Abstract
Background:
Cancer/testis-antigens (CTAs) are specifically expressed in human malignancies and testis tissue, but their molecular functions are poorly understood. CTAs serve as regulators of gene expression, cell cycle and spermatogenesis, as well as targets for immune-based therapies. The CTA PRAME is expressed in various cancers, antagonises retinoic acid signalling and is regulated by DNA methylation and histone acetylation.
Methods:
We analysed the molecular function of the CTA PRAME in primordial germ cells (PGC) and testicular germ cell cancers (GCC). GCCs arise from a common precursor lesion termed germ cell neoplasia in situ (GCNIS), which itself is thought to originate from a defective PGC. GCNIS cells eventually develop into unipotent seminomas or totipotent embryonal carcinomas (ECs), which are capable of differentiation into teratomas, yolk-sac tumours and choriocarcinomas.
Results:
PRAME is, like the master regulator of PGCs SOX17 expressed in human PGCs, GCNIS and seminomas but absent in ECs. shRNA-mediated knockdown of PRAME in seminomatous TCam-2 cells left SOX17 levels unchanged, but resulted in downregulation of pluripotency- and PGC-related genes (LIN28, PRDM14, ZSCAN10), whereas somatic and germ cell differentiation markers were upregulated. So, PRAME seems to act downstream of SOX17 by mediating the regulation of the germ cell differentiation and pluripotency programme. Endoderm differentiation is triggered in somatic cells by SOX17, suggesting that in PGCs, PRAME represses this programme and modulates SOX17 to function as a PGC-master regulator. Surprisingly, knockdown of PRAME in TCam-2 cells did not render the cells sensitive towards retinoic acid, despite the fact that PRAME has been described to antagonise retinoic acid signalling. Finally, we demonstrate that in non-seminomas PRAME expression is silenced by DNA methylation, which can be activated by formation of euchromatin via histone-deacetylase-inhibitors.
Conclusions:
We identified the CTA PRAME as a downstream factor of SOX17 and LIN28 in regulating pluripotency and suppressing somatic/germ cell differentiation in PGC, GCNIS and seminomas.
Similar content being viewed by others
Main
Testicular type II germ cell cancers (GCCs) arise from a developmentally arrested primordial germ cell (PGC), which develops into a precursor lesion, termed as germ cell neoplasia in situ (GCNIS) (Oosterhuis and Looijenga, 2005b; Sonne et al, 2009; Berney et al, 2016). GCNIS cells eventually into a seminoma or non-seminoma. PGCs, GCNIS and seminomas are highly similar with regard to morphology and gene expression. They express the germ cell markers SOX17, cKIT, TFAP2C and PRDM1, as well as the pluripotency markers NANOG, OCT3/4 and LIN28A (Biermann et al, 2007). Seminomas grow uniformly as flat, big, roundish cells with a big nucleus and present as weakly eosinophilic. The stem cell-like population of the non-seminomas is termed embryonal carcinoma (EC), which is pluri- to totipotent and able to differentiate into teratomas (composed of cells of all three germ layers), as well as yolk-sac tumours and choriocarcinomas (extraembryonic tissues; Oosterhuis and Looijenga, 2005b). In contrast to PGCs, GCNIS and seminomas, ECs express the pluripotency factor SOX2 instead of SOX17 (de Jong et al, 2008). SOX17 is a key determinant of murine PGC specification and controls expression of PGC-associated genes, like Prdm1 (Blimp1), Prdm14 and Tfap2c (Schemmer et al, 2013; Irie et al, 2015; Tang et al, 2015). In murine embryonic stem cells, the transcription factor SOX2 partners with OCT3/4 and binds to a canonical binding motif, thereby regulating expression of pluripotency factors (Aksoy et al, 2013). Interestingly, overexpression of Sox17 in murine embryonic stem cells leads to a shift of binding of OCT3/4 from SOX2 to SOX17, resulting in targeting a compressed motif within genes, which are associated with endodermal differentiation (Aksoy et al, 2013). Interestingly, human PGCs, GCNIS and seminoma cells are not prone to spontaneous differentiation, although expressing SOX17.
About 225 genes are classified as cancer/testis-antigens (CTA), which are exclusively expressed in cancer cells or testis tissue, but not in any other somatic cell type (Hofmann et al, 2008). The molecular roles of CTAs are poorly understood, but due to their unique expression pattern CTAs are discussed to serve as modulators of gene expression and cell cycle in tumour cells, as well as regulators of spermatogenesis. In addition, CTA might be targets for immune-based therapies (Whitehurst, 2014).
The CTA PRAME (preferentially expressed antigen in melanoma) is found overexpressed in various cancer types, including melanoma, neuroblastoma and breast cancers (van Baren et al, 1998; van de Vijver et al, 2002; Oberthuer et al, 2004).
In a previous study, we found PRAME highly expressed in the seminoma cell line TCam-2, whereas the EC cell line 2102EP lacked PRAME expression (Nettersheim et al, 2015). In addition, during in vivo reprogramming of TCam-2 to an EC-like cell fate PRAME and SOX17 are downregulated (Nettersheim et al, 2015). Furthermore, in GCNIS cells, seminomas and PGCs PRAME expression correlates to SOX17 expression. So, we reasoned that expression of PRAME is a general feature of PGC, GCNIS and seminomas and its function might be linked to SOX17.
Expression of PRAME has been shown to be regulated by DNA methylation and the chromatin state (Schenk et al, 2007). PRAME inhibits retinoic acid (RA) signalling by binding the RAR/RXR receptors, thereby preventing ligand-induced receptor dimerisation and downstream effector activation (Epping et al, 2005). In melanoma cells, RA-mediated differentiation is restored after RNAi-mediated knockdown of PRAME (Epping et al, 2005). The seminoma-like cell line TCam-2 is resistant towards RA-induced differentiation (Nettersheim et al, 2011b), whereas EC cell lines differentiate in response to RA into cells of all three germ layers (Strickland and Mahdavi, 1978; Nettersheim et al, 2011b). We found that the CTA PRAME is differentially methylated (5mC) between seminoma-like TCam-2 (PRAME 5mC low) and EC-like 2102EP (PRAME 5mC high; Nettersheim et al, 2015). Thus, we further speculate that PRAME might be required for RA resistance of PGCs/seminomas.
In this study, we analysed molecular function of PRAME and its link to SOX17 in PGC and GCC biology. Further, we correlated PRAME expression to DNA methylation and RA responsiveness and analysed how histone deacetylation affects PRAME expression in GCC cell lines.
