Abstract
Background:
The Tbx3 transcription factor is over-expressed in breast cancer, where it has been implicated in proliferation, migration and regulation of the cancer stem cell population. The mechanisms that regulate Tbx3 expression in cancer have not been fully explored. In this study, we demonstrate that Tbx3 is repressed by the tumour suppressor miR-206 in breast cancer cells.
Methods:
Bioinformatics prediction programmes and luciferase reporter assays were used to demonstrate that miR-206 negatively regulates Tbx3. We examined the impact of miR-206 on Tbx3 expression in breast cancer cells using miR-206 mimic and inhibitor. Gene/protein expression was examined by quantitative reverse-transcription–PCR and immunoblotting. The effects of miR-206 and Tbx3 on apoptosis, proliferation, invasion and cancer stem cell population was investigated by cell-death detection, colony formation, 3D-Matrigel and tumorsphere assays.
Results:
In this study, we examined the regulation of Tbx3 by miR-206. We demonstrate that Tbx3 is directly repressed by miR-206, and that this repression of Tbx3 is necessary for miR-206 to inhibit breast tumour cell proliferation and invasion, and decrease the cancer stem cell population. Moreover, Tbx3 and miR-206 expression are inversely correlated in human breast cancer. Kaplan–Meier analysis indicates that patients exhibiting a combination of high Tbx3 and low miR-206 expression have a lower probability of survival when compared with patients with low Tbx3 and high miR-206 expression. These studies uncover a novel mechanism of Tbx3 regulation and identify a new target of the tumour suppressor miR-206.
Conclusions:
The present study identified Tbx3 as a novel target of tumour suppressor miR-206 and characterised the miR-206/Tbx3 signalling pathway, which is involved in proliferation, invasion and maintenance of the cancer stem cell population in breast cancer cells. Our results suggest that restoration of miR-206 in Tbx3-positive breast cancer could be exploited for therapeutic benefit.
Similar content being viewed by others
Main
In 2004, Tbx3 was characterised as an oncogene when it was found capable of immortalising and transforming mouse embryo fibroblasts and found to be significantly over-expressed in a subset of human breast cancer cell lines (Fan et al, 2004). Subsequently, Tbx3 was found to be over-expressed in the epithelium of primary human breast cancers (Yarosh et al, 2008), and analysis of gene expression data sets confirmed that Tbx3 was significantly over-expressed in human breast cancer (Fillmore et al, 2010). Transgenic over-expression of Tbx3 in the mouse mammary gland was found to increase ductal branching, enhance mammary epithelial cell proliferation, promote mammary stem cell expansion and induce focal hyperplasia (Liu et al, 2011). Of note, Tbx3 was recently found to be required for oestrogen-dependent survival of breast cancer stem cells, and its over-expression was shown both necessary and sufficient for breast cancer stem cell expansion (Fillmore et al, 2010).
Tbx3’s oncogenic effects have historically been attributed to its ability to promote tumour cell proliferation, in part through the transcriptional repression of the p21CIP1 and p14ARF growth suppressors (Brummelkamp et al, 2002; Yarosh et al, 2008). However, others have recently reported that rather than promoting proliferation, Tbx3 functions largely in a pro-migratory capacity, both in normal breast (Li et al, 2013) and breast cancer (Peres et al, 2010) cells, and is essential for the anti-proliferative/pro-migratory functions of TGF-β, a cytokine with prominent roles in mammary development and tumorigenesis (Moses and Barcellos-Hoff, 2011). Importantly, high Tbx3 levels are strongly correlated with shorter metastasis-free survival in breast cancer (Fillmore et al, 2010).
microRNAs (miRs) are small, non-coding single-stranded RNAs that bind to the 3′ untranslated region (UTR) of specific mRNAs leading to either mRNA degradation or translational inhibition. miRs are frequently deregulated in cancer, and both gain and loss of function have been reported (Bader et al, 2010). miRs are functionally categorised as either tumour suppressors or oncogenes (oncomiRs), determined by the balance of pathways they regulate. Tumour suppressor miRs often target and downregulate oncogenes, suppressing cancer formation; while oncomiRs often target and downregulate tumour suppressor mRNAs leading to cancer formation. The tumour suppressor miR-206 is downregulated in the majority (>90%) of human breast cancers and low miR-206 is an independent marker of poor survival (Kondo et al, 2008; Li et al, 2013; Zhou et al, 2013). miR-206 acts downstream of ER-α and has an important role in the regulation of genes related to mammary gland development and breast cancer (Lee et al, 2013). Importantly, expression of miR-206 in breast cancer cells decreases cell proliferation and colony formation (Zhou et al, 2013). However, only a handful of validated miR-206 targets are currently known, including ANXA2 and KRAS (Keklikoglou et al, 2015), Cx43 (Fu et al, 2015), KLF4 (Lin et al, 2015), MRTF-A (Chen et al, 2015), ER-α (Adams et al, 2007), c-Met (Yan et al, 2009), Notch-3 (Song et al, 2009), the actin-binding protein Coronin 1C (Wang et al, 2014), Cyclin D1 (Alteri et al, 2013; Elliman et al, 2014) and Cyclin D2 (Zhou et al, 2013). miR target prediction tools suggest that the 3′ UTR of Tbx3 is subject to regulation by various miRs, but few, with the exception of miR-17-92 (Wang et al, 2013, 2014), miR-106b-25 (Wang et al, 2014) and miR-137 (Jiang et al, 2013), have been validated. In this study, we demonstrate that Tbx3 is repressed by the tumour suppressor miR-206 in breast cancer cells, and this repression abrogates the cancer phenotype. Given the collective evidence that Tbx3 has an important functional role in breast cancer, our study lays the foundation for future therapeutic approaches for Tbx3-positive breast cancer.
Materials and methods
Antibodies and reagents
For western blotting analysis, anti-Tbx3 antibody was purchased from Novus Biologicals (H00006926-A01; Littleton, CO, USA); anti-PTEN (9559), anti-Bcl-2 (2876) and anti-phospho-Akt (9271) were all purchased from Cell Signaling Biotechnology (Danvers, MA, USA). Anti-flag tag antibody (F7425) and anti-beta-actin (clone AC-15) were from Sigma-Aldrich (St. Louis, MO, USA). Synthetic miR-206 mimic, antagomiR-206 (αmiR-206) and miR scrambled control (NC) were purchased from Qiagen (Valencia, CA, USA). pMIR-REPORT Luciferase vector and control vector were purchased from Applied Biological Materials (ABM MT-h25249; Richmond, BC, Canada), SMART pool: ON-TARGETplus, Tbx3-siRNA (LU-012197-00-0005) and set of four individual siRNA (LU-012197-000002) were purchased from Dharmacon (Lafayette, CO, USA). Human Tbx3 flag-tagged cDNA (RC220990) was from OriGene Technologies, Inc. (Rockville, MD, USA).
