
Notably, the PanINs of the KPC model occurred predominantly in context of extensive acinar-to-ductal metaplasia, which was not a prominent feature in the human samples. These differences suggest that although the KPC mouse is a good model of the human disease, it also displays features not seen in human cases, especially with regard to tumour initiation. Our findings suggest that HMGA2 is differentially expressed in morphologically divergent precursor lesions in mice and humans. Further studies are therefore needed to determine at which stage(s) of human tumour formation induction of HMGA2 is occurring.
Although several studies have described intrinsic signalling pathways that regulate HMGA2 expression, there are only a few studies that have investigated potential connections between the tumour microenvironment and HMGA2-positivity. These studies have focused on extracellular matrix components, including collagen 1, rather than cell subsets of the tumour microenvironment (Dangi-Garimella et al, 2011, 2013). Notably, in PDAC and AAC the tumour stroma is a major part of the total tumour mass (Lunardi et al, 2014a; Rucki and Zheng, 2014; Pan et al, 2015). Our study provides experimental and correlative findings that link PDGFRβ-defined fibroblasts to HMGA2 status in malignant cells. These correlative data, obtained both at case-level and through double IHC analyses, strongly suggest that paracrine stroma-epithelial signaling contributes to HMGA2 expression (Figure 4A and B). These findings are in agreement with experimental and translational studies which have emphasised the ability of cancer-associated fibroblasts to control cancer cell stemness, EMT and metastasis through paracrine signalling (Vermeulen et al, 2010; Lonardo et al, 2011; Capparelli et al, 2012; Li et al, 2012; Malanchi et al, 2011; Schulte et al, 2012). One very recent study nicely demonstrated, that the specific heterotypic cell–cell interactions between tumour cells carrying oncogenic KRASG12D and surrounding stroma cells are crucial to expand tumour cell signalling beyond cell-autonomous signalling pathways in PDAC (Tape et al, 2016). Another group of tissue-based studies on different solid tumours provided evidence for the existence of distinct fibroblast subpopulations, marked, for example, by PDGFRβ or increased stromal expression of SNAIL, which correlate with the expression of certain EMT markers in the adjacent tumour cells (Schulte et al, 2012; Galván et al, 2015; Senol et al, 2016). Importantly, a specific link between CAFs, or any other stroma cell subtype, and cancer cell expression of HMGA2 has not previously been demonstrated.
In earlier studies, the abundance of PDGFRβ-defined stroma cells has been associated with poor prognosis in breast, prostate and ovarian cancer (Paulsson et al, 2009; Hägglöf et al, 2010; Frings et al, 2013; Corvigno et al, 2016). Furthermore, experimental therapy studies have shown anti-metastatic effects of stroma-targeted therapies using PDGF inhibitors (Pietras et al, 2002, 2003a, b; Baranowska-Kortylewicz et al, 2005). Our findings thus extend and support the few earlier clinical correlative studies, in other tumour types, which have detected significant correlations between certain fibroblast features and cancer cell expression of stem cell or EMT markers (Schulte et al, 2012).
Earlier studies have demonstrated that TGF-β as well as EGF signalling can be inducers of HMGA2 (Thuault et al, 2006; Voon et al, 2013) and we could confirm in our in vitro spheroid culture system, that TGF-β treatment induces HMGA2 expression in Panc-1 cells (Supplementary Figure 3B). Members of PDGF ligands are unlikely to be part of the CAF-derived secretome that induces HMGA2, since we found most PDAC cell lines as well as tumour cells on tissue specimens to not express PDGFRs (data not shown). Taken together, these findings motivate extended experimental investigations on potential links between PDGFRβ-defined fibroblasts and TGF-β or EGF production. Especially the question if stromal PDGFRβ-signalling is actively involved or if the PDGFRβ is only a marker of a CAF-subpopulation that exhibits a certain secretome inducing tumoral HMGA2 expression, needs to be addressed.