Materials and methods
Ethics statement
The ethics commitee of the Rheinische Friedrich-Wilhelms-Universität Bonn approved the analyses of formalin fixed, paraffin-embedded type-II GCC tissues in context of this study. No personal patient data will be collected or stored. Written permission to use the tissue for scientific purposes was obtained from the patients and was approved by the ‘Ethik-Kommission für klinische Versuche am Menschen und epidemiologische Forschung mit personenbezogenen Daten der Medizinischen Fakultät der Rheinischen Friedrich-Wilhelms-Universität Bonn’ (the ethics committee for clinical trials on humans and epidemiological research with patient-related data of the medical faculty of the Rheinischen-Friedrich-Wilhelms-University Bonn; 065/12 to GK).
Cell culture
All GCC cell lines used in this study were cultured as described previously (Nettersheim et al, 2013, 2015). Briefly, TCam-2 and NCCIT were cultured in RPMI medium and 2102EP, NT2/D1, 833KE, H12, GCT27, 1777N, JAR, JEG-3 in DMEM. MPAF, ARZ and EMF (provided by Dr Michael Peitz (Life & Brain, Department of Reconstructive Neurobiology, Bonn, Germany)) as well as FS1 (provided by Dr Valerie Schumacher (Harvard Medical School, Department of Pediatrics, Boston, MA, USA) (Schumacher et al, 2008) were grown in DMEM medium (10% FCS (20% FS1), 1% penicillin/streptomycin, 200 mM L-glutamine, 1 × non-essential amino acids, 100 nM β-mercaptoethanol) at 37 °C and 5% CO2.
Tissue microarrays
Tissue microarrays were assembled and prepared in house after approval by the internal review board. Further information is given in the study by Nettersheim et al (2015). All tumours were classified according to the WHO classification based on their histology. Samples were examined by frozen section to assure a significant tumour cellularity. In total, 69 seminomas and 33 ECs were analysed. Only TFAP2C-positive/SOX2-negative seminomas and SOX2-positive ECs were analysed.
DNA, RNA and protein isolation
DNA, RNA and proteins were isolated as described previously (Nettersheim et al, 2015). DNA was isolated by phenol/chloroform/isoamylalcohol, RNA by the RNeasy Mini kit (Qiagen, Hilden, Germany). Bulk proteins were isolated by RIPA buffer, whereas nuclear and cytoplasmic proteins were separately isolated by the ‘Nuclear Extract Kit’ (Active Motif, La Hulpe, Belgium) according to the manual.
Western blot
Western blots analyses were performed as described previously (Nettersheim et al, 2015). The Mini-PROTEAN Electrophoresis Cell and Trans-Blot Turbo system were used (BioRad, Munich, Germany). Gels were blotted onto PVDF membranes. Chemiluminescent signals were detected using ChemiDoc MP Imaging System (BioRad). β-Actin was used as housekeeper and for data normalisation. See Table 1 for antibody details. Colorimetric analysis of band/expression intensities was performed by Image Lab software (BioRad).
Quantitative RT-PCR
Quantitative RT-PCR (qRT-PCR) was performed as described previously (Nettersheim et al, 2015). For first strand synthesis, the RevertAid First Strand cDNA Synthesis Kit manual (Fermentas, St Leon-Rot, Germany) was used. For PCR, the Maxima SYBR Green qPCR Master Mix (Fermentas, St Leon-Rot, Germany) was used. PCR was performed using the ViiA 7 Real Time PCR System (Applied Biosystems, distributed by Life Technologies, Carlsbad, CA, USA). Each qRT-PCR analysis for each biological replicate was performed in three technical replicates. At the end of each PCR run, a melting point analysis was performed. GAPDH was used as housekeeping gene and for data normalisation. See Table 2 for primer sequences.
Immunohistochemistry and immunofluorescence staining
IHC was performed as published previously (Nettersheim et al, 2015). Tumour tissues were dissected, fixed in 4% formalin overnight and processed in paraffin wax. Signal detection was performed semi-automatically in the Autostainer 480S (Medac, Hamburg, Germany). Nuclei were stained by hematoxylin. IF was performed as described previously (Nettersheim et al, 2011c, 2015). Nuclei were stained by Hoechst 33342. See Table 1 for antibody details and dilution ratios.
PathScan array
A PathScan Intracellular Signaling Array was performed as published and according to the manufacturer’s manual (Cell Signaling Technology, via NEB, Frankfurt am Main, Germany; Nettersheim et al, 2012). In all, 70 μg of total protein was loaded onto the array and incubated at 4 °C overnight. Each sample was analysed in three biological replicates.
Sodium-bisulfite treatment
Sodium-bisulfite sequencing was performed as described previously (Nettersheim et al, 2011a). Briefly, 500 ng of DNA were sodium-bisulfite-converted using the EZ DNA-Methylation Gold kit (Zymo Research, Freiburg, Germany). Each sample was analysed in five replicates. See Table 2 for PCR primer details.
RA and HDI treatment of GCC cell lines
RA treatment was performed as described previously (Nettersheim et al, 2011b). 1 × 105 GCC cells were treated for 8 days (d) with 20 μ M RA (Sigma-Aldrich, Taufkirchen, Germany), as well as for 16 h with 10 nM romidepsin (Celgene, Signal Pharmaceuticals, San Diego, CA, USA) for 24 h with 1 μ M SAHA, for 24 with 1 μ M VPA and for 24 h with 20 nM TSA (all from Sigma-Aldrich). Corresponding solvents were used as controls, that is, DMSO.
Retroviral transduction of PRAME shRNA
The PRAME shRNA oligonucleotides (Table 2) were cloned into the pSUPER.retro.puro-vector (OligoEngine, Seattle, WA, USA) according to the pSUPER RNAi System manual. A shRNA against the GFP sequence was used as unspecific control. A plasmid coding for GFP (pRP-GFP) was utilised to monitor the infection efficiency. Retroviral particles were produced in 1.2 × 106 HEK293 cells by transfecting 2 μg of the retroviral PRAME shRNA plasmid, 2 μg pCMV-gag-pol-plasmid and 220 ng pCMV-VSV-G-plasmid via the calcium phosphate method. The next day, medium was replaced by fresh medium. On day 3, the supernatant was harvested, sterile filtered and applied to the target cells. Stable shRNA-expressing cells were selected by adding 0.5 mg ml−1 puromycin every second day for 1 week.
Expression microarray analysis
Illumina HT-12v4 expression microarray analysis of GCC cell lines and PRAME knockdown of cells were performed as described previously (Nettersheim et al, 2015). All microarray data are publically available (ncbi.nlm.nih.gov/geo/) (GSE71269). Each sample was analysed in three replicates. Affymetrix expression microarray data of GCC tissues have been re-analysed in context of this study (Eckert et al, 2008). In total, 4 normal testis tissues, 3 GCNIS, 4 seminomas, 3 ECs and 3 teratomas were analysed.