Cell culture and treatment
The human immortalised mammary epithelial cell line MCF10A and human breast cancer cell lines MCF7, MDA-MB-231 and ZR75-1 were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured according to the ATCC instructions. For transient transfections cells were plated onto six-well plates 1 day before the transfection and maintained in serum-containing medium without antibiotics. The following day, cells were transfected with either 50 nM of scrambled control (NC), 50 nM miR-206 mimic, 50 nM siRNA-Tbx3 or 100 nM of antagomiR-206 (αmiR-206) using Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA) according to the manufacturer’s instructions.
Dual-luciferase reporter assay
For the validation of Tbx3 as a direct target of miR-206, wild-type and mutant pMIR-REPORT luciferase vectors were purchased from Applied Biological Materials Inc. (ABM MT-h25249). The miR-206 wild-type binding sequence or its mutated form was inserted at the C terminus of the luciferase gene to generate pMIR-TBX3-3′ UTR (containing the full-length 3′ UTR segment of human Tbx3) or pMIR-TBX3-mut-3′ UTR (deleting five nucleotides in the seed region), respectively. Briefly, HEK-293 T (2.5 × 104) cells were seeded into 24-well plates, cultured overnight in antibiotic-free media and co-transfected with 250 ng of pMIR-TBX3-3′ UTR, pMIR-TBX3-mut-3′ UTR or control plasmids, with 50 nM miR scrambled control (NC) or 50 nM miR-206 mimic using Lipofectamine 2000. Forty-eight hours post transfection cells were collected in reporter lysis buffer (Promega) and luciferase activity was measured using Dual-luciferase assay kit (Promega, Madison, WI, USA) according to the manufacturer’s protocol, and detected by Glomax 96 microplate luminometer (Promega). β-galactosidase activity was used for normalisation, and measured by beta-glo Luminescent Assay Kit (Promega).
RNA isolation and quantitative real-time RT–PCR
Total RNA including miRNAs was collected using the miRNeasy Mini Kit (Qiagen, #217004) according to the manufacturer’s protocol. For Tbx3 qPCR, RNA was extracted from breast cancer cells ZR75-1, MCF7 and MDA-MB-231, which had previously been transfected with 50 nM miR scrambled control (NC) or 50 nM miR-206 mimic. Applied Biosystems (Carlsbad, CA, USA) High Capacity cDNA Reverse Transcription Kit (#4368814) was used to convert 500 ng of total RNA into cDNA. For miR-206 qPCR, RNA was extracted from three breast cancer cell lines (ZR75-1, MCF7 and MDA-MB-231) and two normal cell lines (HMLE and MCF10A). Applied Biosystems microRNA Reverse Transcription Kit (#4466596) was used to convert 10 ng RNA into cDNA. Quantitative PCR amplifications were conducted in a BioRad (Hercules, CA, USA) CFX96 real-time PCR system using Applied Biosystems TaqMan gene-specific primers and probes for TBX3 and SsoAdvanced Universal Probe Supermix (BioRad, #1725281), and specific primers and probes for miR-206 and TaqMan Universal Master Mix II (Applied Biosystems #4427983). Message levels for TBX3 and miR-206 were normalised to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels and the U6 small nuclear RNA (RNU6B) control, respectively. Analysis for Tbx3 was performed by the comparative threshold cycle (Ct) method under the following cycling conditions: 3 min at 95 °C; 40 cycles of 10 s at 95 °C and 30 s at 55 °C. Analysis for miR-206 was performed by the comparative Ct method under the following cycling conditions: 2 min at 50 °C; 10 min at 95 °C; 40 cycles of 15 s at 95 °C and 1 min at 60 °C. Relative abundance was determined from the Ct values using the 2−ΔΔCt method after normalisation to GAPDH (for Tbx3) or RNU6B (for miR-206). All experiments were performed 2–4 times.
Western blot analysis
Total protein was extracted from cells 72-h post transfection using the cell lysis buffer (Cell Signaling BioTechnology) supplemented with proteinase and phosphatase inhibitors. After 20 min of incubation on ice, lysates were vigorously vortexed and centrifuged at 13 000 g for 15 min, and protein concentrations in the supernatant were determined using BCA kit (Pierce, Rockford, IL, USA). A unit of 30 μg of lysates were denatured in 2 × SDS sample buffer (50 mmol l−1 Tris-HCl (pH 6.8), 2% SDS, 10% glycerol, 0.25% β-mercaptoethanol and bromophenol blue (1 mg ml−1)) at 100 °C for 5 min, separated on 10% SDS–polyacrylamide gel (BioRad), then transferred to nitrocellulose membrane (Millipore, Billerica, MA, USA). Membranes were blocked with 5% non-fat dry milk in Tris-buffered saline containing 0.05% Tween 20 (TBST), and incubated with specific primary antibodies overnight at 4 °C. After incubation, membranes were washed with TBST and then incubated with horseradish peroxidase-conjugated secondary antibodies. Band signals were visualised using ECL (Pierce) on an AlphaInnotech Imaging System (San Jose, CA, USA).
Detection of apoptosis
The Cell Death Detection ELISA assay (Roche, Indianapolis, IN, USA) was used to detect apoptosis following the manufacturer’s protocol. This assay determines apoptosis by measuring mono- and oligonucleosomes in the lysates of apoptotic cells. Briefly, breast cancer cells MCF7, ZR75-1 and MDA-MB-231 were seeded in 96-well plates and cultured overnight in antibiotic-free media. Cells were then transfected with either 50 nM of miR scrambled control (NC), miR-206 mimic, siRNA-Tbx3 or 100 nM of antagomiR-206 (αmiR-206) using Lipofectamine 2000. Cells were lysed 72 h post transfection. The lysates were placed into a streptavidin-coated microplate and incubated with a mixture of anti-histone-biotin and anti-DNA-peroxidase. The amount of peroxidase retained in the immunocomplex was photometrically determined with ABTS as the substrate. Absorbance was measured at 405 nm. Assays were performed in quadruplicate in three independent experiments.
Colony-formation assay
MCF7, ZR75-1 and MDA-MB-231 cells were transfected with 50 nM of miR scrambled control (NC), miR-206 mimic, siRNA-Tbx3 or 100 nM of antagomiR-206 (αmiR-206). At 72 h post transfection, untransfected and transfected cells were collected by using HBSS (Hank’s balanced salt solution). A total of 1000 cells per well were separately plated in six-well cluster plates in triplicates and incubated for 10–12 days. Growth media was replaced every 3 days with fresh medium. Following incubation, cells were washed with PBS, colonies were counted after fixing and staining in 20% methanol and 0.1% crystal violet solution and analysed for clonogenic survival. Percentage cell survival was determined with untreated cells set at 100%.
