Abstract
Background:
Our previous study found that dysregulated microRNA-146a-5p (miR-146a-5p) is involved in oesophageal squamous cell cancer (ESCC) proliferation. This article aimed to evaluate its detailed mechanisms in ESCC epithelial–mesenchymal transition (EMT) progression.
Methods:
Invasion assay, qRT-PCR and western blotting were used to validate the roles of miR-146a-5p and Notch2 in EMT progression. miRNA target gene prediction databases and dual-luciferase reporter assay were used to validate the target gene.
Results:
miR-146a-5p inhibitor led to increase of invaded ESCC cells, while miR-146a-5p mimics inhibited invasion ability of ESCC cells. Protein level of E-cadherin decreased, whereas those of Snail and Vimentin increased in the anti-miR-146a-5p group, which demonstrated that miR-146a-5p inhibits EMT progression of ESCC cells. miRNA target gene prediction databases indicated the potential of Notch2 as a direct target gene of miR-146a-5p and dual-luciferase reporter assay validated it. Importantly, shRNA-Notch2 restrained EMT and partially abrogated the inhibiting effects of miR-146a-5p on EMT progression of ESCC cells.
Conclusions:
miR-146a-5p functions as a tumour-suppressive miRNA targeting Notch2 and inhibits the EMT progression of ESCC.
Similar content being viewed by others
Main
Oesophageal cancer is one of the lethal cancers worldwide (Siegel et al, 2015). Oesophageal squamous cell cancer (ESCC) accounts for the majority of oesophageal cancer cases worldwide and is predominant in Middle East and central and eastern Asia (Chen et al, 2016). Although the medical technology has improved a lot in recent years, the prognoses of ESCC patients remain to be poor, ranging from 15% to 25%. Epithelial–mesenchymal transition (EMT) is an evolutionarily conserved development process during which epithelial cells lose polarity and develop a mesenchymal phenotype. EMT progression triggers the dissociation of carcinoma cells from primary carcinomas, which subsequently migrate and disseminate to distant sites (Nieto et al, 2016). This progression can be triggered by many signaling pathways, including Notch (Chen et al, 2010), transforming growth factor-β (Chen et al, 2016), epidermal growth factor (Liu et al, 2015), fibroblast growth factor (Du et al, 2015) and PLC-γ (Ji et al, 2015) pathways.
MicroRNAs (miRNAs) are small non-coding regulatory RNA molecules. They could bind to and cleave their target mRNAs or inhibit their translation into proteins. In this way, miRNAs inhibit the expression of specific target genes (Pichler and Calin, 2015). Accumulating evidence has shown that miRNAs can participate in tumourigenesis, progression and metastasis either as oncogenes or tumour suppressors and have potential as diagnostic, prognostic and therapeutics factors in cancer medicine (Sita-Lumsden et al, 2013; Krzeszinski et al, 2014; Wang et al, 2016a).
MiRNA-146a-5p has been widely explored in various cancers. Decreased expression level of miR-146a-5p is found in pancreatic cancer cells (Li et al, 2010), lung (Chen et al, 2013) and gastric carcinomas (Yao et al, 2013). On the other hand, it is upregulated in melanoma (Forloni et al, 2014), squamous cell carcinoma of the cervix (Wang et al, 2008) and thyroid carcinoma (Sun et al, 2015a). In our previous article, we found that miR-146a-5p level was significantly decreased in ESCC tissue and serum and it could act as a promising biomarker for the prognosis and diagnosis of ESCC (Wang et al, 2016a). In this study, we further investigated the mechanisms of miR-146a-5p in EMT progression of ESCC.
Materials and methods
Culture of ESCC cell lines and antibodies
Human ESCC cell lines (Eca109 and EC9706) were provided by Dr. Jiandong Zhang, Qianfoshan Hospital affiliated to Shandong University of China. Both cell lines were cultured in RPMI 1640 (Gibco BRL, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (FBS, Gibco), 100 U ml−1 penicillin G and streptomycin in a 37 °C incubator with humidified atmosphere and 5% CO2.
Anti-Notch2 rabbit polyclonal antibody (ab8926), anti-Snail rabbit monoclonal antibody (ab167609), anti-Vimentin rabbit monoclonal antibody (ab7752), anti-E-cadherin rabbit polyclonal antibody (ab15148) and anti-β-actin mouse monoclonal antibody (ab6276) were purchased from Abcam company (Cambridge, MA, USA).
