Abstract
Background:
Sessile serrated adenomas/polyps (SSA/Ps) are a putative precursor of colon cancer with microsatellite instability (MSI). However, the developmental mechanism of SSA/P remains unknown. We performed genetic analysis and genome-wide DNA methylation analysis in aberrant crypt foci (ACF), SSA/P, and cancer in SSA/P specimens to show a close association between ACF and the SSA/P-cancer sequence. We also evaluated the prevalence and number of ACF in SSA/P patients.
Methods:
ACF in the right-side colon were observed in 36 patients with SSA/Ps alone, 2 with cancers in SSA/P, and 20 normal subjects and biopsied under magnifying endoscopy. B-RAF mutation and MSI were analysed by PCR–restriction fragment length polymorphism (RFLP) and PCR–SSCP, respectively, in 15 ACF, 20 SSA/P, and 2 cancer specimens. DNA methylation array analysis of seven ACF, seven SSA/P, and two cancer in SSA/P specimens was performed using the microarray-based integrated analysis of methylation by isochizomers (MIAMI) method.
Results:
B-RAF mutations were frequently detected in ACF, SSA/P, and cancer in SSA/P tissues. The number of methylated genes increased significantly in the order of ACF<SSA/P<cancer. The most commonly methylated genes in SSA/P were PQLC1, HDHD3, RASL10B, FLI1, GJA3, and SLC26A2. Some of these genes were methylated in ACF, whereas all genes were methylated in cancers. Immunohistochemistry revealed their silenced expression. Microsatellite instability and MLH1 methylation were observed only in cancer. The prevalence and number of ACF were significantly higher in SSA/P patients than in normal subjects. A significant correlation was seen between the numbers of SSA/P and ACF in SSA/P patients.
Conclusions:
Our results suggest that ACF are precursor lesions of the SSA/P-cancer sequence in patients with SSA/P, where ACF arise by B-RAF mutation and methylation of some of the six identified genes and develop into SSA/Ps through accumulated methylation of these genes.
Similar content being viewed by others
Main
Recently, the serrated pathway to colorectal cancer, in which serrated polyps develop into cancers, has received much attention as an alternative pathway in colorectal carcinogenesis. Serrated polyps are categorised into three main subtypes: hyperplastic polyps, sessile serrated adenoma-polyps (SSA/P), and traditional serrated adenoma, according to the latest World Health Organization (WHO) classification (Snover et al, 2010). Of these, SSA/P is reported to develop predominantly in the proximal colon and caecum (right-side colon), and also to harbour frequent B-RAF mutations (Kambara et al, 2004; Higuchi et al, 2005; Spring et al, 2006). Moreover, it has been reported that MINT or p16 genes, markers of a CpG island methylator phenotype (CIMP), are methylated in SSA/P (O'Brien et al, 2006; Kim et al, 2011). As right-side colon cancers frequently show a microsatellite instability (MSI) phenotype owing to methylation of the MLH1 gene, SSA/P is a putative precursor lesion for MSI cancer (Cunningham et al, 1998; Huang et al, 2011; Bettington et al, 2013). However, only several genes, most of which are CIMP markers, have been investigated for methylation analysis in SSA/P tissues. Recently Gaiser et al (2013) performed genome-wide methylation analysis of colorectal polyps including SSA/P specimens and cancers, but they did not show any specific methylated genes that might be involved in SSA/P development. Moreover, it remains unknown whether or not the epi-driver genes are silenced by aberrant methylation and thereby contribute to the development of SSA/P.
Aberrant crypt foci (ACF), methylene blue-stainable crypts in the colorectum, are the earliest precancerous lesion in rodent models of colorectal carcinogenesis (Bird, 1987). We previously observed human ACF using magnifying endoscopy; a significant correlation between the number of adenomas and number/size of rectal ACF in adenoma patients was demonstrated, and the ACF were frequently positive for K-RAS mutation (Takayama et al, 1998, 2001, 2005). Moreover, we and other investigators have shown that the number of rectal ACF is significantly higher in patients with colorectal adenomas and cancers than in normal subjects, suggesting that ACF are a precursor lesion of adenomas and cancers (Takayama et al, 1998; Hurlstone et al, 2005; Seike et al, 2006; Kim et al, 2008), although some contradictory studies have also been reported (Cho et al, 2008; Mutch et al, 2009). To date, there have been only a few studies on ACF in the right-side colon (Shpitz et al, 1998, Drew et al, 2014). However, no studies have investigated ACF in the right-side colon in patients with serrated polyps including SSA/P, and the relationship between ACF and SSA/P is currently unknown.