Three-dimensional morphogenesis assay in Matrigel
MDA-MB-231 and MCF7 cells were cultured on growth factor-reduced Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) using the overlay method with modifications (Debnath et al, 2003). Briefly, a 96-well plate was coated with a thin layer of Matrigel and incubated at 37 °C for 30 min. After the Matrigel solidified, 3–5 × 103 cells, suspended in 100 μl assay medium containing 2% Matrigel, were added drop-wise on top of the Matrigel and allowed to grow overnight. The next day cells were transfected with either 50 nM of miR scrambled control (NC), miR-206 mimic, siRNA-Tbx3 or 100 nM antagomiR-206 (αmiR-206) using Lipofectamine 2000 according to the manufacturer’s instructions. For the rescue experiment, Tbx3 expression level was determined by expressing flag-tagged TBX3 with either miR scrambled control (NC) or miR-206 mimic in HEK-293T cells and total cell lysates were blotted with anti-flag antibody. For the rescue experiment in three-dimensional (3D) cultures, MDA-MB-231 cells were co-transfected with miR scrambled control (NC) or miR-206 mimic with 250 ng of either vector control or TBX3 cDNA and allowed to grow for 5–7 days. Growth media were replaced every 2 days with fresh media, without disturbing the cell/matrix layer, until the experiment was completed. The 3D structures of the cells were analysed and images were taken using × 4 and × 10 magnification with a confocal microscope (Olympus IX71 microscope, Olympus, Shinjuku-ku, Tokyo, Japan). Images were processed with ImageJ (U.S. National Institutes of Health, Bethesda, MD, USA) and analysed/quantified for the quantity of invasive colonies, number of branches in invasive cell stellate and the relative mean area covered by the cells. Quantification of colony area is analysed measuring the diameter of ⩾50 colonies.
Tumorsphere formation assay
MCF7 and MDA-MB-231 cells were reverse-transfected with miR scrambled control (NC), miR-206 mimic or antagomiR-206 (αmiR-206), or co-transfected with miR scrambled control (NC) or miR-206 mimic with 25 ng of either vector control or TBX3 cDNA (for the rescue experiment) directly in ultra-low attachment (ULA) 24-well plates (Corning, Tewksbury, MA, USA) in serum-free, antibiotic-free MammoCult media (Stem Cell Technologies, Vancouver, BC, Canada), at a confluency of 1.0 × 104 cells per well and allowed to grow for 7 days. Seven days after the incubation, primary spheres (larger than 75 μm) were counted and then dissociated into single cells. Cells from dissociated primary spheres were reverse-transfected again, and re-plated in ULA plates and allowed to grow for another 7 days. Seven days after incubation, secondary spheres (larger than 75 μm) were quantified.
Computational analysis of human breast cancer data
The results of computational analysis are in whole or part based on data generated by The Cancer Genome Atlas (TCGA) Research Network: http://cancergenome.nih.gov/. Sequencing read counts were used to determine levels of Tbx3 mRNA expression and miR-206 expression, in adjacent normal vs breast tumour samples. Welch two-sample t-test was used to determine whether the means of the two groups were statistically different. The relationship between Tbx3 and miR-206 for breast tumour and adjacent normal samples was determined using RSEM mRNASeq, RPM miRNASeq and read count data from TCGA. Tbx3 mRNA and miR-206 expression data were matched by patient sample, and correlation was computed using the Spearman rank correlation coefficient method. Data were log2 transformed before plotting. In all, 799 patient samples had undetectable miR-206 expression and were thus excluded from the correlation analysis. Kaplan–Meier survival plots were generated using the ‘survival’ package in R. Data for Kaplan–Meier plots consisted of Tbx3 mRNA read count, miR-206 miRNA RPM and patient information (death status and days to last contact). Patients were ranked based on the descending level of Tbx3 and miR-206 expression they exhibited. For Tbx3, the bottom two-fifth (40%) of patients were placed into the ‘Low-Tbx3’ group. For miR-206, the top one-fourth (25%) of patients were placed into the ‘High-miR-206’ group. Tarone–Ware test (G-rho family of Harrington and Fleming survival test) was used to determine whether the difference in survival probability between groups was statistically different. All graphical representation and statistical analysis of data were performed using the R programming language. In all cases, differences were considered statistically significant when P⩽0.05.
Statistical analysis
All data are presented as mean±s.e.m. of at least three independent experiments. The level of significance between groups was determined using one-way ANOVA with a post hoc Tukey HSD (honestly significant difference) test (GraphPad Prism5, GraphPad Software, Inc., La Jolla, CA, USA). In all cases, differences were considered statistically significant when P<0.05.
Results
Tbx3 mRNA is inversely correlated with miR-206 expression in human breast cancer
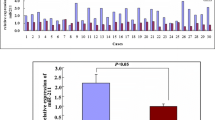
MicroRNAs repress their targets by translational repression and/or mRNA degradation. For the latter, an inverse correlation is expected between the miRNAs and their target mRNA. To examine the relationship between miR-206 and Tbx3 mRNA expression in human breast cancer, adjacent normal and breast tumour samples from the TCGA were analysed. Tbx3 mRNA expression in the TCGA data set is shown in Figure 1A and miR-206 expression is shown in Figure 1B. In all, 573 of 1099 breast tumours (52%) have Tbx3 mRNA expression greater than the mean for normal samples, in agreement with prior reports that Tbx3 is over-expressed in breast cancer. miR-206 expression was decreased in breast tumours, in agreement with prior reports of its downregulation. In addition, we observed an inverse correlation between Tbx3 mRNA expression and miR-206 expression, both in adjacent normal breast samples (Figure 1C) and breast tumours (Figure 1D) from the TCGA data set, suggesting that miR-206 represses Tbx3 expression, at least in part through promoting Tbx3 mRNA degradation. Kaplan–Meier analysis of breast cancer patients was next examined to assess the impact of Tbx3/miR-206 status on disease-free survival. Notably, patients with a combination of low Tbx3 and high miR-206 had a significantly increased survival time (mean survival=5086±440 days) compared with patients with a combination of high Tbx3 and low miR-206 expression (mean survival=3429±227 days; Figure 1E), strongly suggesting that the Tbx3/miR-206 axis is relevant for breast cancer outcomes.