Quantitative real-time PCR (qRT-PCR)
For Notch2. Total RNA was extracted from cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Complementary DNA (cDNA) was generated using the qPCR-RT Kit (Toyobo, Osaka, Japan). Real-time PCR was carried out on the Bio-Rad Single Color Real-Time PCR system (Bio-Rad, Hercules, CA, USA) using gene-specific primers: for Notch2, sense primer: 5′-GGGACCCTGTCATACCCTCT-3′, and anti-sense primer: 5′-GAGCCATGCTTACGCTTTCG-3′; and for β-actin, sense primer: 5′-CAAAGGCCAACAGAGAGAAGAT-3′ and anti-sense primer: 5′-TGAGACACACCATCACCAGAAT-3′ (Sangon Biotech, Shanghai, China). PCR reaction conditions were set as follows: 95 °C for 1 min and 40 cycles of 95 °C for 15 s, 58 °C for 15 s and 72 °C for 45 s. Notch2 expression was calculated according to the mathematical model R=2−ΔΔCq, where ΔCq=CqNotch2−Cqβ-actin, and ΔΔCq=ΔCqtest−ΔCqcontrol.
For miR-146a-5p. Total miRNA was extracted from cells using the miRNeasy Mini Kit (QIAGEN, Duesseldorf, Nordrhein-Westfalen, German) in accordance with the manufacturer’s protocol. cDNA synthesis was performed using an All-in-One TM miRNA qRT-PCR Detection Kit (GeneCopoeia, Rockville, MD, USA). The RT reaction system (25 μl) comprised 1 μl of 2.5 U μl−1 Poly A Polymerase, 1 μl of RTase Mix, 5 μl of 5 × PAP/RT buffer, 2 μg of total RNA templates and RNase/DNase-free ddH2O. The reaction conditions were carried out as follows: 37 °C for 60 min and 85 °C for 5 min. The reaction system (20 μl) for qPCR comprised 10 μl of 2 × qPCR Mix, 2 μl of First-strand cDNA (diluted 1:5), 2 μl of Universal Adaptor PCR Primer, 2 μl of All-in-OneTM miRNA qPCR Primer (miR-146a-5p: HmiRQP0196; U6: HmiRQP9001, GeneCopoeia) and 4 μl of ddH2O. Amplification was performed in a Bio-Rad Single Color Real-Time PCR system (Bio-Rad, Hercules, CA, USA) under the following reaction conditions: 95 °C for 10 min and 40 cycles of 95 °C for 10 s, 60 °C for 20 s and 72 °C for 10 s. RNU6 was used as an endogenous control for normalisation of the data. Relative quantification of miR-146a-5p expression was calculated with 2−ΔΔCq method. All RT-PCRs were performed in triplicate, and the data are presented as the mean±s.d.
Protein extraction and western blotting
RIPA buffer was used for total protein extraction. Prepared protein samples were separated by 10% SDS polyacrylamide gels and transferred onto polyvinylidene fluoride membranes. The membrane was blocked with 5% skimmed milk in TBST and incubated with the appropriate antibody (anti-Notch2, 1 : 500; anti-Snail, 1 : 1000; anti-Vimentin, 1 : 1000; anti-E-cadherin, 1 : 500). Then horseradish peroxidase-conjugated secondary antibodies (1 : 5000) were added. Bands were subsequently visualised using a chemiluminescence detection system (EMD Millipore, Billerica, MA, USA) and the density was determined using ImageJ software (US National Institutes of Health, Bethesda, MD, USA). β-Actin (1 : 5000) was used as a loading control.
Transfection
The EC 9706 and Eca 109 cells were transfected with specific Notch2 shRNA (sequence: CGGTGTACCATTGACATTG; Genechem, Shanghai, China) for 72 h using Lipofectamine reagent in serum-free 1640 medium according to the manufacturer’s instruction. The multiplicity of infection was 20 for Eca 109 and 40 for EC 9706 cells. Untreated cells were used as a negative control. The efficacy of transfection was tested by qRT-PCR and western blotting.