Moreover, our data are also relevant in the context of recent reports that discussed pro- as well as anti-tumoural effects of fibroblasts in pancreatic cancer (Feig et al, 2013; Özdemir et al, 2014; Rhim et al, 2014). Collectively, these studies suggest that multiple functionally distinct marker-defined fibroblasts exist in pancreatic cancer. The findings of the present study strongly support continued efforts to define novel fibroblast subsets for further exploration of their regulation, function and clinical associations.
Change history
27 June 2017
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Abe N, Watanabe T, Suzuki Y, Matsumoto N, Masaki T, Mori T, Sugiyama M, Chiappetta G, Fusco A, Atomi Y (2003) An increased high-mobility group A2 expression level is associated with malignant phenotype in pancreatic exocrine tissue. Br J Cancer 89: 2104–2109.
Baranowska-Kortylewicz J, Abe M, Pietras K, Kortylewicz ZP, Kurizaki T, Nearman J, Paulsson J, Mosley RL, Enke CA, Ostman A (2005) Effect of platelet-derived growth factor receptor-beta inhibition with STI571 on radioimmunotherapy. Cancer Res 65: 7824–7831.
Biankin AV, Morey AL, Lee C-S, Kench JG, Biankin SA, Hook HC, Head DR, Hugh TB, Sutherland RL, Henshall SM (2002) DPC4/Smad4 expression and outcome in pancreatic ductal adenocarcinoma. J Clin Oncol 20: 4531–4542.
Bustin M (1999) Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol 19: 5237–5246.
Bustin M, Reeves R (1996) High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol 54: 35–100.
Califano D, Pignata S, Losito NS, Ottaiano A, Greggi S, De Simone V, Cecere S, Aiello C, Esposito F, Fusco A, Chiappetta G (2014) High HMGA2 expression and high body mass index negatively affect the prognosis of patients with ovarian cancer. J Cell Physiol 229: 53–59.
Capparelli C, Guido C, Whitaker-Menezes D, Bonuccelli G, Balliet R, Pestell TG, Goldberg AF, Pestell RG, Howell A, Sneddon S, Birbe R, Tsirigos A, Martinez-Outschoorn U, Sotgia F, Lisanti MP (2012) Autophagy and senescence in cancer-associated fibroblasts metabolically supports tumor growth and metastasis via glycolysis and ketone production. Cell Cycle 11: 2285–2302.
Cleynen I, Van de Ven WJM (2008) The HMGA proteins: a myriad of functions (review). Int J Oncol 32: 289–305.
Corvigno S, Wisman GBA, Mezheyeuski A, van der Zee AG, Nijman HW, Åvall-Lundqvist E, Östman A, Dahlstrand H (2016) Markers of fibroblast-rich tumor stroma and perivascular cells in serous ovarian cancer: inter- and intra-patient heterogeneity and impact on survival. Oncotarget 7: 18573–18584.
Dangi-Garimella S, Krantz SB, Barron MR, Shields MA, Heiferman MJ, Grippo PJ, Bentrem DJ, Munshi HG (2011) Three-dimensional collagen I promotes gemcitabine resistance in pancreatic cancer through MT1-MMP-mediated expression of HMGA2. Cancer Res 71: 1019–1028.
Dangi-Garimella S, Sahai V, Ebine K, Kumar K, Munshi HG (2013) Three-dimensional collagen I promotes gemcitabine resistance in vitro in pancreatic cancer cells through HMGA2-dependent histone acetyltransferase expression. PloS One 8: e64566.
Feig C, Jones JO, Kraman M, Wells RJB, Deonarine A, Chan DS, Connell CM, Roberts EW, Zhao Q, Caballero OL, Teichmann SA, Janowitz T, Jodrell DI, Tuveson DA, Fearon DT (2013) Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci USA 110: 20212–20217.
Ferlay J, Shin H-R, Bray F, Forman D, Mathers C, Parkin DM (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127: 2893–2917.
Frings O, Augsten M, Tobin NP, Carlson J, Paulsson J, Pena C, Olsson E, Veerla S, Bergh J, Ostman A, Sonnhammer ELL (2013) Prognostic significance in breast cancer of a gene signature capturing stromal PDGF signaling. Am J Pathol 182: 2037–2047.