Tbx3 mRNA and miR-206 in human breast cancer. (A) Tbx3 mRNA expression in adjacent (Adj.) normal (n=104) and breast tumour (n=1100) samples. Box plot depicting mRNA expression levels for Tbx3 in human breast invasive carcinoma and adjacent normal breast tissue. (B) miR-206 expression in adjacent (Adj.) normal (n=103) and breast tumour (n=1101) samples. Box plot depicting expression levels for miR-206 in human breast invasive carcinoma and adjacent normal breast tissue. Correlation between Tbx3 mRNA and miR-206 expression levels in (C) adjacent normal breast samples (n=75) and (D) breast tumours (n=330), in which each dot corresponds to one patient sample. Spearman score shows significant inverse correlation between Tbx3 and miR-206 levels in both normal breast tissue and breast cancer. (E) The combination of Tbx3 and miR-206 expression is significantly associated with patient survival. Kaplan–Meier graph (n=1182) showing patients exhibiting a combination of high Tbx3 and low miR-206 expression had lower probability of survival when compared with patients exhibiting a combination of low Tbx3 and high miR-206 expression. **P<0.01 between indicated groups was calculated by Welch two-sample t-test.
miR-206 directly targets Tbx3 and decreases Tbx3 mRNA and protein expression in breast cancer cells
To examine whether Tbx3 may be a target of miR-206, we utilised the publicly available databases to determine whether the 3′ UTR of Tbx3 contains a putative binding site for miR-206. The complementarity between Tbx3 mRNA and miR-206 is shown in Figure 2A. To determine whether Tbx3 is a direct target of miR-206, a luciferase reporter assay was performed by co-transfecting HEK-293T cells with miR scrambled control (NC) or miR-206, and vectors containing either empty vector, the Tbx3 wild-type binding sequence (pMIR-TBX3-3′ UTR) or its mutated form (pMIR-TBX3-mut-3′ UTR) in the C terminus of the luciferase gene (Figure 2B). Luciferase activity was significantly decreased only when miR-206 was expressed together with the construct containing the wild-type Tbx3-3′ UTR (WT). miR-206 showed no inhibitory effect on the activity of the mutant Tbx3-3′ UTR reporter (Mut) or empty vector (V), demonstrating direct regulation of Tbx3 by miR-206 (Figure 2C). Of note, the predicted miR-206-binding site on Tbx3 is highly conserved across species (Figure 2D). Next, we examined whether miR-206 decreases Tbx3 transcript levels in human breast cancer cells. miR-206 was expressed in three different breast cancer cell lines which express endogenous Tbx3: MDA-MB-231, ZR75-1 and MCF7 cells. miR-206 expression in these cells is shown in Supplementary Figure 1. In all three cell lines, miR-206 expression led to a significant decrease in Tbx3 transcript levels (Figure 2E). To examine the impact of miR-206 on Tbx3 protein expression, miR-206 mimic was expressed in MDA-MB-231 and MCF7 cells. Our result indicated that the expression of miR-206 led to a significant decrease in Tbx3 protein expression in both cell lines (Figure 2F).
Tbx3 is a direct target of miR-206 in breast cancer cells, and restoration of miR-206 decreases Tbx3 mRNA and protein expression in breast cancer cells. (A) Putative miR-206 target site within the human Tbx3 3′ UTR as predicted by TargetScan and PicTar algorithms. (B) The miR-206 wild-type binding sequence or its mutated form was inserted into the C terminus of the luciferase gene to generate pMIR-TBX3-3′ UTR or pMIR-TBX3-mut-3′ UTR, respectively. (C) Bar plots showing relative luciferase activity in HEK-293T cells co-transfected with a luciferase reporter construct containing a fragment of the human Tbx3 3′ UTR encompassing the miR-206 binding site (Tbx3 3′ UTR) or empty vector (V), along with miR scrambled control (NC) or miR-206 for 48 h. Relative luciferase units were normalised to control β-gal reporter vector for transfection efficiency. (D) Sequence alignment showing the evolutionary conservation of the 3′ UTR of Tbx3 target site across different species. (E) Bar plots showing relative Tbx3 mRNA expression levels, in ZR75-1, MCF7 and MDA-MB-231 cells transfected with miR scrambled control (NC) or miR-206. Analysis for the Tbx3 message levels was performed by the comparative Ct method. Relative abundance was determined from the Ct values using the 2−ΔΔCt method after normalisation to GAPDH. Results are representative of at least three independent experiments. (F) Western blot analysis of MDA-MB-231 and MCF7 cells transfected with miR scrambled control (NC), miR-206 or αmiR-206 for 72 h. Beta-actin served as a loading control. Results are representative of three to five independent experiments. Means with error bars representing±s.e.m. **P<0.01; ***P<0.001.
miR-206 expression impacts Tbx3 targets
As Tbx3 has been reported to repress PTEN expression in head-and-neck cancer cells (Burgucu et al, 2012), we next examined the impact of miR-206 on PTEN expression in breast cancer cells. In both MDA-MB-231 and MCF7 cells, miR-206 expression led to a downregulation of Tbx3 and a concomitant upregulation of PTEN (Figure 3A). This increase in PTEN expression was accompanied by a decrease in phosphorylation of Akt and a decrease in the expression of the pro-survival protein, Bcl-2, suggesting that miR-206-mediated repression of Tbx3 leads to significant changes in Tbx3-regulated pathways. To investigate this further, we examined the impact of Tbx3 depletion, with siRNA, in MDA-MB-231 and ZR75-1 cells. Depletion of Tbx3 led to an upregulation of PTEN and a downregulation of phosphorylated Akt and Bcl-2, recapitulating the effects of miR-206 (Figure 3B). Similar results of Tbx3 depletion were observed in HEK-293T cells treated with four individual Tbx3 siRNAs (Supplementary Figure 2A). As the PTEN/Akt/Bcl-2 pathway is involved in survival signalling, we next examined the impact of miR-206 on breast cancer cell proliferation and apoptosis. We observed that miR-206 expression and Tbx3 depletion with siRNA resulted in a similar decrease in colony-forming ability in MDA-MB-231, ZR75-1 and MCF7 cells (Figure 3C and Supplementary Figure 2B). In addition, both miR-206 expression and Tbx3 depletion led to an increase in apoptosis in MDA-MB-231, ZR75-1 and MCF7 cells, suggesting that Tbx3 is a functionally relevant target of miR-206 (Figure 3D). The impact of miR-206 and siRNA on Tbx3 expression was in all cases, confirmed by western blot analysis. A representative blot is shown in Figure 3E. We acknowledge that regulation of targets in addition to Tbx3 may contribute to miR-206’s effects on proliferation and apoptosis. As reported by Lim et al, 2005, microRNAs have the capacity to regulate a large number of targets simultaneously. However, our rescue experiment (see below) suggests that repression of Tbx3 is required for the full anti-oncogenic activity of miR-206.
miR-206-mediated downregulation of Tbx3 impacts its targets, inhibits proliferation and promotes apoptosis. (A) Western blot analysis of MCF7 and MDA-MB-231 cells transfected with miR scrambled (NC), miR-206 or αmiR-206 for 72 h. Beta-actin was used as a protein-loading control. (B) Western blot analysis of ZR75-1 and MDA-MB-231 cells transfected with miR scrambled control (NC) or siRNA-Tbx3 (si-Tbx3). Beta-actin was used as a loading control. (C) Bar plots showing per cent colony formation of MDA-MB-231, ZR75-1 and MCF7 cells transfected with miR scrambled control (NC), miR-206, αmiR-206, siRNA-Tbx3 (si-Tbx3) or left untreated (UT). Per cent colony formation was determined with untreated set at 100%. (D) Bar plots showing relative DNA fragmentation in MDA-MB-231, ZR75-1 and MCF7 cells transfected with miR scrambled control (NC), miR-206, αmiR-206, siRNA-Tbx3 (si-Tbx3) or left untreated (UT). Untreated (UT) cells and miR scrambled control (NC) were used as controls. The results are representative of three independent experiment performed in triplicates. Means with error bars representing±s.e.m. *P<0.05; **P<0.01; ***P<0.001.