Chromatin-immunoprecipation followed by sequencing analysis
Chromatin-immunoprecipation followed by sequencing (ChIP-seq) analysis was performed by Active Motif (Carlsbad, CA, USA) in context of a different study and was re-analysed in this study with regard to PRAME. Genomic DNA of TCam-2 cells, including Drosophila DNA as spike-in control were used as input control. The ChIP-seq data are publically available via GEO (GSE78262).
STRING and DAVID analysis and Venn diagrams
STRING protein–protein interaction prediction were performed online using default settings (string-db.org; Szklarczyk et al, 2015). DAVID-based functional annotation analysis was also performed online using highest classification stringency setting (david.ncifcrf.gov; Huang et al, 2009). Venn diagrams were generated using ‘Venny’ (bioinfogp.cnb.csic.es/tools/venny).
Results
CTAs are expressed in testis and cancer tissues. Using expression microarray analysis, we first compared expression of CTAs in GCC cell lines (Supplementary Data S1A). We excluded all CTAs with expression intensity similar or lower than that of SOX2 in TCam-2 cells (log27.5). We found members of the MAGE (A3, A6, A8, A12) and GAGE (2B, 2E, 4, 5, 6, 7,12B, 12C, 12F, 12G, 12H, 12I, 12J) family highly expressed in choriocarcinoma-like JAR cells, whereas seminomatous TCam-2 cells strongly express PRAME and XAGE family members (1, 1A, 1B, 1C, 2B (also high in JAR); Supplementary Figure S1A). Seven other CTAs, that is, MAGE members (D1, D2, E1, F1), SPA17, MORC2 and TDRD1 are commonly expressed in all analysed GCC cell lines (Supplementary Figure S1A). Expression of none of the CTA members reached significance level in both EC cell lines (2102EP, NCCIT; Supplementary Figure S1A).
Next, we compared expression of CTAs in normal adult testis tissues and GCC tissues (Supplementary Data S1B). We excluded all CTAs with expression intensity equivalent or lower than that of SOX2 in seminomas (log29.7). A total of 53 CTAs were found significantly expressed in normal adult testis tissues, 33 in GCNIS, 25 in seminomas and 23 in ECs (Supplementary Data S1B). Again, we found the CTAs PRAME and XAGE1 strongly expressed in GCNIS/seminomas, but not in ECs.
Type II GCC are thought to originate from a developmentally arrested PGC (Oosterhuis and Looijenga, 2005a). GCNIS/seminomas resemble PGCs in terms of gene expression and morphology, whereas ECs are similar to embryonic stem cells. To analyse, if PRAME is also expressed in PGCs, we performed a meta-analysis of RNA-sequencing data of human PGCs (male and female; weeks 5.5, 7, 9, 19 of gestation), human PGC-like cells, human embryonic stem cells, gonadal somatic cells and TCam-2 performed by Guo et al (2015), Irie et al (2015) and Tang et al (2015). PRAME is expressed in human PGCs of all gestational weeks analysed and seminoma (TCam-2) cells, but not detectable in somatic cells, human embryonic stem cells or human PGC-like cells (Supplementary Figure S1B). Of note, aside expression of PRAME in TCam-2, no other PRAME family member (PRAMEF1–22) is expressed in GCC cell lines, fibroblasts (MPAF) or Sertoli cells (FS1; Supplementary Figure S1A).
These data indicate that the CTA PRAME is expressed in PGC/GCNIS/seminoma and demarcates these entities from ECs. Further, we observed downregulation of PRAME in seminomatous TCam-2 after xenografting into the somatic microenvironment of the murine flank, where they reprogram to an EC-like fate (Nettersheim et al, 2015). This reprogramming process is associated by a rapid induction of SOX2 and downregulation of SOX17 (Nettersheim et al, 2015).
Our data indicate that PRAME is expressed in PGCs, GCNIS and seminomas, and like SOX17, can be utilised to discriminate GCNIS/seminomas from ECs/non-seminomas. Next, we compared PRAME expression with SOX2 and SOX17 expression in normal adult testis tissues, GCNIS, seminomas, ECs and teratomas. We demonstrated that PRAME expression is high in normal adult testis tissues, GCNIS and seminomas, but low in ECs (Figure 1A; de Jong et al, 2008). In GCC tissues, PRAME expression correlated to SOX17 and inversely correlated to SOX2 expression, suggesting that in GCNIS/seminomas PRAME might be functionally linked to SOX17 (Figure 1A).
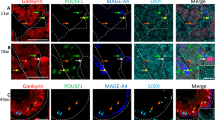
PRAME expression in GCC tissues and cell lines. (A) Correlation of SOX2 and SOX17 expression to PRAME expression in normal adult testis tissue (NATT) and indicated GCC tissues. (B) qRT–PCR analysis of PRAME expression in indicated GCC tissues and corresponding cell lines as well as fibroblasts (Fibro) and Sertoli cells (Sert). (C) Western blot analysis of PRAME expression in seminoma and EC tissues, as well as GCC cell lines and human fibroblasts. (D) Pie diagrams summarising cellular localisation of PRAME in seminoma and EC tissues as determined by IHC. (E) IHC staining of PRAME in seminomas and ECs. Scale-bars: 200 μm. (F) Western blot analysis of PRAME expression in the nuclear and cytoplasmic extract of TCam-2 cells. Efficient separation of nuclear and cytoplasmic fraction was demonstrated by OCT3/4 and β-actin detection. Sem=seminomas; EC=embryonal carcinoma; Cc=choriocarcinoma.