In this study, therefore, we performed analyses of B-RAF and K-RAS mutations, MSI, and genome-wide DNA methylation array using ACF in the right-side colon, SSA/P, and cancer in SSA/P specimens to clarify the molecular mechanism of the assumed ACF-SSA/P-cancer sequence. We also investigated the prevalence and number of ACF in the right-side colon of SSA/P patients compared with normal subjects to show a close association between SSA/P and ACF in the right-side colon.
Materials and methods
Subjects and study design
This study was approved by the ethics committee of Tokushima University Hospital (Tokushima, Japan). We first enrolled 20 patients with SSA/P (without cancer) and 2 patients with cancers in SSA/P from January 2012 through March 2013. All the patients had been known or suspected to have SSA/P lesions in the colon and were referred to our hospital for endoscopic removal. We investigated the number of ACF in the ascending colon and caecum (defined as the right-side colon in this study) and biopsied them under magnifying endoscopy, after removal of the SSA/P lesions by endoscopic mucosal resection (EMR). The histological diagnosis of SSA/P was made independently by two pathologists (TF and KS) according to the criteria of WHO (Snover et al, 2010). Only lesions diagnosed as SSA/P concordantly by the pathologists were used. We also investigated B-RAF and K-RAS mutations, and MSI in 15 ACF, 20 SSA/P, and 2 cancer in SSA/P specimens, and performed genome-wide DNA methylation array analysis of seven ACF, seven SSA/P, and two cancer in SSA/P specimens. An additional 16 patients with SSA/P and 20 normal subjects were enrolled from August 2013 through February 2014 for assessment of ACF. Normal subjects were defined as subjects who were referred to our hospital for colonoscopy because of symptoms such as abdominal discomfort, distention, or a feeling of tightness on defecation, but with no apparent lesions of the colon observable by colonoscopy. Witten informed consent was obtained from all patients who had been known or suspected to have SSA/P before colonoscopy. For normal subjects, consent was obtained before colonoscopy to undergo ACF observation if no apparent lesion was identified. The baseline characteristics of patients with SSA/P are shown in Supplementary Table 1. The mean age and sex ratio (male/female) among patients with SSA/P and normal subjects were 62.1±13.0 years and 21 out of 17 vs 62.5±10.4 years and 11 out of 9, respectively.
ACF observation by magnifying endoscopy
A magnifying endoscope (model EC-L590ZW, FUJIFILM Holdings Corp., Tokyo, Japan) that magnifies objects by a factor of 135, equipped with an autofocusing device, was used throughout the examination. All subjects underwent total colonoscopy. In patients with SSA/P, after the SSA/P lesion was removed, the right-side colon was examined for ACF as previously described (Takayama et al, 1998, 2001). It was washed thoroughly with water, sprayed with 0.25% methylene blue solution, washed again thoroughly with water, and ACF were carefully identified using magnifying endoscopy. In normal subjects, after total colonoscopy, the right-side colon was examined for ACF using the same procedure. Regarding the accuracy of our ACF counting method, we previously reported that the inter-rater agreement rates and Cronbach’s alpha were sufficiently high (Takayama et al, 2011). All procedures were recorded on videotape and evaluated by two independent observers who were unaware of the subjects’ clinical histories. ACF were defined as minute lesions identifiable under magnifying chromoendoscopy in which crypts were more darkly stained with methylene blue than normal crypts (Roncucci et al, 1991; Takayama et al, 1998).