Galván JA, Zlobec I, Wartenberg M, Lugli A, Gloor B, Perren A, Karamitopoulou E (2015) Expression of E-cadherin repressors SNAIL, ZEB1 and ZEB2 by tumour and stromal cells influences tumour-budding phenotype and suggests heterogeneity of stromal cells in pancreatic cancer. Br J Cancer 112: 1944–1950.
Hägglöf C, Hammarsten P, Josefsson A, Stattin P, Paulsson J, Bergh A, Ostman A (2010) Stromal PDGFRbeta expression in prostate tumors and non-malignant prostate tissue predicts prostate cancer survival. PloS One 5: e10747.
Haselmann V, Kurz A, Bertsch U, Hübner S, Olempska-Müller M, Fritsch J, Häsler R, Pickl A, Fritsche H, Annewanter F, Engler C, Fleig B, Bernt A, Röder C, Schmidt H, Gelhaus C, Hauser C, Egberts J-H, Heneweer C, Rohde AM, Böger C, Knippschild U, Röcken C, Adam D, Walczak H, Schütze S, Janssen O, Wulczyn FG, Wajant H, Kalthoff H, Trauzold A (2014) Nuclear death receptor TRAIL-R2 inhibits maturation of let-7 and promotes proliferation of pancreatic and other tumor cells. Gastroenterology 146: 278–290.
Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA (2005) Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 7: 469–483.
Howe JR, Klimstra DS, Moccia RD, Conlon KC, Brennan MF (1998) Factors predictive of survival in ampullary carcinoma. Ann Surg 228: 87–94.
Hristov AC, Cope L, Reyes MD, Singh M, Iacobuzio-Donahue C, Maitra A, Resar LMS (2009) HMGA2 protein expression correlates with lymph node metastasis and increased tumor grade in pancreatic ductal adenocarcinoma. Mod Pathol 22: 43–49.
Hu Y, Smyth GK (2009) ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 347: 70–78.
Iacobuzio-Donahue CA, Fu B, Yachida S, Luo M, Abe H, Henderson CM, Vilardell F, Wang Z, Keller JW, Banerjee P, Herman JM, Cameron JL, Yeo CJ, Halushka MK, Eshleman JR, Raben M, Klein AP, Hruban RH, Hidalgo M, Laheru D (2009) DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol 27: 1806–1813.
Jesnowski R, Fürst D, Ringel J, Chen Y, Schrödel A, Kleeff J, Kolb A, Schareck WD, Löhr M (2005) Immortalization of pancreatic stellate cells as an in vitro model of pancreatic fibrosis: deactivation is induced by matrigel and N-acetylcysteine. Lab Invest 85: 1276–1291.
Kawase T, Yasui Y, Nishina S, Hara Y, Yanatori I, Tomiyama Y, Nakashima Y, Yoshida K, Kishi F, Nakamura M, Hino K (2015) Fibroblast activation protein-α-expressing fibroblasts promote the progression of pancreatic ductal adenocarcinoma. BMC Gastroenterol 15: 109.
Kraman M, Bambrough PJ, Arnold JN, Roberts EW, Magiera L, Jones JO, Gopinathan A, Tuveson DA, Fearon DT (2010) Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science 330: 827–830.
Lee J, Ha S, Jung C-K, Lee HH (2015) High-mobility-group A2 overexpression provokes a poor prognosis of gastric cancer through the epithelial-mesenchymal transition. Int J Oncol 46: 2431–2438.
Lepage C, Capocaccia R, Hackl M, Lemmens V, Molina E, Pierannunzio D, Sant M, Trama A, Faivre J EUROCARE-5 Working Grouph (2015) Survival in patients with primary liver cancer, gallbladder and extrahepatic biliary tract cancer and pancreatic cancer in Europe 1999-2007: Results of EUROCARE-5. Eur J Cancer 51: 2169–2178.
Li D, Xie K, Wolff R, Abbruzzese JL (2004) Pancreatic cancer. Lancet 363: 1049–1057.