We next compared PRAME expression and protein level between seminomas and ECs as well as corresponding cell lines by qRT-PCR and western blot analysis (HPA045153, Table 1; Figure 1B and C). We found high levels of PRAME in seminomas/TCam-2, whereas expression was negligible in EC, choriocarcinoma cell lines, fibroblast and Sertoli cells. (Figure 1B and C). By use of a second PRAME antibody (H-10, Table 1), we verified PRAME expression in TCam-2 cells and absence in 2102EP (Supplementary Figure S2A). In addition, we performed IHC of GCC tissue microarrays containing 69 pure seminoma and 33 EC tissues (Figure 1D). Only TFAP2C-positive and SOX2-negative seminomas, and SOX2-positive ECs were analysed (Supplementary Figure S2B). Seminomas displayed a strong signal of PRAME in the cytoplasm and the nucleus (Figure 1D, 45%; E, PRAME 1) or in the cytoplasm only (Figure 1D, 55%; E, PRAME 2), whereas all ECs displayed only very weak to negligible cytoplasmic staining (Figure 1D, 100%; E, PRAME 3/4). In ECs, no nuclear PRAME staining could be detected (Figure 1D and E, PRAME 3/4). This antibody staining (HPA045153, Table 1) was verified by the use of a second antibody detecting PRAME (ab89097, Table 1) and yielded comparable results, that is, nuclear and/or cytoplasmic staining in seminomas (Supplementary Figure S2C, PRAME 5/6) and only weak cytoplasmic staining of PRAME in ECs (Supplementary Figure S2C, PRAME 7/8). Cytoplasmic/membranous and nuclear localisation of PRAME in TCam-2 cells was confirmed by immunofluorescence (IF) staining (Supplementary Figure S2D). As negative control, we stained human placenta tissue for PRAME and found a weak signal in the cytoplasm of trophoblast cells. So, in IHC the antibodies seem to give rise to an unspecific cytoplasmic staining (Supplementary Figure S2C, PRAME 9/10). To test, whether a specific cytoplasmic PRAME signal can be detected by western blotting, we isolated cytoplasmic and nuclear proteins of TCam-2 cells. Efficient separation of both fractions was shown by detection of OCT3/4 (nuclear) and β-Actin (cytoplasmic; Figure 1F). A signal at 58 kDa corresponding to the expected size of the PRAME protein was detectable in both, the cytoplasm and the nucleus (Figure 1F). As in IF, a more prominent signal was detected in the nucleus. Thus, in western blotting the antibody is able to specifically detect PRAME, but IHC staining is biased by detection of additional unspecific signals in the cytoplasm.
To analyse the molecular consequences of a PRAME knockdown in TCam-2 cells, we established a shRNA knockdown using retroviral vectors. The fact that >95% of the cells were positive for GFP after infection of TCam-2 cells with GFP-coding viral particles demonstrates an efficient transduction procedure (Figure 2A; pRP-GFP). In TCam-2, qRT-PCR and western blot analyses demonstrated that the PRAME knockdown level was on average 81% (shRNA1.1: 80%, 1.2: 80%, 1.3: 83%; Figure 2B and C). To screen for dose-dependent effects, a further PRAME shRNA (shRNA 2) was used that reduced PRAME mRNA levels to ∼50% (Figure 2D). One week post infection and antibiotic selection, stable PRAME shRNA-expressing clones emerged. These clones had changed morphology to a more flattened, roundish shape with an increased cell size, suggesting that they had initiated differentiation (Figure 2A).
PRAME shRNA-mediated knockdown in TCam-2 cells. (A) Detection of a GFP signal in nearly all pRP-GFP-infected TCam-2 (pRP-GFP) demonstrates high transduction efficiency. Compared with mock infected TCam-2 (mock), PRAME-knockdown cells (PRAME shRNA 1) show a change in morphology, that is, increase in cell size. (B, C) qRT–PCR and western blot analysis of PRAME expression in PRAME shRNA-infected and puromycin-selected TCam-2 cells. In western blotting, mock-infected, GFP shRNA-infected and empty vector-infected TCam-2 served as controls.
In order to reveal the molecular effects of a PRAME knockdown, we performed expression microarray analyses.
The in vivo reprogramming of TCam-2 to an EC-like state is accompanied by rapid downregulation of PRAME, which is suggested to antagonise RA signalling by binding and blocking RA receptors (Epping et al, 2005; Nettersheim et al, 2015). During the reprogramming, we observed upregulation of RA-signalling-associated factors (RBP1, CRABP1/2, FABP5 and CYP16A1; Nettersheim et al, 2015). In addition, RNAi-mediated knockdown of PRAME in melanoma cells restores RA sensitivity (Epping et al, 2005). Thus, knockdown of PRAME might render TCam-2 cells sensitive toward RA. Therefore, we analysed the TCam-2-knockdown cells±RA.
We sorted the data for genes significantly deregulated (threshold log21.5) in PRAME shRNA TCam-2 cells (Figure 3A, Supplementary Data S1C) and utilised the DAVID algorithm to predict enriched gene categories and found 12 clusters enriched, when analysing the upregulated genes (Supplementary Data S1D). With the highest enrichment score (⩾0.86), categories associated with reproductive structure development, development of primary sexual characteristics and sex differentiation were enriched. In addition, categories related to cell adhesion and migration, regulation of kinase activity/phosphorylation and muscle development were detected. The DAVID analysis suggests that PRAME is normally involved in repression of these processes.
Analysis of gene expression dynamics and regulatory networks in TCam-2 PRAME shRNA cells±RA. (A) Expression microarray data of probes deregulated in PRAME shRNA cells treated with or without RA and in RA-treated TCam-2. (B, C) STRING-based protein–protein interaction prediction of all probes upregulated (B) or downregulated (C) in PRAME shRNA cells. (D) qRT–PCR analysis of indicated marker genes in RA-treated TCam-2 and TCam-2 PRAME shRNA cells with and without RA application. (E) qRT–PCR analysis of indicated marker genes in shRNA 1- and 2-infected TCam-2 cells, showing a 80% and 50% PRAME knockdown. (F) Co-IP analysis using either a PRAME (IP PRAME) or a LIN28A (IP LIN28) antibody. Ten percent of protein lysate was used as input control (input). Dynabeads incubated with protein lysate but without any antibody were used as negative control (no ab). (G) PathScan analysis of indicated signalling pathway activities in PRAME shRNA cells (left side). A western blot analysis confirmed increased phosphorylation of ERK1/2 in PRAME shRNA cells (right side). Western blot raw data is given in Supplementary Figure S4C.
We found 14 clusters, when analysing the downregulated genes (Supplementary Data S1E). With a highest enrichment score (⩾1.83), categories related to leucine-zipper transcription factors and regulation of apoptosis were enriched (Supplementary Data S1E).
We used the STRING-algorithm to predict protein-interactions between all genes up- or downregulated after PRAME knockdown (Figure 3B and C). STRING-predicted interaction of LIN28A and PRDM14 with ZSCAN10 (Figure 3C, downregulated). Like LIN28A and PRDM14, ZSCAN10 (zinc-finger and SCAN domain-containing 10, aka ZFP206) is a pluripotency-associated factor and involved in the regulation of Oct3/4, Nanog, Sox2/Sox17 and Sall4 in murine embryonic stem cells (Zhang et al, 2006; Wang et al, 2007). So, three regulators of pluripotency (LIN28A, PRDM14 and ZSCAN10) are downregulated upon PRAME knockdown, suggesting that PRAME normally contributes to maintenance of the pluripotency programme in TCam-2 cells.