miR-206 expression and Tbx3 depletion yield similar phenotypes in breast cancer cells
We next examined the growth and morphology of MDA-MB-231 cells in 3D culture (Figure 4A). MDA-MB-231 cells are highly invasive and grow as stellate structures in 3D-Matrigel culture (Kenny et al, 2007). As expected, untreated cells, cells transfected with miR scrambled control (NC), or cells treated with antagomiR-206 (αmiR-206) formed stellate structures in Matrigel. Conversely, cells treated with miR-206 mimic or siRNA-Tbx3 (si-Tbx3) showed a striking loss of invasive colony-forming ability, branching morphology and decreased colony area (Figure 4A and B). With respect to colony area, similar results were observed in MCF7 (top panel) and ZR75-1 (bottom panel) cells grown in 3D culture (Supplementary Figure 3).
miR-206 expression and Tbx3 knock-down inhibits the 3D growth of breast cancer cells in Matrigel. Tbx3 lacking the 3′ UTR is not repressed by miR-206. (A) Phase-contrast images of MDA-MB-231 cells in 3D culture treated with miR-206, αmiR-206 or siRNA-Tbx3 (si-Tbx3). Untreated (UT) and miR scrambled control (NC) treated cells were used as controls. (B) Bar plots represent the quantification of the number of invasive colonies, mean colony area and number of branching stellate cells. (C) Western blot analysis of MDA-MB-231 cells co-transfected with miR scrambled control with either empty vector (NC+V) or with flag-tagged TBX3 expression vector (NC+TBX3), and with miR-206 with empty vector (miR-206+V) or with flag-tagged TBX3 expression vector (miR-206+TBX3). Beta-actin was used as a loading control. (D) Phase-contrast images of MDA-MB-231 cells in 3D cultures and co-transfected with miR scrambled control (NC) or miR-206 with either empty vector (V) control or TBX3 expression vector (TBX3). (E) Bar plots showing the reduction in invasive phenotype of MDA-MB-231 cells as quantified by counting invasive colonies, mean colony area and the number of branching stellate cells. The results are representatives of three independent experiments. Scale bar: × 4 images, 200 μm; × 10 images, 50 μm. Means with error bars representing±s.e.m. *P<0.05; **P<0.01; ***P<0.001.
Re-expression of Tbx3 reverses the effects of miR-206
Our results suggest that Tbx3 is a functionally relevant target of miR-206. To further explore the interaction between Tbx3 and miR-206, we performed a ‘rescue’ experiment. We first examined whether the expression of Tbx3, driven by a cDNA lacking the 3′ UTR, was downregulated by miR-206. Flag-tagged TBX3 was expressed with either miR scramble control (NC) or miR-206 mimic in MDA-MB-231 cells, and total cell lysates were blotted with anti-flag and anti-Tbx3 antibodies. Beta-actin was used as a loading control (Figure 4C). As expected, miR-206 had no effect on the expression of Tbx3 lacking a 3′ UTR/miR-206-binding site (Flag-Tbx3, compare lanes 3 and 4). However, endogenous Tbx3 expression was suppressed by miR-206, as expected (Tbx3, compare lanes 1 and 2). Importantly, ectopic expression of TBX3 rescued Tbx3 expression in the face of miR-206, allowing us to address whether Tbx3 repression is necessary for miR-206-mediated effects on MDA-MB-231 cell morphology (Figure 4D). As expected, MDA-MB-231 cells transfected with miR scramble control and vector control (NC+V) formed stellate structures in 3D-Matrigel culture (Figure 4D, upper left). MDA-MB-231 cells transfected with miR scramble control and ectopic TBX3 (Figure 4D, NC+TBX3, upper right) also formed stellate structures. In addition, colony invasion, branching and area were augmented by ectopic TBX3 expression (all results for this figure are quantified in Figure 4E). Conversely, cells transfected with miR-206 mimic and vector control (miR-206+V) demonstrated a loss of invasive morphology, as well as decreased branching and colony area (Figure 4D, bottom left). However, cells expressing miR-206 in the face of ectopic TBX3 (no Tbx3 repression) retained the characteristics of control cells (Figure 4D, miR-206+TBX3, lower right) and formed stellate structures with robust invasion. These results demonstrate that Tbx3 repression is an essential mechanism by which miR-206 inhibits the colony size, stellate branching and invasiveness of breast cancer cells. With respect to colony area, similar results were observed in a rescue experiment performed in non-invasive MCF7 cells (Supplementary Figure 4).
As Tbx3 has been implicated in regulation of the breast cancer stem cell populations (Fillmore et al, 2010), we next examined the impact of Tbx3 manipulation on tumorsphere formation. Sphere formation is a well-accepted functional assay for the quantification of tumour-initiating cells, also known as cancer stem cells (Dontu et al, 2003; Liao et al, 2007; Liu et al, 2007; Mani et al, 2008; Cicalese et al, 2009; Kondratyev et al, 2012). Both miR-206 and siRNA-Tbx3 (si-Tbx3) led to a decrease in tumorsphere formation in MCF7 (Figure 5A and B) and MDA-MB-231 cells (Figure 5D and E). However, cells expressing miR-206 in the face of ectopic TBX3 (Flag-TBX3 cDNA, as described for Figure 4) retain the sphere-forming ability of control cells. Furthermore, western blot analysis of MCF7 and MDA-MB-231 cells indicated that endogenous Tbx3 expression is decreased upon miR-206 expression, but ectopic expression of TBX3 restores Tbx3 expression, in the face of miR-206 expression (Figure 5C and F). These results demonstrate that Tbx3 repression is an essential mechanism by which miR-206 limits the breast cancer stem cell population.
miR-206 exerts a tumour suppressive function and inhibits tumorsphere formation of breast cancer cells. Phase-contrast × 4 images of tumorspheres of (A) MCF7 and (B) MDA-MB-231 cells reverse-transfected with miR scrambled control (NC), miR-206, αmiR-206, siRNA-Tbx3 (si-Tbx3), miR-206 plus empty vector (miR-206+V) or miR-206 plus TBX3 expression vector (miR-206+TBX3). Bar plots showing the relative sphere number of (B) MCF7 and (E) MDA-MB-231 cells corresponding to A and B, respectively. Relative sphere number was normalised to miR scrambled control (NC). Images are representative of spheres under the mentioned treatments at the secondary sphere stage. Western blot analysis of (C) MCF7 and (F) MDA-MB-231 cells co-transfected with miR scrambled control (NC), miR-206, αmiR-206, siRNA-Tbx3 (si-Tbx3), miR-206 plus empty vector (miR-206+V) or miR-206 plus TBX3 expression vector (miR-206+TBX3). Beta-actin was used a loading control. Scale bar: 200 μm. Means with error bars representing±s.e.m. *P<0.05; **P<0.01.