Li X, Ma Q, Xu Q, Liu H, Lei J, Duan W, Bhat K, Wang F, Wu E, Wang Z (2012) SDF-1/CXCR4 signaling induces pancreatic cancer cell invasion and epithelial-mesenchymal transition in vitro through non-canonical activation of Hedgehog pathway. Cancer Lett 322: 169–176.
Lonardo E, Hermann PC, Mueller M-T, Huber S, Balic A, Miranda-Lorenzo I, Zagorac S, Alcala S, Rodriguez-Arabaolaza I, Ramirez JC, Torres-Ruíz R, Garcia E, Hidalgo M, Cebrián DÁ, Heuchel R, Löhr M, Berger F, Bartenstein P, Aicher A, Heeschen C (2011) Nodal/Activin signaling drives self-renewal and tumorigenicity of pancreatic cancer stem cells and provides a target for combined drug therapy. Cell Stem Cell 9: 433–446.
Longati P, Jia X, Eimer J, Wagman A, Witt M-R, Rehnmark S, Verbeke C, Toftgård R, Löhr M, Heuchel RL (2013) 3D pancreatic carcinoma spheroids induce a matrix-rich, chemoresistant phenotype offering a better model for drug testing. BMC Cancer 13: 95.
Lunardi S, Muschel RJ, Brunner TB (2014) The stromal compartments in pancreatic cancer: are there any therapeutic targets? Cancer Lett 343: 147–155.
Madison BB, Jeganathan AN, Mizuno R, Winslow MM, Castells A, Cuatrecasas M, Rustgi AK (2015) Let-7 Represses carcinogenesis and a stem cell phenotype in the intestine via regulation of Hmga2. PLoS Genet 11: e1005408.
Malanchi I, Santamaria-Martínez A, Susanto E, Peng H, Lehr H-A, Delaloye J-F, Huelsken J (2011) Interactions between cancer stem cells and their niche govern metastatic colonization. Nature 481: 85–89.
Malvezzi M, Carioli G, Bertuccio P, Rosso T, Boffetta P, Levi F, La Vecchia C, Negri E (2016) European cancer mortality predictions for the year 2016 with focus on leukaemias. Ann Oncol 27: 725–731.
Motoyama K, Inoue H, Nakamura Y, Uetake H, Sugihara K, Mori M (2008) Clinical significance of high mobility group A2 in human gastric cancer and its relationship to let-7 microRNA family. Clin Cancer Res 14: 2334–2340.
Ostman A, Augsten M (2009) Cancer-associated fibroblasts and tumor growth—bystanders turning into key players. Curr Opin Genet Dev 19: 67–73.
Özdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu C-C, Simpson TR, Laklai H, Sugimoto H, Kahlert C, Novitskiy SV, De Jesus-Acosta A, Sharma P, Heidari P, Mahmood U, Chin L, Moses HL, Weaver VM, Maitra A, Allison JP, LeBleu VS, Kalluri R (2014) Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell 25: 719–734.
Pan B, Liao Q, Niu Z, Zhou L, Zhao Y (2015) Cancer-associated fibroblasts in pancreatic adenocarcinoma. Future Oncol 11: 2603–2610.
Paulsson J, Sjöblom T, Micke P, Pontén F, Landberg G, Heldin C-H, Bergh J, Brennan DJ, Jirström K, Ostman A (2009) Prognostic significance of stromal platelet-derived growth factor beta-receptor expression in human breast cancer. Am J Pathol 175: 334–341.
Pietras K, Rubin K, Sjöblom T, Buchdunger E, Sjöquist M, Heldin C-H, Ostman A (2002) Inhibition of PDGF receptor signaling in tumor stroma enhances antitumor effect of chemotherapy. Cancer Res 62: 5476–5484.
Pietras K, Sjöblom T, Rubin K, Heldin C-H, Ostman A (2003a) PDGF receptors as cancer drug targets. Cancer Cell 3: 439–443.
Pietras K, Stumm M, Hubert M, Buchdunger E, Rubin K, Heldin C-H, McSheehy P, Wartmann M, Ostman A (2003b) STI571 enhances the therapeutic index of epothilone B by a tumor-selective increase of drug uptake. Clin Cancer Res 9: 3779–3787.