For the manual alteration of miR-146a-5p expression, miRNA mimics (sense: UGAGAACUGAAUUCCAUGGGUU; antisense: CCCAUGGAAUUCAGUUCUCAUU) and miRNA inhibitor (AACCCAUGGAAUUCAGUUCUCA) and miRNA-NC (sense: UUCUCCGAACGUGUCACGUdTdT; antisense: ACGUGACACGUUCGGAGAAdTdT) (Genechem) were transfected into EC 9706 and Eca 109 cells using Lipofectamine 2000 Reagent (Invitrogen) according to the manufacturer’s instructions.
Dual-luciferase reporter assay
EC 9706 and Eca 109 cells were transfected with luciferase vectors (a luciferase vector containing the wild-type target gene’s 3′-UTR and a luciferase vector containing the mutant-type target gene’s 3′-UTR) for Notch2 (NCBI Reference Sequence: NM_024408.3; 3′UTR region: 7714–11474) together with miR-146a-5p mimics or inhibitor via Lipofectamine 2000 Reagent (Invitrogen). After 72 h, the luciferase activity was measured by using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). Data were presented as the ratios between the firefly and Renilla fluorescence activities.
Invasion assay. Invasion ability of ESCC cells was measured by the number of cells invading through matrigel-coated transwell inserts (Corning, Corning, New York, USA). Briefly, transwell inserts with 8-mm pores were coated with matrigel (matrigel/DMEM=8 : 1; 60 μl per well; BD Bioscience, Franklin Lakes, NJ, USA). EC 9706 or Eca109 cells after transfection were seeded at a density of 1 × 105 per well in the upper chambers with 250 μl of 1640 medium supplemented without FBS. In all, 700 μl 1640 medium with 20% FBS were added to the 24-well plate. After 48 h of incubation, cells that had invaded to lower surface of the matrigel-coated membrane were fixed with methanol, stained with 0.1% crystal violet and counted in five randomly selected fields under a light microscope (Olympus BX51, Olympus, Tokyo, Japan).
Statistical analysis
The quantitative data are expressed as mean±s.d. Comparison between two or more groups was subjected to a two-tailed Student’s t-test or ANOVA when appropriate. Differences were considered significant for P-values <0.05. All statistical analyses were performed with the SPSS 17.0 statistical software (SPSS Inc., Chicago, IL, USA) and the GraphPad Prism 5 software (GraphPad Software Inc, San Diego, CA, USA).
Results
miR-146a-5p inhibits EMT process
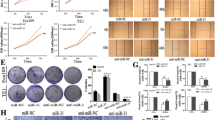
Expression level of miR-146a-5p was downregulated in ESCC tissues compared with the adjacent normal tissues and it could predict overall survival (OS) and progression-free survival (PFS) of ESCC patients, which has been reported in our previous article (Wang et al, 2016a). The invasiveness of ESCC cells (EC 9706 and Eca 109) were examined after transfection. In the miR-146a-5p mimics group, miR-146a-5p was overexpressed (Figure 1A) and the invasiveness of ESCC decreased significantly (both P<0.001) (Figure 1B). On the contrary, knockdown of miR-146a-5p via miRNA-inhibitor transfection (Figure 1A) evidently promoted invasion of ESCC cells (both P<0.001) (Figure 1C). Expression level of E-cadherin was upregulated (P=0.002 for EC 9706, P<0.001 for Eca 109) while Snail (P=0.004 for EC 9706, P<0.001 for Eca 109) and Vimentin were downregulated (P=0.003 for EC 9706, P<0.001 for Eca 109) in the miR-146a-5p mimics group by western blotting. On the other hand, after inhibition of miR-146a-5p, the protein level of E-cadherin was reduced (P=0.007 for EC 9706, P<0.001 for Eca 109) while Snail (P=0.005 for EC 9706, P=0.017 for Eca 109) and Vimentin (P<0.001 for EC 9706 and Eca 109) were increased (Figure 1D and E).
miR-146a-5p regulates EMT of ESCC. (A) The relative expression levels of miR-146a-5p after transfection. (B, C) The effects of miR-146a-5p mimics and inhibitor on invasion ability of ESCC. (D, E) miR-146a-5p mimics and inhibitor led to the changes of EMT markers’ expression. *P<0.05; **P<0.01; ***P<0.001.