Discussion
Despite their profound role in influencing cellular fate, transcription factors have historically been overlooked as ‘druggable’ targets because they lack enzymatic activity (Kondratyev et al, 2012). Recent strategies under development for targeting oncogenic transcription factors include complex decoy oligodeoxynucleotides (Yan and Higgins, 2013), ‘stapled’ peptides and siRNA delivery (Wang et al, 2013). MicroRNA-based strategies are especially promising as microRNA mimics and antagomiRs are not thought to have insurmountable delivery hurdles. Furthermore, microRNAs are known to target entire pathways rather than single components of a pathway, providing the advantage of simultaneous regulation of multiple key players in a pathway. While the exquisite selectivity of targeted therapies was once considered a strength, this selectivity also enables rapid adaptation and the emergence of therapeutic resistance (Bader et al, 2010). Of note, microRNA-based therapies are rapidly entering clinical translation (Redmond and Carroll, 2009; https://clinicaltrials.gov/).
Although microRNA-based therapies hold great promise, genetic alterations in cancer may enable tumours to evade such therapies. For example, single-nucleotide polymorphisms (SNPs) and both somatic and germline mutations have been identified in microRNAs and their targets, altering their functional interaction (Ziebarth et al, 2012; Hydbring and Badalian-Very, 2013; Mulrane et al, 2013). There are 97 annotated SNPs (60 validated) that occur in the 3′ UTR of TBX3 (http://www.ncbi.nlm.nih.gov/snp). Using the miRNASNP database (Gong et al, 2015), we identified a SNP (rs141350926A->U) in the 3′ UTR of TBX3, which is predicted to disrupt miR-206 binding (Supplementary Figure 5). The functional consequences of that SNP will require further study. Besides SNPs/mutations, other mechanisms that disrupt regulation by microRNAs are known. For example, selective loss of the 3′ UTR has been reported for the Cyclin D1 mRNA in prostate cancer cells, rendering it insensitive to miR-206-mediated regulation (Elliman et al, 2014). Alterations in the machinery responsible for processing miRs have also been reported in cancer, with loss of Dicer expression linked to breast cancer recurrence (Sun et al, 2009; Dedes et al, 2011; Yan et al, 2012). Competing endogenous RNAs (ceRNAs, Khoshnaw et al, 2012) may also factor into the efficacy of miR-based therapies, adding another layer of complexity to RNA crosstalk (Mulrane et al, 2013). ceRNAs contain multiple binding sites for microRNAs and act as ‘endogenous decoys’ for microRNAs (Salmena et al, 2011). A well-known example is found in the tumour suppressor PTEN. PTEN expression was shown to be modulated by protein-coding mRNAs, which were revealed to have the unexpected ability to sequester PTEN-regulating microRNAs (Tay et al, 2011).
In this study, we demonstrate that the Tbx3 oncogene is directly regulated by the tumour suppressor microRNA miR-206. Tbx3 is known to be over-expressed in breast cancer (Yarosh et al, 2008), whereas miR-206 is downregulated in breast cancer (Kondo et al, 2008; Li et al, 2013; Zhou et al, 2013). Recent reports have indicated that over-expression of Tbx3 is correlated with epithelial to mesenchymal transition in colorectal cancer (Wang et al, 2015) and is a predictor of poor prognosis in colorectal and pancreatic carcinoma (Shan et al, 2014; Wang et al, 2015). Our work demonstrates an important functional relationship between Tbx3 and miR-206. Our findings also suggest that miR-206 dysregulation may contribute to Tbx3 over-expression. We demonstrate that miR-206 directly targets Tbx3, that miR-206 and Tbx3 mRNA are inversely correlated in human breast cancer and that Tbx3 repression is necessary to elicit a significant functional effect of miR-206 in breast cancer cells.
Notably, miR-206 is unable to restrict the cancer stem cell population and inhibit the invasive growth of breast cancer cells in the face of ectopic Tbx3 expression. miR-206 was first identified as a skeletal muscle-specific microRNA (McCarthy, 2008) and its re-expression in rhabdomyosarcoma cells was shown to promote myogenic differentiation and inhibit tumour growth (Mishra and Merlino, 2009; Taulli et al, 2009; Yan et al, 2009). miR-206 has also been shown to have growth inhibitory properties in other tumour types such as lung cancer (Wang et al, 2011; Chen et al, 2015), endometroid adenocarcinoma (Chen et al, 2012), gastric cancer (Yang et al, 2013; Zhang et al, 2013; Ren et al, 2014), melanoma (Georgantas et al, 2014), ovarian cancer (Li et al, 2014), laryngeal squamous cell carcinoma (Zhang et al, 2011), oral squamous cell carcinoma (Lin et al, 2014) and thyroid cancer (Zhang et al, 2015) demonstrating its potentially wide utility as a cancer therapeutic. Of note, we report that patients exhibiting a combination of high Tbx3 and low miR-206 expression have a lower probability of disease-free survival, indicating that the Tbx3/miR-206 axis has an important role in breast cancer recurrence and patient survival. It is well established that recurrence and relapse is a major obstacle in effective breast cancer treatment as early failure results in worse outcomes and fewer therapeutic options. Identification of miR-206/Tbx3 status may have prognostic potential. Essential to moving miR-206 from ‘bench to bedside’ is a comprehensive understanding of its targets. Our work identifies Tbx3 as a functionally important target of miR-206 in breast cancer.
Change history
10 May 2016
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Adams BD, Furneaux H, White BA (2007) The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol 21 (5): 1132–1147.
Alteri A, De Vito F, Messina G, Pompili M, Calconi A, Visca P, Mottolese M, Presutti C, Grossi M (2013) Cyclin D1 is a major target of miR-206 in cell differentiation and transformation. Cell Cycle 12 (24): 3781–3790.
Bader AG, Brown D, Winkler M (2010) The promise of microRNA replacement therapy. Cancer Res 70 (18): 7027–7030.
Brummelkamp TR, Kortlever RM, Lingbeek M, Trettel F, MacDonald ME, van Lohuizen M, Bernards R (2002) TBX-3, the gene mutated in Ulnar-Mammary Syndrome, is a negative regulator of p19ARF and inhibits senescence. J Biol Chem 277 (8): 6567–6572.
Burgucu D, Guney K, Sahinturk D, Ozbudak IH, Ozel D, Ozbilim G, Yavuzer U (2012) Tbx3 represses PTEN and is over-expressed in head and neck squamous cell carcinoma. BMC Cancer 12: 481.
Chen QY, Jiao DM, Yan L, Wu YQ, Hu HZ, Song J, Yan J, Wu LJ, Xu LQ, Shi JG (2015) Comprehensive gene and microRNA expression profiling reveals miR-206 inhibits MET in lung cancer metastasis. Mol Biosyst 11 (8): 2290–2302.