Among the genes upregulated after PRAME knockdown are several factors related to endodermal and/or mesodermal differentiation processes, such as IGFBP5, OSR1, NR2F2, TGFB1, GPC3, SOX21, CD34 and TP63 (Figure 3A). In line, our DAVID analysis detected enrichment of genes associated with muscle development (mesoderm; Supplementary Data S1D). These findings suggest that PRAME shRNA cells initiate differentiation into the somatic lineage.
Furthermore, knockdown of PRAME led to upregulation of genes associated with reproductive development and germ cell differentiation (CCND1, DMRT1, OSR1, TP63, SPRY4), indicating that PRAME suppresses this programme (Supplementary Data S1D).
In summary, our data demonstrate that in TCam-2 PRAME supports the pluripotency network and suppresses a somatic and germ cell differentiation process.
We hypothesised that a PRAME knockdown might render TCam-2 cells sensitive towards RA. However, the expression levels of all deregulated genes were highly comparable between TCam-2 PRAME shRNA cells with or without RA treatment, suggesting that RA has minor effects on expression of indicated genes (Figure 3A). In total, we found only 19 microarray probes (coding for 14 annotated genes) differentially expressed between RA-treated and -untreated PRAME shRNA cells (Supplementary Data S1C). Among them, RA-signalling-related genes CYP26A1, CYP26B1, HOXA5 and RARB. Interestingly, genes upregulated in PRAME shRNA cells showed also a trend towards upregulation in RA-treated TCam-2 cells, but expression intensities were mostly below our significance threshold level (Figure 3A). In contrast, expression of the majority of genes downregulated in PRAME shRNA cells were not affected in RA-treated TCam-2 cells (Figure 3A). After RA treatment of TCam-2 cells, we found only 23 genes significantly deregulated in expression (22 upregulated, 1 downregulated; Supplementary Data S1C), including RA-signalling-related genes CYP26A1, CYP26B1, HOXA5, RARB, which were also upregulated in RA-treated PRAME shRNA cells.
We verified that the RA concentration applied in our experiments (20 μM) is sufficient to induce differentiation in the EC cell lines NT2/D1 and NCCIT. A qRT-PCR analysis 8 days (d) after RA treatment demonstrates that both cell lines downregulated typical EC markers and pluripotency factors (NANOG, OCT3/4, SOX2, GDF3, DNMT3B, DNMT3L) and induced expression of mesodermal differentiation factors PAX6 and HAND1, as well as RA receptors RARA and RARB (Supplementary Figure S3A).
These findings demonstrate that a PRAME knockdown does not sensitise TCam-2 cells towards a RA-induced differentiation process, although upregulation of RA-response genes, like RARB demonstrated a cellular response toward RA.
Using qRT-PCR, we verified selected deregulations found by the microarray analysis. As already demonstrated, in PRAME shRNA cells we detected downregulation of PRAME, LIN28A, DNMT3L, PRDM14, ATF3 and GAL, whereas DUSP6, DMRT1 and CCDN1 were upregulated (Figure 3D). By western blotting, we confirmed downregulation of PRDM14 in PRAME shRNA cells (Supplementary Figure S3B).
We screened for expression of further pluripotency and epigenetic reprogramming-related factors, because downregulation of LIN28A, PRDM14 and ZSCAN10 might influence expression of other pluripotency factors. We found downregulation of pluripotency factors NANOG, OCT3/4, SALL4, ZIC3, DPPA3, DPPA5, REX1 as well as of the de novo DNA methyltransferase DNMT3A (Figure 3D). Expression of the maintenance DNA methyltransferase DNMT1 was not considerably changed (Figure 3D). The qRT-PCR analysis confirmed that addition of RA had no significant impact on the expression of the analysed pluripotency-, PGC- and differentiation-associated genes (Figure 3D). Although, RARB was strongly induced in RA-treated samples, indicating that RA indeed stimulates RAR receptor expression, but obviously without any effect on the differentiation status of TCam-2 cells (Figure 3D).
To analyse whether the observed effects are dose-dependent, we analysed the expression of pluripotency factors in TCam-2 cells showing 80% and 50% knockdown of PRAME (Figures 2B and 3E). In a dose-dependent manner, we demonstrated the downregulation of NANOG, OCT3/4, LIN28A, SALL4, REX1 and PRDM14, as well as PGC markers DPPA3, PRDM1 and TFAP2C. Downregulation of OCT3/4, LIN28A and TFAP2C was verified by western blotting (Supplementary Figure S3C).
The deregulations in expression of pluripotency/PGC factors were also detected by our microarray analysis, but expression intensities were slightly below our set significance threshold. Nevertheless, the same trend in downregulation of pluripotency factors was found (Supplementary Figure S3D).
During human PGC specification, PRDM1 and TFAP2C are regulated by SOX17, whereas in murine embryonic stem cells SOX17 drives endodermal differentiation (Aksoy et al, 2013; Irie et al, 2015). Upon PRAME knockdown expression of SOX17 did not change significantly, whereas PGC markers, PRDM1, PRDM14, TFAP2C and DPPA3 were downregulated, and markers for germ cell and somatic differentiation were upregulated. This demonstrates that expression of SOX17 does not depend on PRAME. However, PRAME is required to maintain PGC-state in repressing somatic and further germ cell differentiation in TCam-2.
Furthermore, our analysis of the PRAME shRNA cells pointed at a regulatory link between PRAME and the RNA-binding protein LIN28A. By Co-IP analysis using either a LIN28A or a PRAME antibody, we demonstrated that indeed PRAME and LIN28A interact with each other (Figure 3F). Thus, the influence of PRAME on the pluripotency programme is at least in part a result of the interaction with LIN28A, maybe by binding and guiding LIN28A to its target RNAs.
Next, we asked if a PRAME knockdown affects activity of signalling pathways in TCam-2 cells (Supplementary Figure S4A). A PathScan intracellular signalling array analysis detected a significantly increased phosphorylation of ERK1/2, P38 and AMPK-α in PRAME shRNA cells, suggesting that PRAME normally limits activity of the corresponding pathways (Figure 3G, Supplementary Figure S4B). By western blotting, we confirmed increased ERK1/2 signalling activity in PRAME shRNA cells (Figure 3G, right side; Supplementary Figure S4C). So, knockdown of PRAME leads to an increase in MAPK signalling. In line, our DAVID analysis predicted enrichment of genes associated with regulation of kinase activity/phosphorylation, and by microarray analysis we found upregulation of marker genes associated with ERK1/2/P38-MAPK signalling, that is, CYP26A1, IGFBP5, THBS1, CXCL14, GPC3, CCND1, TGFB1 as well as the negative regulators of MAPK signalling, SPRY4 and DUSP6, which might limit MAPK signalling again.