Notch2 is a direct target of miR-146a-5p in ESCC
To investigate the target gene of miR-146a-5p in the EMT process, we first performed a miRNA target gene prediction with miRwalk, miRmap, miRanda, miRBase and Targetscan databases. We found that Notch2 exhibits miR-146a-5p-binding sequences in its 3′-UTR regions (nucleotides 1884–1891, Figure 2A). Luciferase assays were performed to obtain direct evidence that Notch2 is a target of miR-146a-5p. As expected, the luciferase activity was decreased with miR-146a-5p overexpression in the wt Notch2-3’-UTR group (P=0.003 for EC 9706 and P<0.001 for Eca 109) (Figure 2B), compared with the mu Notch2-3′-UTR group (both P>0.05), suggesting that miR-146a-5p reduced the luciferase activity of wt Notch2-3′-UTR but had no effect on mu Notch2-3′-UTR. Meanwhile, an increase in the luciferase activity of wt Notch2-3′-UTR was observed after miR-146a-5p-inhibitor transfection (P=0.002 for EC 9706 and P<0.001 for Eca 109) (Figure 2B). To confirm that Notch2 acts as a miR-146a-5p target, we examined Nocth2 protein levels in miR-146a-5p mimics or inhibitor transfected ESCC cells. Notch2 expression level was decreased (P=0.004 for EC 9706 and P=0.006 for Eca 109) (Figures 2C and D) after miR-146a-5p mimics was transfected, compared with the control group. Reciprocally, the miR-146a-5p knockdown was accompanied by an increase in the Notch2 expression in EC 9706 and Eca 109 cells (P=0.008 for EC 9706 and P=0.007 for Eca 109) (Figure 2C and D).
The prediction and validation of miR-146a-5p target gene. (A) Notch2 exhibits miR-146a-5p-binding sequences in its 3′-UTR regions. (B) Luciferase assays validated that Notch2 is a target of miR-146a-5p. (C, D) Further evidence of miR-146a-5p targeting Notch2 by western blotting. **P<0.01; ***P<0.001.
Notch2 promotes EMT process of ESCC
Previously, we found that Notch2 was upregulated in ESCC cancerous tissues compared with adjacent normal tissues, and it was significantly associated with the patients’ OS and PFS. Besides, Notch2 inhibition with shRNA decreased ESCC cell proliferation and survival ability (Wang et al, 2016b). The EC 9706 and Eca 109 cells were transfected with specific Notch2 shRNA using Lipofectamine reagent. After transfection, mRNA expression level of Notch2 was significantly decreased according to qRT-PCR results (both P<0.001, Figure 3A). We used invasion assay and western blotting to examine the effect of Notch2 on EMT process. After transfection, invasion ability of EC 9706 and Eca 109 were decreased significantly (both P<0.001, Figure 3B and C). According to western blotting, the expression levels of Snail (both P<0.001) and Vimentin (both P<0.001) were decreased while E-cadherin was increased (P<0.001 for EC 9706 and P=0.002 for Eca 109) (Figure 3D and E) in the anti-Notch2 group.
miR-146a-5p regulates EMT process in ESCC cells via targeting Notch2
To investigate whether miR-146a-5p targets Notch2 to inhibit EMT process, we set four groups including miR-146a-5p-NC together with Notch2-NC, anti-miR-146a-5p together with anti-Notch2, anti-miR-146a-5p together with Notch2-NC and miR-146a-5p-NC together with anti-Notch2. As shown in Figure 4A and B, miR-146a-5p inhibitor increased ESCC cells' invasive ability, whereas shRNA-Notch2 could abolish this change (all P<0.001). Besides, miR-146a-5p inhibitor led to changes of EMT-related markers’ expression (increase of Snail and Vimentin and decrease of E-cadherin) in the EC 9706 and Eca 109 cell lines, whereas co-transfection with shRNA-Notch2 partially abolished these changes at the protein level (all P<0.05, Figure 4C and D). These findings suggest that miR-146a-5p regulates EMT progression by suppressing Notch2 expression.
The miR-146a-5p -mediated EMT process depends on Notch2 . (A, B) miR-146a-5p inhibitor caused an increase in ESCC cells’ invasive ability, whereas shRNA-Notch2 could abolish this change. (C, D) miR-146a-5p inhibitor led to increase of Snail and Vimentin and decrease of E-cadherin, whereas co-transfection with shRNA-Notch2 partially abolished these changes. *P<0.05; **P<0.01; ***P<0.001.