Chen X, Yan Q, Li S, Zhou L, Yang H, Yang Y, Liu X, Wan X (2012) Expression of the tumor suppressor miR-206 is associated with cellular proliferative inhibition and impairs invasion in ERα-positive endometrioid adenocarcinoma. Cancer Lett 314 (1): 41–53.
Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, Giulini B, Brisken C, Minucci S, Di Fiore PP, Pelicci PG (2009) The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell 138 (6): 1083–1095.
Debnath J, Muthuswamy SK, Brugge JS (2003) Morphogenesis and Oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30: 256–268.
Dedes KJ, Natrajan R, Lambros MB, Geyer FC, Lopez-Garcia MA, Savage K, Jones RL, Reis-Filho JS (2011) Down-regulation of the miRNA master regulators Drosha and Dicer is associated with specific subgroups of breast cancer. Eur J Cancer 47 (1): 138–150.
Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS (2003) In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev 17 (10): 1253–1270.
Elliman SJ, Howley BV, Mehta DS, Fearnhead HO, Kemp DM, Barkley LR (2014) Selective repression of the oncogene cyclin D1 by the tumor suppressor miR-206 in cancers. Oncogenesis 3: e113.
Fan W, Huang X, Chen C, Gray J, Huang T (2004) TBX3 and its isoform TBX3+2a are functionally distinctive in inhibition of senescence and are overexpressed in a subset of breast cancer cell lines. Cancer Res 64 (15): 5132–5139.
Fillmore CM, Gupta PB, Rudnick JA, Caballero S, Keller PJ, Lander ES, Kuperwasser C (2010) Estrogen expands breast cancer stem-like cells through paracrine FGF/Tbx3 signaling. Proc Natl Acad Sci USA 107 (50): 21737–21742.
Fu Y, Shao ZM, He QZ, Jiang BQ, Wu Y, Zhuang ZG (2015) Hsa-miR-206 represses the proliferation and invasion of breast cancer cells by targeting Cx43. Eur Rev Med Pharmacol Sci 19 (11): 2091–2104.
Georgantas RW 3rd, Streicher K, Luo X, Greenlees L, Zhu W, Liu Z, Brohawan P, Morehouse C, Higgs BW, Richman L, Jallal B, Yao Y, Ranade K (2014) MicroRNA-206 induces G1 arrest in melanoma by inhibition of CDK4 and Cyclin D. Pigment Cell Melanoma Res 27 (2): 275–286.
Gong J, Liu C, Liu W, Wu Y, Ma Z, Chen H, Guo AY (2015) An update of miRNASNP database for better SNP selection by GWAS data, miRNA expression and online tools. Database 2015: bav029.
Hydbring P, Badalian-Very G (2013) Clinical applications of microRNAs. F1000Research 2: 136.
Jiang K, Ren C, Nair VD (2013) MicroRNA-137 represses Klf4 and Tbx3 during differentiation of mouse embryonic stem cells. Stem Cell Res 11 (3): 1299–1313.
Keklikoglou I, Hosaka K, Bender C, Bott A, Koerner C, Mitra D, Will R, Woerner A, Muenstermann E, Wilhelm H, Cao Y, Wiemann S (2015) MicroRNA-206 functions as a pleiotropic modulator of cell proliferation, invasion and lymphangiogenesis in pancreatic adenocarcinoma by targeting ANXA2 and KRAS genes. Oncogene 34 (37): 4867–4878.
Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Peterson OW, Gray JW, Bissell MJ (2007) The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol 1 (1): 84–96.
Khoshnaw SM, Rakha EA, Abdel-Fatah TM, Nolan CC, Hodi Z, Macmillan DR, Ellis IO, Green AR (2012) Loss of Dicer expression is associated with breast cancer progression and recurrence. Breast Cancer Res Treat 135 (2): 403–413.
Kondo N, Toyama T, Sugiura H, Fujii Y, Yamashita H (2008) miR-206 Expression is down-regulated in estrogen receptor alpha-positive human breast cancer. Cancer Res 68 (13): 5004–5008.
Kondratyev M, Kreso A, Hallett RM, Girgis-Gabardo A, Barcelon ME, Ilieva D, Ware C, Majumder PK, Hassell JA (2012) Gamma-secretase inhibitors target tumor-initiating cells in a mouse model of ERBB2 breast cancer. Oncogene 31 (1): 93–103.
Lee MJ, Yoon KS, Cho KW, Kim KS, Jung HS. (2013) Expression of miR-206 during the initiation of mammary gland development. Cell Tissue Res 353 (3): 425–433.
Li J, Weinberg MS, Zerbini L, Prince S (2013) The oncogenic TBX3 is a downstream target and mediator of the TGF-beta1 signaling pathway. Mol Biol Cell 24 (22): 3569–3576.
Li S, Li Y, Wen Z, Kong F, Guan X, Liu W (2014) microRNA-206 overexpression inhibits cellular proliferation and invasion of estrogen receptor alpha-positive ovarian cancer cells. Mol Med Rep 9 (5): 1703–1708.
Li Y, Hong F, Yu Z (2013) Decreased expression of microRNA-206 in breast cancer and its association with disease characteristics and patient survival. J Int Med Res 41 (3): 596–602.
Liao MJ, Zhang CC, Zhou B, Zimonjic DB, Mani SA, Kaba M, Gifford A, Reinhardt F, Popescu NC, Guo W, Eaton EN, Lodish HF, Weinberg RA (2007) Enrichment of a population of mammary gland cells that form mammospheres and have in vivo repopulating activity. Cancer Res 67 (17): 8131–8138.
Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433 (7027): 769–773.
Lin CC, Sharma SB, Farrugia MK, McLaughlin SL, Ice RJ, Loskutov YV, Pugacheva EN, Brundage KM, Chen D, Ruppert JM (2015) Kruppel-like factor 4 signals through microRNA-206 to promote tumor initiation and cell survival. Oncogenesis 8 (4): e155.
Lin F, Yao L, Xiao J, Liu D, Ni Z (2014) MiR-206 functions as a tumor suppressor and directly targets K-Ras in human oral squamous cell carcinoma. Onco Targets Ther 7: 1583–1591.
Liu J, Esmailpour T, Shang X, Gulsen G, Liu A, Huang T (2011) TBX3 over-expression causes mammary gland hyperplasia and increases mammary stem-like cells in an inducible transgenic mouse model. BMC Dev Biol 11: 65.
Liu JC, Deng T, Lehal RS, Kim J, Zacksenhaus E (2007) Identification of tumorsphere- and tumor-initiating cells in HER2/Neu-induced mammary tumors. Cancer Res 67 (18): 8671–8681.
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA (2008) The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133 (4): 704–715.
McCarthy JJ (2008) MicroRNA-206: the skeletal muscle-specific myomiR. Biochim Biophys Acta 1779 (11): 682–691.