Two-PCR and RFLP for detection of B-RAF codon 600 and K-RAS codon 12 and 13 mutations
B-RAF codon 600 and K-RAS codon 12 and 13 mutations were detected using a 2-step PCRRFLP method, as previously described (Miyanishi et al, 2001; Dote et al, 2004; Nagasaka et al, 2004). In brief, cellular DNA was extracted from EMR or biopsy specimens of SSA/P or ACF and used as a template for PCR. The PCR products were amplified using mismatched primers and analysed by RFLP to detect point mutations in B-RAF codon 600 and in K-RAS codons 12 and 13. The cancer portion of the cancer in SSA/P tissue was macrodissected and DNA was extracted for PCR–RFLP analysis.
MSI analysis
Microsatellite instability analysis was performed using cellular DNA as a template for PCR. The pentaplex PCR system that includes primer pairs for five microsatellite targets (BAT-25, BAT-26, D2S123, D5S346, and D17S250) was used according to the method of You et al (2010) with minor modification. Tumours with instability at ⩾2 markers were classified as high-degree microsatellite instability (MSI-H), at 1 marker as low-degree microsatellite instability (MSI-L), and at 0 markers as microsatellite stable (MSS).
DNA methylation array analysis
A genome-wide DNA methylation array analysis was performed using the microarray-based integrated analysis of methylation by isochizomers (MIAMI) method, as previously described (Hatada et al, 2006; Horii et al, 2009; Kobayashi et al, 2012). In brief, this method utilises two isochizomers, Hpa II and Msp I, which recognise the same DNA sequence (CCGG). Genomic DNA was first digested with Hpa II, a methylation-sensitive restriction enzyme that only cleaves unmethylated DNA, and then adaptor-ligated and amplified by PCR with the primers for adaptor sequences. They were then digested with Msp I, a methylation-insensitive enzyme that digests CCGG sites irrespective of their methylation status, followed by amplification with the same set of primers (Hpa II-Msp I treatment). The second digestion with Msp I only yielded products from unmethylated DNA fragments. Therefore, only HpaII-cleavable unmethylated DNA fragments were amplified. The amplified products were labelled with Cy3 or Cy5 and co-hybridised to a microarray spotted with 38172 oligonucleotides covering the vicinity of the transcription start sites of 15883 genes (Agilent ChIP-on-Chip Custom Microarray, Agilent Technologies, Santa Clara, CA, USA). After hybridisation, the membranes were scanned, and the fluorescence intensities were quantified and normalised. The same samples were digested first with Msp I instead of Hpa II (Msp I—Msp I treatment) and analysed on a duplicate array to correct for false-positives.
Methylation-specific PCR
The bisulphite-modified DNA samples were used as a template for methylation-specific PCR (MSP). The methylation status of the sites in the PQLC1, HDHD3, RASL10B, FLI1, GJA3, and SLC26A2 genes identified by methylation array analysis was investigated by MSP, as previously described (Brinkhuizen et al, 2012). The primers used for MSP are described in Supplementary Table 2.
Immunohistochemistry
Immunohistochemical staining was performed using the streptavidin-biotin-peroxidase method with labelled streptavidin-biotin (LSAB, Dako, Kyoto, Japan), according to the manufacturer’s instructions. Briefly, paraffin-embedded sections were deparaffinised in xylene and hydrated in graded ethanol solutions and phosphate-buffered saline. Endogenous peroxidase was inactivated by incubation with 0.3% H2O2–MeOH. Subsequently, the slides were heated in 0.01 M citrate buffer in a water bath at 95 °C (pH=6.0) for 15 min. Rabbit anti-human FLI1 polyclonal antibody (Kubo et al, 2003) (diluted 1 : 150, Santa Cruz Biotechnology, Santa Cruz, CA, USA), rabbit anti-human GJA3 polyclonal antibody (Banerjee et al, 2010) (diluted 1 : 100, Funakoshi Co., Ltd Tokyo, Japan), and goat anti-human SLA26A2 polyclonal antibody (Haila et al, 2001) (diluted 1 : 250, Sigma-Aldrich), rabbit anti-human PQLC1 polyclonal antibody (HPA051666) (diluted 1 : 150, Sigma-Aldrich, St Louis, MO, USA), rabbit anti-human HDHD3 polyclonal antibody (HPA024158) (diluted 1 : 100, Sigma-Aldrich) and rabbit anti-human RASL10B polyclonal antibody (HPA046842) (diluted 1 : 100, Sigma-Aldrich) were used as primary antibodies. Detailed data for the HPA antibodies are listed at the website http://www.proteinatlas.org/. The sections were incubated with primary antibodies, washed with PBS, and incubated with secondary biotinylated antibody from an LSAB+ peroxidase kit (Dako). Subsequently, the sections were incubated with streptavidin-horseradish peroxidase (HRP) conjugate and visualised with DAB chromogen (3', 3-diaminobenzidine, Dako). Finally, the sections were counterstained with Mayer's hematoxylin.