Discussion
Accumulating studies have demonstrated that miRNAs regulate the expression of oncogene or tumour suppressor (Png et al, 2012; Krzeszinski et al, 2014; Cheng et al, 2015), which suggest a new mechanism involved in the initiation and development of ESCC. Aberrant expression of miR-146a-5p in other human cancers (Sun et al, 2014; Shi et al, 2015; Sun et al, 2015b; Cui et al, 2016; Lerner et al, 2016) promoted us to determine its expression status in ESCC. In previous article, we found that miR-146a-5p was downregulated in ESCC cancerous tissues as well as serum, which indicated its potential antitumour function (Wang et al, 2016a). In this article, we demonstrated its role in EMT progression. It inhibits EMT progression of EC 9706 and Eca 109 cells depending on Notch2.
miR-146a-5p plays important roles in carcinogenesis and development of tumour. In this article, our gain- and loss-of-function experiments demonstrated that miR-146a-5p inhibitor led to the increase of invaded ESCC cells, whereas miR-146a-5p mimics inhibited the invasion ability. Meanwhile, the protein level of E-cadherin was reduced, whereas Snail and Vimentin were increased in the anti-miR-146a-5p group. These results indicate that miR-146a-5p inhibits EMT progression of ESCC cells. It is well established that miRNAs perform their function by regulating the expression of a target gene. Therefore, we decided to identify the functional target gene for miR-146a-5p that was involved in EMT regulation. MiRNA target gene prediction databases indicated the potential of Notch2 as a direct target of miR-146a-5p in ESCC and the dual-luciferase reporter assay validated it. Accordingly, Notch signal pathway has been considered as a crucial regulator of EMT (Espinoza and Miele, 2013; Ishida et al, 2013; Yuan et al, 2014; Zoni et al, 2015). Furthermore, the overexpression level and oncogenic role of Notch2 have been observed in numerous human cancer types, such as lung adenocarcinoma (Mimae et al, 2012), glioma (Yu et al, 2015), cervical cancer (Zhang et al, 2014), hepatoblastoma (Litten et al, 2011) and salivary adenoid cystic carcinoma (Qu et al, 2016). We have revealed that protein level of Notch2 was upregulated in ESCC cancerous cancer and it could promote the proliferation and survival ability of EC 9706 and Eca 109 (Wang et al, 2016b). Herein, it indicated that knockdown of Notch2 decreased EMT progression and invasion ability of ESCC cells. Importantly, the inhibiting effect of miR-146a-5p on EMT progression and cell invasion were partially abrogated by Notch2 knockdown in ESCC cells. Thus we propose that miR-146a-5p inhibits ESCC EMT by suppressing Notch2.
As is known to us, both miR-146a-5p and miR-146a-3p derive from pre-miR-146a. Their sequences and roles are different from each other (Gysler et al, 2016). MiR-146a-5p is wildly investigated, whereas researches on miR-146a-3p are not sufficient enough. To verify the specificity of the mimics and inhibitor used in our experiment, we blasted the sequences of our mimics and inhibitor via miRBASE database. Results indicated that the sequences match well with miR-146a-5p (both E-value<0.001), while similarity sequence test’s E-values were both 5.1 for miR-146a-3p, which revealed that the mimics and inhibitor could unlikely change the level of miR-146a-3p.
In conclusion, we suggest that miR-146a-5p functions as a tumour-suppressive miRNA and inhibits the EMT progression of ESCC via suppressing Notch2.
Change history
06 December 2016
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Chen G, Umelo IA, Lv S, Teugels E, Fostier K, Kronenberger P, Dewaele A, Sadones J, Geers C, De Greve J (2013) miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PLoS One 8: e60317.
Chen J, Imanaka N, Chen J, Griffin JD (2010) Hypoxia potentiates Notch signaling in breast cancer leading to decreased E-cadherin expression and increased cell migration and invasion. Br J Cancer 102: 351–360.
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J (2016) Cancer statistics in China, 2015. CA Cancer J Clin 66: 115–132.
Cheng CJ, Bahal R, Babar IA, Pincus Z, Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM, Saltzman WM, Slack FJ (2015) MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature 518: 107–110.