Piscuoglio S, Zlobec I, Pallante P, Sepe R, Esposito F, Zimmermann A, Diamantis I, Terracciano L, Fusco A, Karamitopoulou E (2012) HMGA1 and HMGA2 protein expression correlates with advanced tumour grade and lymph node metastasis in pancreatic adenocarcinoma. Histopathology 60: 397–404.
Reeves R (2001) Molecular biology of HMGA proteins: hubs of nuclear function. Gene 277: 63–81.
Reeves R (2015) High mobility group (HMG) proteins: modulators of chromatin structure and DNA repair in mammalian cells. DNA Repair 36: 122–136.
Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, Dekleva EN, Saunders T, Becerra CP, Tattersall IW, Westphalen CB, Kitajewski J, Fernandez-Barrena MG, Fernandez-Zapico ME, Iacobuzio-Donahue C, Olive KP, Stanger BZ (2014) Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell 25: 735–747.
Rizzi C, Cataldi P, Iop A, Isola M, Sgarra R, Manfioletti G, Giancotti V (2013) The expression of the high-mobility group A2 protein in colorectal cancer and surrounding fibroblasts is linked to tumor invasiveness. Hum Pathol 44: 122–132.
Rostain F, Hamza S, Drouillard A, Faivre J, Bouvier A-M, Lepage C (2014) Trends in incidence and management of cancer of the ampulla of Vater. World J Gastroenterol 20: 10144–10150.
Rucki AA, Zheng L (2014) Pancreatic cancer stroma: understanding biology leads to new therapeutic strategies. World J Gastroenterol 20: 2237–2246.
Ryan DP, Hong TS, Bardeesy N (2014) Pancreatic adenocarcinoma. N Engl J Med 371: 1039–1049.
Sarhadi VK, Wikman H, Salmenkivi K, Kuosma E, Sioris T, Salo J, Karjalainen A, Knuutila S, Anttila S (2006) Increased expression of high mobility group A proteins in lung cancer. J Pathol 209: 206–212.
Schulte J, Weidig M, Balzer P, Richter P, Franz M, Junker K, Gajda M, Friedrich K, Wunderlich H, Östman A, Petersen I, Berndt A (2012) Expression of the E-cadherin repressors Snail, Slug and Zeb1 in urothelial carcinoma of the urinary bladder: relation to stromal fibroblast activation and invasive behaviour of carcinoma cells. Histochem Cell Biol 138: 847–860.
Senol S, Sayar I, Ceyran AB, Ibiloglu I, Akalin I, Firat U, Kosemetin D, Engin Zerk P, Aydin A (2016) Stromal clues in endometrial carcinoma: loss of expression of β-catenin, epithelial-mesenchymal transition regulators, and estrogen-progesterone receptor. Int J Gynecol Pathol 35: 238–248.
Siegel RL, Miller KD, Jemal A (2016) Cancer statistics, 2016. CA Cancer J Clin 66: 7–30.
Strell C, Rundqvist H, Ostman A (2012) Fibroblasts—a key host cell type in tumor initiation, progression, and metastasis. Ups J Med Sci 117: 187–195.
Tape CJ, Ling S, Dimitriadi M, McMahon KM, Worboys JD, Leong HS, Norrie IC, Miller CJ, Poulogiannis G, Lauffenburger DA, Jørgensen C (2016) Oncogenic KRAS regulates tumor cell signaling via stromal reciprocation. Cell 165: 1818.
Tascilar M, Skinner HG, Rosty C, Sohn T, Wilentz RE, Offerhaus GJ, Adsay V, Abrams RA, Cameron JL, Kern SE, Yeo CJ, Hruban RH, Goggins M (2001) The SMAD4 protein and prognosis of pancreatic ductal adenocarcinoma. Clin Cancer Res 7: 4115–4121.
Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, Hartwell K, Onder TT, Gupta PB, Evans KW, Hollier BG, Ram PT, Lander ES, Rosen JM, Weinberg RA, Mani SA (2010) Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA 107: 15449–15454.