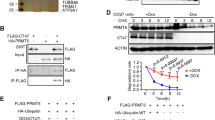
Finally, we asked how expression of PRAME is regulated in GCC cells? In human malignancies, like acute myeloid leukaemia, PRAME expression is regulated by DNA methylation (Schenk et al, 2007). In addition, it has been shown that PRAME expression can be de-repressed by treating acute myeloid leukaemia cells with HDAC inhibitors (HDI), suggesting that PRAME is regulated by DNA methylation and histone acetylation (Yao et al, 2013). By a ChIP-seq analysis of TCam-2 cells against pan-acetylated histone 3 (H3ac) and illustrating the data in the Genome Browser, we found high enrichment of H3ac around the transcription start sites (TSS) (Figure 4A, blue box). Thus, we assume that euchromatin at the PRAME TSS allows for expression in TCam-2.
Epigenetic regulation of PRAME expression by DNA methylation and histone acetylation. (A) Illustration of pan-H3ac ChIP-seq data in TCam-2 and genomic features of the PRAME locus in the Genome Browser. (B) Sodium-bisulfite sequencing of the PRAME promotor in GCC tissues and corresponding cell lines, as well as during in vivo reprogramming of TCam-2 to an EC-like state. (C) qRT–PCR analysis of PRAME expression in HDI (romidepsin)-treated GCC cell lines, human fibroblasts (MPAF, ARZ) and human Sertoli cells (FS1). Fold change (romidepsin-treated versus solvent control) is indicated above bars. (D) Western blot analysis of PRAME and pan-Histone H3 acetylation (pan-H3ac) in selected HDI-treated samples. (E) qRT–PCR analysis of PRAME expression in HDI (SAHA, VPA, TSA)-treated 2102EP and NCCIT cells.
Within the first PRAME TSS, a CpG-island was found (Figure 4A, blue box). We used sodium-bisulfite sequencing to analyse the DNA methylation within this region in GCCs and corresponding cell lines. Seminomas and TCam-2 cells harbour a hypomethylated PRAME promotor region (<20%), whereas ECs and corresponding cell lines (2102EP, NCCIT) displayed PRAME hypermethylation (>80%; Figure 4B). As previously demonstrated by us, PRAME methylation increased during the in vivo reprogramming of TCam-2 to an EC-like cell fate, which is accompanied by PRAME downregulation (Figure 4B; TCam-2 in vivo 2 w, 6 w; Nettersheim et al, 2015). Thus, PRAME expression inversely correlates to the DNA methylation status of the PRAME promoter in GCNIS/seminomas and ECs, suggesting that PRAME expression is silenced by DNA methylation in non-seminomas.
It has been shown that PRAME expression can be de-repressed by treating acute myeloid leukaemia cells with HDI (Yao et al, 2013). In order to check whether PRAME negative GCC cell lines would behave similarly we treated GCC cell lines with the HDI romidepsin and analysed PRAME expression levels by qRT-PCR analysis (Figure 4C). Adult (MPAF) and neonatal (ARZ) fibroblasts, as well as Sertoli cells (FS1) were used as controls. PRAME expression was upregulated in two EC cell lines (2102EP: 8.7-fold; NCCIT: 3.2-fold) and two choriocarcinoma cell lines (JAR: 1.9-fold (HDI vs solvent control), JEG-3: 18.0-fold), whereas PRAME was slightly downregulated in TCam-2 and remained nearly unchanged in fibroblasts (MPAF: 1.3-fold, ARZ: 0.7-fold), as well as Sertoli cells (FS1: 1.2-fold) (Figure 4C). We verified upregulation of PRAME in 2102EP and JAR cells, as well as nearly unchanged PRAME expression in TCam-2 and MPAF by western blotting (Figure 4D). In addition, we confirmed efficacy of the HDI treatment by demonstrating hyperacetylation of histone H3 (Figure 4D). Furthermore, we treated 2102EP and NCCIT cells with the HDI SAHA (1 μ M), VPA (1 μ M) and TSA (20 nM) for 24 h (Figure 4E). Both cell lines induced PRAME expression after the treatment, with 2102EP showing a stronger response than NCCIT (Figure 4E). In conclusion, PRAME expression can be restored in non-seminomatous GCC cell lines by inhibition of HDACs. So, formation of euchromatin around the PRAME locus seems to override the repressive DNA methylation mark, leading to de-repression of PRAME expression.
Discussion
In this study, we analysed expression of the CTA PRAME in human testicular GCCs and corresponding cell lines, and demonstrated that PRAME is expressed in PGCs/GCNIS/seminomas and is absent in ECs. Knocking down PRAME in seminomatous TCam-2 leads to downregulation of pluripotency and PGC markers, as well as activation of MAPK signalling. In addition, the cells displayed increased cell size and appeared as big flat roundish cells indicative of differentiation. Expression microarray analyses revealed upregulation of genes suggestive for endodermal/mesodermal and germ cell differentiation. Thus, in seminomas (and probably in human PGCs) expression of PRAME leads to a fixation of the PGC fate by suppressing germ cell and somatic differentiation.
So, why do TCam-2 cells need PRAME? In the murine system, depending on the binding partner the transcription factor OCT3/4 binds to different motifs. When SOX2 is present it partners with OCT3/4 and binds to canonical motifs that are associated with pluripotency, whereas in the presence of SOX17, OCT3/4/SOX17 bind to compressed motifs which trigger endodermal differentiation (Aksoy et al, 2013; Viotti et al, 2014). In addition, during specification of human PGCs, SOX17 has been described as a master regulator transactivating the PGC markers, PRDM1, PRDM14 and TFAP2C (Irie et al, 2015; Tang et al, 2015). These data suggest that in seminomas SOX17 is able to bind the canonical motif and thus, supports the pluripotency programme. Hence, in this case, SOX17 can replace SOX2 in the pluripotency cluster.
How is this achieved? Knockdown of PRAME does not affect the level of SOX17, whereas a downregulation of SOX17 during the in vivo reprogramming is followed by the loss of PRAME expression (Nettersheim et al, 2015). This indicates that SOX17 is upstream of PRAME in the cascade of PGC genes.
Furthermore, knockdown of PRAME results in downregulation of pluripotency/PGC factors (LIN28A, PRDM14, ZSCAN10, DPPA3, PRDM1, TFAP2C) and upregulation of genes indicative for germ cell differentiation, CCND1, DMRT1, OSR1, TP63, SPRY4. This suggests that PRAME expression leads to a fixation of the SOX17-induced PGC fate.