Statistics
All data were analysed using STATA version 8 software (Stata Corp., College Station, TX, USA). ANOVA was used to assess differences in the number of methylated genes among ACF, SSA/P, and cancer in SSA/P specimens. Scheffe’s test was used to compare the numbers of methylated genes between the groups. The correlation between the number of SSA/P and the number of ACF was evaluated by Spearman’s test. A P-value <0.05 was considered significant.
Results
Endoscopic appearance of SSA/P and ACF
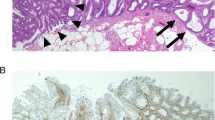
Figure 1A shows a representative endoscopic view of SSA/P with a sessile isochromatic appearance in the ascending colon. Histological examination of this lesion revealed distorted and dilated crypts near the base with serrated architecture and no cytological dysplasia, consistent with SSA/P (Figure 1B) (Higuchi et al, 2005; Snover et al, 2010). Figure 1C shows a representative endoscopic view of ACF in the right-side colon of the same case. ACF could be identified as a focus consisting of abnormal crypts darkly stained with methylene blue. Histological examination revealed a serrated structure and distortion in some of the crypts but no cytological dysplasia (Figure 1D).
Endoscopic and histologic features of SSA/P ( A and B ) and ACF ( C and D ) in the right-side colon. (A) Representative endoscopic view of SSA/P with 10 mm diameter. The size was estimated using biopsy forceps. (B) Histological examination showed serration in the lower crypt, and distorted, dilated, or anchor-shaped crypts (H&E staining). (C) Representative endoscopic view of ACF in the right-side colon from the same patient (case 7). ACF were identified by methylene blue staining under magnifying endoscopy. ACF consisted of larger and more darkly stained crypts than normal crypts. (D) Histological examinations showed serration and distortion of the crypts (H&E staining).
B-RAF and K-RAS mutations in ACF, SSA/P, and cancer in SSA/P
As B-RAF mutations at codon 600 are frequently positive in SSA/P tissues (Kambara et al, 2004; Spring et al, 2006), we first examined B-RAF mutations in 15 ACF, 20 SSA/P, and 2 cancer in SSA/P specimens. B-RAF codon 600 mutations were detected in 16 out of 20 (80%) SSA/P specimens, consistent with previous reports (Kambara et al, 2004; Spring et al, 2006). They were also detected in 10 out of 15 (66.7%) ACF and 2 out of 2 (100%) cancer in SSA/P specimens (Figure 2A). Thus, B-RAF mutations were frequently present in ACF, SSA/P, and cancer in SSA/P specimens, raising the possibility that the ACF in patients with SSA/P are precursor lesions of the SSA/P-cancer sequence.
Analysis for B-RAF codon 600 and K-RAS codon 12 mutations in ACF, SSA/P, and cancer in SSA/P. Point mutations of B-RAF codon 600 and K-RAS codon 12 were examined using the 2-step PCR–RFLP method. (A) B-RAF mutations in ACF, SSA/P, and cancer in SSA/P. The HT-29 colon cancer cell line, which is known to have a B-RAF mutation, was used as a positive control. Normal colonic mucosa was used as a negative control. (B) K-RAS mutations in ACF, SSA/P, and cancer in SSA/P. The LS174T colon cancer cell line, which is known to have a K-RAS mutation, was used as a positive control. A1 represents ACF from case 1. S1 and C18 represent SSA/P and cancer in SSA/P from case 1 and case 18, respectively.