Mishra PJ, Merlino G (2009) MicroRNA reexpression as differentiation therapy in cancer. J Clin Invest 119 (8): 2119–2123.
Moses H, Barcellos-Hoff MH (2011) TGF-beta biology in mammary development and breast cancer. Cold Spring Harb Perspect Biol 3 (1): a003277.
Mulrane L, McGee SF, Gallagher WM, O’Connor DP (2013) miRNA dysregulation in breast cancer. Cancer Res 73 (22): 6554–6662.
Peres J, Davis E, Mowla S, Bennett DC, Li JA, Wansleben S, Prince S (2010) The highly homologous T-box transcription factors, TBX2 and TBX3, have distinct roles in the oncogenic process. Genes Cancer 1 (3): 272–282.
Redmond AM, Carroll JS (2009) Defining and targeting transcription factors in cancer. Genome Biol 10 (7): 311.
Ren J, Huang HJ, Gong Y, Yue S, Tang LM, Cheng SY (2014) MicroRNA-206 suppresses gastric cancer cell growth and metastasis. Cell Biosci 4: 26.
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146 (3): 353–358.
Shan ZZ, Yan XB, Yan LL, Tian Y, Meng QC, Qiu WW, Zhang Z, Jin ZM (2014) Overexpression of Tbx3 is correlated with Epithelial-Mesenchymal Transition phenotype and predicts poor prognosis of colorectal cancer. Am J Cancer Res 5 (1): 344–353.
Song G, Zhang Y, Wang L (2009) MicroRNA-206 targets notch3, activates apoptosis, and inhibits tumor cell migration and focus formation. J Biol Chem 284 (46): 31921–31927.
Sun G, Yan J, Noltner K, Feng J, Li H, Sarkis DA, Sommer SS, Rossi JJ (2009) SNPs in human miRNA genes affect biogenesis and function. RNA 15 (9): 1640–1651.
Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M, Tuschl T, Ponzetto C (2009) The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest 119 (8): 2366–2378.
Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, Lieberman J, Rigoutsos I, Pandolfi PP (2011) Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147 (2): 344–357.
Wang HC, Meng QC, Shan ZZ, Yuan Z, Huang XY (2015) Overexpression of Tbx3 predicts poor prognosis of patients with resectable pancreatic carcinoma. Asian Pac J Cancer Prev 16 (4): 1397–1401.
Wang J, Bai Y, Li N, Ye W, Zhang M, Greene SB, Tao Y, Chen Y, Wehrens XH, Martin JF (2014) Pitx2-microRNA pathway that delimits sinoatrial node development and inhibits predisposition to atrial fibrillation. Proc Natl Acad Sci USA 111 (25): 9181–9186.
Wang J, Tsouko E, Jonsson P, Bergh J, Hartman J, Aydogdu E, Williams C (2014) miR-206 inhibits cell migration through direct targeting of the actin-binding protein Coronin 1C in triple-negative breast cancer. Mol Oncol 8 (8): 1690–1702.
Wang X, Ling C, Bai Y, Zhao J (2011) MicroRNA-206 is associated with invasion and metastasis of lung cancer. Anat Rec (Hoboken) 294 (1): 88–92.
Wang X, Liu Q, Hou B, Zhang W, Yan M, Jia H, Li H, Yan D, Zheng F, Ding W, Yi C, Hai Wang (2013) Concomitant targeting of multiple key transcription factors effectively disrupts cancer stem cells enriched in side population of human pancreatic cancer cells. PloS One 8 (9): e73942.
Yan C, Higgins PJ (2013) Drugging the undruggable: transcription therapy for cancer. Biochim Biophys Acta 1835 (1): 76–85.
Yan D, Dong Xda E, Chen X, Wang L, Lu C, Wang J, Qu J, Tu L (2009) MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J Biol Chem 284 (43): 29596–29604.
Yan M, Huang HY, Wang T, Wan Y, Cui SD, Liu ZZ, Fan QX (2012) Dysregulated expression of dicer and drosha in breast cancer. Pathol Oncol Res 18 (2): 343–348.
Yang Q, Zhang C, Huang B, Li H, Zhang R, Huang Y, Wang J (2013) Downregulation of microRNA-206 is a potent prognostic marker for patients with gastric cancer. Eur J Gastroenterol Hepatol 25 (8): 953–957.
Yarosh W, Barrientos T, Esmailpour T, Lin L, Carpenter PM, Osann K, Anton-Culver H, Huang T (2008) TBX3 is overexpressed in breast cancer and represses p14 ARF by interacting with histone deacetylases. Cancer Res 68 (3): 693–6999.
Zhang L, Liu X, Jin H, Guo X, Xia L, Chen Z, Bai M, Liu J, Shang X, Wu K, Pan Y, Fan D (2013) miR-206 inhibits gastric cancer proliferation in part by repressing cyclinD2. Cancer Lett 332 (1): 94–101.
Zhang T, Liu M, Wang C, Lin C, Sun Y, Jin D (2011) Down-regulation of MiR-206 promotes proliferation and invasion of laryngeal cancer by regulating VEGF expression. Anticancer Res 31 (11): 3859–3863.
Zhang WL, Lv W, Sun SZ, Wu XZ, Zhang JH (2015) miR-206 inhibits metastasis-relevant traits by degrading MRTF-A in anaplastic thyroid cancer. Int J Oncol 47 (1): 133–142.
Zhou J, Tian Y, Li J, Lu B, Sun M, Zou Y, Kong R, Luo Y, Shi Y, Wang K, Ji G (2013) miR-206 is down-regulated in breast cancer and inhibits cell proliferation through the up-regulation of cyclinD2. Biochem Biophys Res Commun 433 (2): 207–212.
Ziebarth JD, Bhattacharya A, Cui Y (2012) Integrative analysis of somatic mutations altering microRNA targeting in cancer genomes. PloS One 7 (10): e47137.
Acknowledgements
This work was supported by the University of California Cancer Research Coordinating Committee grant 3-444952-36240 (Sweeney) and CA166412 (Carraway). M Umeh-Garcia is supported by T32 GM007377.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Amir, S., Simion, C., Umeh-Garcia, M. et al. Regulation of the T-box transcription factor Tbx3 by the tumour suppressor microRNA-206 in breast cancer. Br J Cancer 114, 1125–1134 (2016). https://doi.org/10.1038/bjc.2016.73
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2016.73
- Springer Nature Limited
Keywords
This article is cited by
-
MicroRNA-1 suppresses proliferation, migration and invasion by targeting Notch2 in esophageal squamous cell carcinoma
Scientific Reports (2018)
-
Novel HDAC5-interacting motifs of Tbx3 are essential for the suppression of E-cadherin expression and for the promotion of metastasis in hepatocellular carcinoma
Signal Transduction and Targeted Therapy (2018)
-
Overexpression of suppressive microRNAs, miR-30a and miR-200c are associated with improved survival of breast cancer patients
Scientific Reports (2017)