Moreover, lowering the levels of PRAME led to upregulation of endoderm markers. As mentioned above, PRAME seems required for binding of the SOX17/OCT3/4 dimers to pluripotency markers. Reduction of PRAME leads to alteration in binding-site selection, allowing (as demonstrated in murine embryonic stem cells) the SOX17/OCT3/4 dimers to bind to somatic differentiation genes.
Of note, a knockdown of PRAME leads to increased activity of MAPK signalling (ERK1/2, P38). It is known that repression of MAPK signalling by MEK inhibition is required to maintain ES cells in pluripotency and repress somatic differentiation. It remains to be elucidated whether PRAME itself represses MAPK signalling, or whether the upregulation of MAPK signals is a secondary process due to the upregulation of somatic differentiation markers.
Treatment of the PRAME-knockdown TCam-2 cells with RA had no considerable effect on expression of pluripotency and PGC markers, suggesting that RA has little role in PRAME-knockdown-induced reduction of pluripotency and loss of a seminoma-like cell state. In PRAME-knockdown cells, the RA-metabolising enzyme CYP26A1 was strongly upregulated, which might catabolise RA to excreted oxoderivatives (4-OH RA, 4-oxo RA, 18-OH RA). This leads to the suppression of RA-induced differentiation.
We found that PRAME expression inversely correlated to DNA methylation in the PRAME promoter, and that in non-seminomatous cell lines repressed PRAME expression can be restored by HDI treatment. In contrast to non-seminomas, seminomas respond very well to DNA-damaging treatments, that is, cisplatin-based therapies. In future experiments it would be of interest to find out if PRAME has a role in this sensitivity and if restoring PRAME expression by HDI treatment in (chemotherapy-resistant) non-seminomas might re-sensitise these cells towards a treatment with DNA-damaging agents.
We combined our findings and hypotheses in a model (Figure 5). In PGC/GCNIS/seminomas, the PRAME promoter is unmethylated and encircled by euchromatin, allowing for PRAME expression (Figure 5A). In the presence of PRAME, SOX17/OCT3/4 dimers bind to canonical motifs, usually occupied by SOX2/OCT3/4 dimers. This leads to expression of the pluripotency factors LIN28A, PRDM14 and ZSCAN10. Furthermore, PRAME is downstream of SOX17 in mediating the activation of PGC factors PRDM1, TFAP2C and DPPA3. This leads to a fixation of the PGC cell fate. Upon knockdown of PRAME, SOX17 binds to compressed motifs, resulting in induction of somatic and germ cell differentiation programs (Figure 5B).
Model of the cellular mechanisms of PRAME. (A) Regulation of pluripotency, PGC-ness and differentiation by PRAME, SOX17 and OCT3/4 in PGCs, GCNIS and seminomas. (B) A knockdown of PRAME leads to suppression of pluripotency- and PGC-associated factors, whereas expression of somatic and germ cell differentiation markers is induced via binding of SOX17/OCT3/4 to compressed binding motifs.
Change history
09 August 2016
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Aksoy I, Jauch R, Chen J, Dyla M, Divakar U, Bogu GK, Teo R, Ng CKL, Herath W, Lili S, Hutchins AP, Robson P, Kolatkar PR, Stanton LW (2013) Oct4 switches partnering from Sox2 to Sox17 to reinterpret the enhancer code and specify endoderm. EMBO J 32: 938–953.
Berney DM, Looijenga L, Idrees M, Oosterhuis JW, Rajpert-De Meyts E, Ulbright TM, Skakkebaek NE (2016) Germ cell neoplasia in situ (GCNIS): evolution of the current nomenclature for testicular pre-invasive germ cell malignancy. Histopathology 69 (1): 7–10.
Biermann K, Heukamp LC, Steger K, Zhou H, Franke FE, Sonnack V, Brehm R, Berg J, Bastian PJ, Muller SC, Wang-Eckert L, Buettner R (2007) Genome-wide expression profiling reveals new insights into pathogenesis and progression of testicular germ cell tumors. Cancer Genomics Proteomics 4: 359–367.
de Jong J, Stoop H, Gillis AJM, van Gurp RJHLM, van de Geijn G-JM, de Boer M, Hersmus R, Saunders PTK, Anderson RA, Oosterhuis JW, Looijenga LHJ (2008) Differential expression of SOX17 and SOX2 in germ cells and stem cells has biological and clinical implications. J Pathol 215: 21–30.
Eckert D, Nettersheim D, Heukamp LC, Kitazawa S, Biermann K, Schorle H (2008) TCam-2 but not JKT-1 cells resemble seminoma in cell culture. Cell Tissue Res 331: 529–538.
Epping MT, Wang L, Edel MJ, Carlée L, Hernandez M, Bernards R (2005) The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell 122: 835–847.
Guo F, Yan L, Guo H, Li L, Hu B, Zhao Y, Yong J, Hu Y, Wang X, Wei Y, Wang W, Li R, Yan J, Zhi X, Zhang Y, Jin H, Zhang W, Hou Y, Zhu P, Li J, Zhang L, Liu S, Ren Y, Zhu X, Wen L, Gao YQ, Tang F, Qiao J (2015) The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell 161: 1437–1452.
Hofmann O, Caballero OL, Stevenson BJ, Chen Y-T, Cohen T, Chua R, Maher CA, Panji S, Schaefer U, Kruger A, Lehvaslaiho M, Carninci P, Hayashizaki Y, Jongeneel CV, Simpson AJG, Old LJ, Hide W (2008) Genome-wide analysis of cancer/testis gene expression. Proc Natl Acad Sci USA 105: 20422–20427.
Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57.
Irie N, Weinberger L, Tang WWC, Kobayashi T, Viukov S, Manor YS, Dietmann S, Hanna JH, Surani MA (2015) SOX17 is a critical specifier of human primordial germ cell fate. Cell 160: 253–268.
Nettersheim D, Biermann K, Gillis AJM, Steger K, Looijenga LHJ, Schorle H (2011a) NANOG promoter methylation and expression correlation during normal and malignant human germ cell development. Epigenetics 6: 114–122.
Nettersheim D, Gillis A, Biermann K, Looijenga LHJ, Schorle H (2011b) The seminoma cell line TCam-2 is sensitive to HDAC inhibitor depsipeptide but tolerates various other chemotherapeutic drugs and loss of NANOG expression. Genes Chromosom Cancer 50: 1033–1042.