We next examined K-RAS mutations at codon 12 and 13 because these mutations are frequently seen in rectal ACF, adenoma, and cancer (Takayama et al, 1998, 2001). However, K-RAS mutations at codon 12 were only detected in 2 out of 15 (13.3%) ACF, 2 out of 20 (10.0%) SSA/P, and 0 out of 2 (0%) cancerous portions in SSA/P specimens (Figure 2B). No K-RAS mutations at codon 13 were detected in any of the ACF, SSA/P, or cancer in SSA/P specimens (Supplementary Figure 1).
MSI in ACF, SSA/P, and cancer in SSA/P
As colorectal cancers with MSI develop predominantly in the right-side colon, we next examined MSI status in 15 ACF, 20 SSA/P, and 2 cancer in SSA/P specimens. All 15 ACF specimens were MSS, and there were no MSI-L, or MSI-H phenotypes. Of the 20 SSA/P specimens, 17 were MSS and 3 were MSI-L; however, none were MSI-H. While one of the cancer in SSA/P specimens was MSI-H (case 36) and the other was MSS (case 18). The latter cancer was positive for p53 mutation (Supplementary Figure 2). These results for the cancer in SSA/P specimens were consistent with previous reports (Jass et al, 2006; Fujita et al, 2011; Maeda et al, 2011; Ban et al, 2014). The representative results of MSI analysis in ACF, SSA/P, and cancer in SSA/P are shown in Supplementary Figure 3.
DNA methylation array analysis of ACF, SSA/P, and cancer in SSA/P
Genome-wide DNA methylation analysis of 7 ACF, 7 SSA/P, and 2 cancer in SSA/P specimens was performed using the MIAMI method in comparison with the corresponding normal colonic epithelia. As a majority of these three lesions were positive for B-RAF mutations, in this particular methylation analysis, all lesions with B-RAF mutations were analysed except for one ACF sample. Representative scatter plots of the signals from each probe in ACF (case 5), SSA/P (case 3), and cancer in SSA/P specimens (case 18) are shown in Figure 3. The values for log ((HpaII intensity) lesion/(HpaII intensity) normal) are plotted on the x-axis, representing the relative methylation changes of each lesion. The values for the log ((Msp I intensity) lesion/(Mspl intensity) normal) are plotted on the y-axis, representing the control for the enzyme effects at sample digestion. The threshold values were determined according to the original MIAMI method described by Hatada et al (Hatada et al, 2006). Dots located within the upper and lower green lines (± log 2, respectively) and on the right side of the yellow line at log 5 of the horizontal distance from the regression line of the plots represent hypermethylated genes in each lesion compared with paired normal colorectal epithelium: 9 genes were determined to be methylated in the ACF specimen. Likewise, 32 genes and 165 genes were methylated in SSA/P and cancer in SSA/P specimens, respectively. The mean number of methylated genes in ACF, SSA/P, and cancer in SSA/P specimens were 11.3±7.7, 37.0±17.3, and 193±39, respectively, showing a significant stepwise increment from ACF to SSA/P and then to cancer in SSA/P (Figure 4, P<0.05).
Scatter plots of signals for each probe based on the microarray-based integrated analysis of methylation by isochizomers (MIAMI) method in ACF ( A ), SSA/P ( B ), and cancer in SSA/P ( C). Green lines represent y=± log2, blue lines represent x=± log5, and yellow lines are located at±log5 horizontal distance from the regression line (red line) of the plots in accordance with the original MIAMI method of Hatada et al (2006). Dots located within the two green lines and on the right side of the yellow line were determined to be hypermethylated.
On the basis of the results of the methylation array analysis, we searched for common methylated genes among the seven SSA/P tissues and found that PQLC1 and HDHD3 genes were the most commonly methylated genes; they were methylated in six out of seven cases (86%) (Figure 5). In addition, RASL10B gene was methylated in five out of seven cases (71%) and FLI1, GJA3, and SLC26A2 genes were methylated in four out of seven cases (57%) respectively. All six genes were commonly methylated in the two cancer in SSA/P cases. In ACF tissues, PQLC1 was methylated in three out of seven cases (43%); SLC26A2, RASL10B and FLI1 genes were methylated in two out of seven cases (29%); and GJA3 was methylated in one out of seven cases (14%). The methylation status of these genes, and B-RAF, K-RAS mutations and MSI status in each lesion are summarised in Figure 5.