Thuault S, Valcourt U, Petersen M, Manfioletti G, Heldin C-H, Moustakas A (2006) Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. J Cell Biol 174: 175–183.
Vermeulen L, De Sousa E, Melo F, van der Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M, Merz C, Rodermond H, Sprick MR, Kemper K, Richel DJ, Stassi G, Medema JP (2010) Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol 12: 468–476.
Voon DC-C, Wang H, Koo JKW, Chai JH, Hor YT, Tan TZ, Chu Y-S, Mori S, Ito Y (2013) EMT-induced stemness and tumorigenicity are fueled by the EGFR/Ras pathway. PloS One 8: e70427.
Wagner M, Redaelli C, Lietz M, Seiler CA, Friess H, Büchler MW (2004) Curative resection is the single most important factor determining outcome in patients with pancreatic adenocarcinoma. Br J Surg 91: 586–594.
Wang LM, Silva MA, D’Costa Z, Bockelmann R, Soonawalla Z, Liu S, O’Neill E, Mukherjee S, McKenna WG, Muschel R, Fokas E (2016) The prognostic role of desmoplastic stroma in pancreatic ductal adenocarcinoma. Oncotarget 7: 4183–4194.
Watanabe S, Ueda Y, Akaboshi S, Hino Y, Sekita Y, Nakao M (2009) HMGA2 maintains oncogenic RAS-induced epithelial-mesenchymal transition in human pancreatic cancer cells. Am J Pathol 174: 854–868.
Weiland A, Roswall P, Hatzihristidis TC, Pietras K, Ostman A, Strell C (2012) Fibroblast-dependent regulation of the stem cell properties of cancer cells. Neoplasma 59: 719–727.
Wend P, Runke S, Wend K, Anchondo B, Yesayan M, Jardon M, Hardie N, Loddenkemper C, Ulasov I, Lesniak MS, Wolsky R, Bentolila LA, Grant SG, Elashoff D, Lehr S, Latimer JJ, Bose S, Sattar H, Krum SA, Miranda-Carboni GA (2013) WNT10B/β-catenin signalling induces HMGA2 and proliferation in metastatic triple-negative breast cancer. EMBO Mol Med 5: 264–279.
Acknowledgements
Members of the AÖ group are acknowledged for support and constructive criticism throughout the study. The AÖ group is supported by grants from the Swedish Research Council (STARGET network), the Breast Cancer Theme Center (BRECT) at Karolinska Institutet, Swedish Cancer Society, the research funds at Radiumhemmet and Stockholm County Council (ALF). The JP group is supported by the Swedish Research Council grant nr 2011-5389, the JML group is supported by the VetenskapsrÍdet (VR) grant nr K2013-67X22322-01-3 and RLH is supported by Cancerfonden (CAN 2013/780).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Strell, C., Norberg, K., Mezheyeuski, A. et al. Stroma-regulated HMGA2 is an independent prognostic marker in PDAC and AAC. Br J Cancer 117, 65–77 (2017). https://doi.org/10.1038/bjc.2017.140
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2017.140
- Springer Nature Limited
Keywords
This article is cited by
-
Characterization of extrachromosomal circular DNAs in plasma of patients with clear cell renal cell carcinoma
World Journal of Urology (2024)
-
Prognostic significance of high mobility group A2 (HMGA2) in pancreatic ductal adenocarcinoma: malignant functions of cytoplasmic HMGA2 expression
Journal of Cancer Research and Clinical Oncology (2021)
-
A novel pancreatic tumour and stellate cell 3D co-culture spheroid model
BMC Cancer (2020)
-
Knockdown of HMGA2 regulates the level of autophagy via interactions between MSI2 and Beclin1 to inhibit NF1-associated malignant peripheral nerve sheath tumour growth
Journal of Experimental & Clinical Cancer Research (2019)
-
The critical roles of activated stellate cells-mediated paracrine signaling, metabolism and onco-immunology in pancreatic ductal adenocarcinoma
Molecular Cancer (2018)