Nettersheim D, Gillis AJ, Looijenga LH, Schorle H (2011c) TGF-beta1, EGF and FGF4 synergistically induce differentiation of the seminoma cell line TCam-2 into a cell type resembling mixed non-seminoma. Int J Androl 34: e189–e203.
Nettersheim D, Heukamp LC, Fronhoffs F, Grewe MJ, Haas N, Waha A, Honecker F, Waha A, Kristiansen G, Schorle H (2013) Analysis of TET expression/activity and 5mC oxidation during normal and malignant germ cell development. PLoS One 8: e82881.
Nettersheim D, Jostes S, Sharma R, Schneider S, Hofmann A, Ferreira HJ, Hoffmann P, Kristiansen G, Esteller MB, Schorle H (2015) BMP inhibition in seminomas initiates acquisition of pluripotency via NODAL signaling resulting in reprogramming to an embryonal carcinoma. PLoS Genet 11: e1005415.
Nettersheim D, Westernströer B, Haas N, Leinhaas A, Brüstle O, Schlatt S, Schorle H (2012) Establishment of a versatile seminoma model indicates cellular plasticity of germ cell tumor cells. Genes Chromosom Cancer 51: 717–726.
Oberthuer A, Hero B, Spitz R, Berthold F, Fischer M (2004) The tumor-associated antigen PRAME is universally expressed in high-stage neuroblastoma and associated with poor outcome. Clini Cancer Res 10: 4307–4313.
Oosterhuis JW, Looijenga LH (2005a) Testicular germ-cell tumours in a broader perspective. Nat Rev Cancer 5: 210–222.
Oosterhuis JW, Looijenga LHJ (2005b) Testicular germ-cell tumours in a broader perspective. Nat Rev Cancer 5: 210–222.
Schemmer J, Araúzo-Bravo MJ, Haas N, Schäfer S, Weber SN, Becker A, Eckert D, Zimmer A, Nettersheim D, Schorle H (2013) Transcription factor TFAP2C regulates major programs required for murine fetal germ cell maintenance and haploinsufficiency predisposes to teratomas in male mice. PLoS One 8: e71113.
Schenk T, Stengel S, Goellner S, Steinbach D, Saluz HP (2007) Hypomethylation of PRAME is responsible for its aberrant overexpression in human malignancies. Genes Chromosom Cancer 46: 796–804.
Schumacher V, Gueler B, Looijenga LHJ, Becker JU, Amann K, Engers R, Dotsch J, Stoop H, Schulz W, Royer-Pokora B (2008) Characteristics of testicular dysgenesis syndrome and decreased expression of SRY and SOX9 in Frasier syndrome. Mol Reprod Dev 75: 1484–1494.
Sonne SB, Almstrup K, Dalgaard M, Juncker AS, Edsgard D, Ruban L, Harrison NJ, Schwager C, Abdollahi A, Huber PE, Brunak S, Gjerdrum LM, Moore HD, Andrews PW, Skakkebaek NE, Meyts ERD, Leffers H (2009) Analysis of gene expression profiles of microdissected cell populations indicates that testicular carcinoma in situ is an arrested gonocyte. Cancer Res 69: 5241–5250.
Strickland S, Mahdavi V (1978) The induction of differentiation in teratocarcinoma stem cells by retinoic acid. Cell 15: 393–403.
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, Mering von C (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43: D447–D452.
Tang WWC, Dietmann S, Irie N, Leitch HG, Floros VI, Bradshaw CR, Hackett JA, Chinnery PF, Surani MA (2015) A unique gene regulatory network resets the human germline epigenome for development. Cell 161: 1453–1467.
van Baren N, Chambost H, Ferrant A, Michaux L, Ikeda H, Millard I, Olive D, Boon T, Coulie PG (1998) PRAME, a gene encoding an antigen recognized on a human melanoma by cytolytic T cells, is expressed in acute leukaemia cells. Br J Haematol 102: 1376–1379.
van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AAM, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R (2002) A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347: 1999–2009.
Viotti M, Nowotschin S, Hadjantonakis A-K (2014) SOX17 links gut endoderm morphogenesis and germ layer segregation. Nat Cell Biol 16: 1146–1156.
Wang Z-X, Kueh JLL, Teh CH-L, Rossbach M, Lim L, Li P, Wong K-Y, Lufkin T, Robson P, Stanton LW (2007) Zfp206 is a transcription factor that controls pluripotency of embryonic stem cells. Stem Cells 25: 2173–2182.
Whitehurst AW (2014) Cause and consequence of cancer/testis antigen activation in cancer. Annu Rev Pharmacol Toxicol 54: 251–272.
Yao Y, Zhou J, Wang L, Gao X, Ning Q, Jiang M, Wang J, Wang L, Yu L (2013) Increased PRAME-specific CTL killing of acute myeloid leukemia cells by either a novel histone deacetylase inhibitor chidamide alone or combined treatment with decitabine. PLoS ONE 8: e70522.
Zhang W, Walker E, Tamplin OJ, Rossant J, Stanford WL, Hughes TR (2006) Zfp206 regulates ES cell gene expression and differentiation. Nucleic Acids Res 34: 4780–4790.
Acknowledgements
We kindly thank Blanca Randel, Yvonne Schleehuber, Gaby Beine, Susanne Steiner, Barbara Reddemann and Alena Heimsoeth for technical assistance. Romidepsin was provided by Gloucester Pharmaceuticals (Celgene, Signal Pharmaceuticals, LLC, San Diego, CA, USA; MTA ID # CC0488464).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Nettersheim, D., Arndt, I., Sharma, R. et al. The cancer/testis-antigen PRAME supports the pluripotency network and represses somatic and germ cell differentiation programs in seminomas. Br J Cancer 115, 454–464 (2016). https://doi.org/10.1038/bjc.2016.187
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2016.187
- Springer Nature Limited
Keywords
This article is cited by
-
Therapeutical interference with the epigenetic landscape of germ cell tumors: a comparative drug study and new mechanistical insights
Clinical Epigenetics (2022)
-
Sequential enhancer state remodelling defines human germline competence and specification
Nature Cell Biology (2022)
-
Subcellular localization of the mouse PRAMEL1 and PRAMEX1 reveals multifaceted roles in the nucleus and cytoplasm of germ cells during spermatogenesis
Cell & Bioscience (2021)
-
Application of miRNAs in the diagnosis and monitoring of testicular germ cell tumours
Nature Reviews Urology (2020)
-
CDK4/6 inhibition presents as a therapeutic option for paediatric and adult germ cell tumours and induces cell cycle arrest and apoptosis via canonical and non-canonical mechanisms
British Journal of Cancer (2020)