Commonly methylated genes in SSAP tissues. The most commonly methylated genes in seven SSA/P tissues were PQLC1, HDHD3, RASL10B, FLI1, GJA3, and SLC26A2. The methylation status of these six genes, and B-RAF, K-RAS mutations and microsatellite instability (MSI) status in seven ACF, seven SSA/P, and two cancer in SSA/P specimens are summarised. MSS, microsatellite stable; MSI-H, MSI-high.
Validation of methylation array analysis by MSP
To validate the results of methylation array analysis, we assessed the methylation status of the six genes by MSP. Bands of 500 bp representing methylation of PQLC1 gene were detected in SSA/P tissues from cases 1–4, 6, and 7; ACF tissue from cases 5–7; and cancer tissues from cases 1 and 2 (Figure 6). However, no methylation bands were detected in SSA/P specimens that did not exhibit methylation in the methylation array analysis (data not shown). Thus, the methylation of PQLC1 gene was validated by MSP. Likewise, methylation bands of HDHD3 gene (450 bp), RASL10B gene (600 bp), FLI1 gene (550 bp), GJA3 gene (360 bp), and SLC26A2 gene (350 bp) were confirmed in the corresponding specimens that exhibited methylation in the methylation array analysis. These data indicate that the methylation of all six genes was validated by MSP.
Expression of six genes in SSA/P tissues
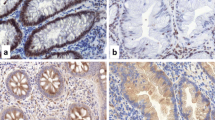
To determine whether the expression of the six genes was silenced by aberrant methylation, we performed immunohistochemical staining on SSA/P tissues that showed methylation of the six genes; three SSA/P tissues were stained for expression of each gene. Representative staining patterns are shown in Figure 7. PQLC1, an unknown protein, showed staining in the cytoplasm and nucleus of normal epithelial cells. However, its staining was clearly diminished in the SSA/P cells (Figure 7A). HDHD3, also an unknown protein, was appreciably stained in the cytoplasm of normal epithelial cells, but its staining was markedly reduced in the SSA/P cells (Figure 7B). RASL10B, a small GTPase protein, was intensely stained in the cytoplasm of normal epithelial cells, but no such staining was present in the SSA/P cells (Figure 7C). FLI1 stained mainly the membrane of normal epithelial cells, whereas it was not stained in the SSA/P cells (Figure 7D). GJA3, a membrane protein, was stained predominantly in the membrane of normal epithelial cells, but was almost negative in the SSA/P tissue (Figure 7E). SLC26A2, an anion transporter, was present in the luminal side of normal epithelial cells. However, its staining was essentially negative in the SSA/P cells (Figure 7F). Thus, protein expression of the six genes was markedly decreased or silenced in all SSA/P tissues examined. These results strongly suggest that aberrant methylation of these genes causes silencing or a decrease of protein expression by inhibition of transcription.
Immunohistochemical staining for PQLC1, HDHD3, RASL10B, FLI1, GJA3, and SLC26A2 in SSA/P tissues. Representative staining patterns of PQLC1 (A) HDHD3 (B) RASL10B (C) FLI1 (D) GJA3 (E) and SLC26A2 (F) are shown. Original magnification; ×100. High-magnification images of each boxed area are shown in the inset (× 400).
Prevalence and number of ACF in patients with SSA/P and normal subjects
We also investigated the prevalence and number of ACF in the right-side colon of 38 SSA/P patients compared with 20 normal subjects using magnifying endoscopy to strengthen the hypothesis that ACF are a precursor lesion of the SSA/P-cancer sequence. The prevalence of ACF in SSA/P patients was 37out of 38 (97.4%), which was significantly higher than that in normal subjects (2 out of 20, 10.0%). The mean number of ACF in SSA/P patients was 3.79±2.11, which was significantly higher than that in normal subjects (0.10±0.33) (P<0.01). Moreover, there was a significant positive correlation between the number of ACF and the number of SSA/P (P<0.05) (Supplementary Figure 4). These data, in combination with epigenetic and genetic findings of ACF, suggest that ACF in the right-side colon are precursor lesions of the SSA/P-cancer sequence.