Cui Y, She K, Tian D, Zhang P, Xin X (2016) miR-146a inhibits proliferation and enhances chemosensitivity in epithelial ovarian cancer via reduction of SOD2. Oncol Res 23: 275–282.
Du F, Li S, Wang T, Zhang HY, Li DT, Du ZX, Wang HQ (2015) Implication of Bcl-2-associated athanogene 3 in fibroblast growth factor-2-mediated epithelial-mesenchymal transition in renal epithelial cells. Exp Biol Med (Maywood) 240: 566–575.
Espinoza I, Miele L (2013) Deadly crosstalk: Notch signaling at the intersection of EMT and cancer stem cells. Cancer Lett 341: 41–45.
Forloni M, Dogra SK, Dong Y, Conte D Jr., Ou J, Zhu LJ, Deng A, Mahalingam M, Green MR, Wajapeyee N (2014) miR-146a promotes the initiation and progression of melanoma by activating Notch signaling. Elife 3: e01460.
Gysler SM, Mulla MJ, Guerra M, Brosens JJ, Salmon JE, Chamley LW, Abrahams VM (2016) Antiphospholipid antibody-induced miR-146a-3p drives trophoblast interleukin-8 secretion through activation of Toll-like receptor 8. Mol Hum Reprod 22: 465–474.
Ishida T, Hijioka H, Kume K, Miyawaki A, Nakamura N (2013) Notch signaling induces EMT in OSCC cell lines in a hypoxic environment. Oncol Lett 6: 1201–1206.
Ji J, Jia S, Jia Y, Ji K, Hargest R, Jiang WG (2015) WISP-2 in human gastric cancer and its potential metastatic suppressor role in gastric cancer cells mediated by JNK and PLC-gamma pathways. Br J Cancer 113: 921–933.
Krzeszinski JY, Wei W, Huynh H, Jin Z, Wang X, Chang TC, Xie XJ, He L, Mangala LS, Lopez-Berestein G, Sood AK, Mendell JT, Wan Y (2014) miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature 512: 431–435.
Lerner C, Wemmert S, Bochen F, Kulas P, Linxweiler M, Hasenfus A, Heinzelmann J, Leidinger P, Backes C, Meese E, Urbschat S, Schick B (2016) Characterization of miR-146a and miR-155 in blood, tissue and cell lines of head and neck squamous cell carcinoma patients and their impact on cell proliferation and migration. J Cancer Res Clin Oncol 142: 757–766.
Li Y, Vandenboom TG 2nd, Wang Z, Kong D, Ali S, Philip PA, Sarkar FH (2010) miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res 70: 1486–1495.
Litten JB, Chen TT, Schultz R, Herman K, Comstock J, Schiffman J, Tomlinson GE, Rakheja D (2011) Activated NOTCH2 is overexpressed in hepatoblastomas: an immunohistochemical study. Pediatr Dev Pathol 14: 378–383.
Liu Q, Sheng W, Dong M, Dong X, Dong Q, Li F (2015) Gli1 promotes transforming growth factor-beta1- and epidermal growth factor-induced epithelial to mesenchymal transition in pancreatic cancer cells. Surgery 158: 211–224.
Mimae T, Okada M, Hagiyama M, Miyata Y, Tsutani Y, Inoue T, Murakami Y, Ito A (2012) Upregulation of notch2 and six1 is associated with progression of early-stage lung adenocarcinoma and a more aggressive phenotype at advanced stages. Clin Cancer Res 18: 945–955.
Nieto MA, Huang RY, Jackson RA, Thiery JP (2016) EMT: 2016. Cell 166: 21–45.
Pichler M, Calin GA (2015) MicroRNAs in cancer: from developmental genes in worms to their clinical application in patients. Br J Cancer 113: 569–573.
Png KJ, Halberg N, Yoshida M, Tavazoie SF (2012) A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature 481: 190–194.
Qu J, Song M, Xie J, Huang XY, Hu XM, Gan RH, Zhao Y, Lin LS, Chen J, Lin X, Zheng DL, Lu YG (2016) Notch2 signaling contributes to cell growth, invasion, and migration in salivary adenoid cystic carcinoma. Mol Cell Biochem 411: 135–141.
Shi Z, Johnson JJ, Jiang R, Liu Y, Stack MS (2015) Decrease of miR-146a is associated with the aggressiveness of human oral squamous cell carcinoma. Arch Oral Biol 60: 1416–1427.
Siegel RL, Miller KD, Jemal A (2015) Cancer statistics, 2015. CA Cancer J Clin 65: 5–29.
Sita-Lumsden A, Dart DA, Waxman J, Bevan CL (2013) Circulating microRNAs as potential new biomarkers for prostate cancer. Br J Cancer 108: 1925–1930.
Sun M, Fang S, Li W, Li C, Wang L, Wang F, Wang Y (2015a) Associations of miR-146a and miR-146b expression and clinical characteristics in papillary thyroid carcinoma. Cancer Biomark 15: 33–40.
Sun Q, Zhao X, Liu X, Wang Y, Huang J, Jiang B, Chen Q, Yu J (2014) miR-146a functions as a tumor suppressor in prostate cancer by targeting Rac1. Prostate 74: 1613–1621.
Sun X, Zhang J, Hou Z, Han Q, Zhang C, Tian Z (2015b) miR-146a is directly regulated by STAT3 in human hepatocellular carcinoma cells and involved in anti-tumor immune suppression. Cell Cycle 14: 243–252.
Wang C, Guan S, Liu F, Chen X, Han L, Wang D, Nesa EU, Wang X, Bao C, Wang N, Cheng Y (2016a) Prognostic and diagnostic potential of miR-146a in oesophageal squamous cell carcinoma. Br J Cancer 114: 290–297.
Wang C, Li Q, Liu F, Chen X, Liu B, Nesa EU, Guan S, Han L, Tan B, Wang N, Wang X, Song Q, Jia Y, Wang J, Lu M, Cheng Y (2016b) Notch2 as a promising prognostic biomarker for oesophageal squamous cell carcinoma. Sci Rep 6: 25722.
Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, Zheng ZM (2008) Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One 3: e2557.
Yao Q, Cao Z, Tu C, Zhao Y, Liu H, Zhang S (2013) MicroRNA-146a acts as a metastasis suppressor in gastric cancer by targeting WASF2. Cancer Lett 335: 219–224.
Yu HP, Qi ST, Feng WF, Zhang GZ, Zhang HP, Tian JJ (2015) Interference of Notch 2 inhibits the progression of gliomas and induces cell apoptosis by induction of the cell cycle at the G0/G1 phase. Mol Med Rep 11: 734–738.
Yuan X, Wu H, Han N, Xu H, Chu Q, Yu S, Chen Y, Wu K (2014) Notch signaling and EMT in non-small cell lung cancer: biological significance and therapeutic application. J Hematol Oncol 7: 87.
Zhang P, Li H, Yang B, Yang F, Zhang LL, Kong QY, Chen XY, Wu ML, Liu J (2014) Biological significance and therapeutic implication of resveratrol-inhibited Wnt, Notch and STAT3 signaling in cervical cancer cells. Genes Cancer 5: 154–164.
Zoni E, van der Pluijm G, Gray PC, Kruithof-de Julio M (2015) Epithelial plasticity in cancer: unmasking a microRNA network for TGF-beta-, Notch-, and Wnt-mediated EMT. J Oncol 2015: 198967.
Acknowledgements
This work was supported by National Natural Science Foundation of China (no. 81572958) and Science and Technology Development Planning Project of Shandong Province (2014GSF118058).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Wang, C., Zhang, W., Zhang, L. et al. miR-146a-5p mediates epithelial–mesenchymal transition of oesophageal squamous cell carcinoma via targeting Notch2. Br J Cancer 115, 1548–1554 (2016). https://doi.org/10.1038/bjc.2016.367
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2016.367
- Springer Nature Limited
Keywords
This article is cited by
-
miR-146a-5p-mediated suppression on trophoblast cell progression and epithelial-mesenchymal transition in preeclampsia
Biological Research (2021)
-
MicroRNA-1 suppresses proliferation, migration and invasion by targeting Notch2 in esophageal squamous cell carcinoma
Scientific Reports (2018)
-
Expression profile of microRNA-146a along HPV-induced multistep carcinogenesis: a study in HPV16 transgenic mice
Journal of Cancer Research and Clinical Oncology (2018)