Discussion
In this study, we found frequent B-RAF mutations in ACF of the right-side colon, SSA/P, and cancer in SSA/P, and also a stepwise increment of methylated genes in this order. Moreover, the number of methylated genes in ACF of right-side colon was 11.3±7.7 (range, 4–28), whereas it was only 1.3±1.0 (range, 0–2) in rectal ACF (Supplementary Table 3). Previously, we and other investigators showed that rectal (and sigmoidal) ACF are frequently positive for K-RAS mutations but not B-RAF mutations. These results suggest that ACF in the right-side colon is genetically distinct from rectal ACF and is a putative precursor lesion of the SSA/P-cancer sequence. Our results also suggest that B-RAF mutation is an early event associated with DNA methylation in colon carcinogenesis via SSA/P.
One out of the two cancer (in SSA/P) tissues showed an MSI-H phenotype with MLH1 methylation, whereas the other one showed an MSS phenotype with p53 mutation. Although the number of cancer in SSA/P tissues examined was small, these results were consistent with previous reports indicating that there are two mechanistic pathways involved in the SSA/P-(dysplasia-) cancer sequence; one through MLH1 methylation and the other through p53 mutation (Jass et al, 2006; Fujita et al, 2011; Maeda et al, 2011; Ban et al, 2014).
Our methylation array analysis revealed that six novel genes (PQLC1, HDHD3, RASL10B, FLI1, GJA3, and SLC26A2) were most frequently methylated, and their protein expression was suppressed in SSA/P tissues. The statistical power calculation revealed significant positivity of methylation in PQLC1 (P<0.01), HDHD3 (P<0.01), and RASL10B (P<0.05) by Fisher’s exact test. Similarly, there was marginal positivity of methylation in FLI1, GJA3, and SLC26A2 (P=0.069). Moreover, all six genes were methylated in cancers in SSA/P tissues (100%), whereas, only some of them were methylated in ACF tissues. These results suggest that epigenetic silencing of these six genes has an important role in the development of SSA/P. Thus, our findings suggest an ACF-SSA/P-cancer sequence where ACF arise by B-RAF mutation and methylation of some of the six identified genes, then develop into SSA/Ps through accumulated methylation of these genes, and probably progress to cancer by additional epigenetic or genetic alterations. Interestingly, the expressions of these six genes were frequently suppressed in the right-side colon cancers with no apparent SSA/P as well, whereas they were not suppressed in most of the left-side colon cancers and adenomas as revealed by immunohistochemistry (Supplementary Figure 5). These results raise the possibility that these six genes have an important role in the right-side colon carcinogenesis.
Our clinical study revealed numerous ACF in the right-side colon of SSA/P patients, whereas there were only few ACF in the right-side colon of normal subjects. There was also a significant correlation between the number of ACF and the number of SSA/P in SSA/P patients. These results support the hypothesis that ACF in the right-side colon are a precursor lesion of the SSA/P-cancer sequence. It has been hypothesised that SSA/Ps develop through hyperplastic polyps, particularly through microvesicular hyperplastic polyps (MVHPs). As histological examination of ACF in the right-side colon revealed microvesicles in the crypts (Figure 1D), which is one of the characteristics of MVHP, we may assume a pathway from ACF to MVHP and subsequently to SSA/P. In the present study, however, we did not investigate ACF in the right-side colon from patients with other types of polyps including adenomas and traditional serrated adenomas. Therefore, it is not yet clear whether ACF in the right-side colon are specific to SSA/P or not. It will be important in the future to investigate ACF in these patients in comparison with ACF in patients with SSA/P.
It has been reported that marker genes for a CIMP such as MINT1, MINT3, FGFBP3, or SLIT2 are methylated in SSA/P tissues (Kambara et al, 2004; O'Brien et al, 2006; Kaji et al, 2012; Beggs et al, 2013). In our study, however, methylation of these genes was seen in only one to two of the two SSA/P specimens, suggesting that these genes do not have a pivotal role in the development of SSA/P. In addition, the methylation sites of two genes identified (PQLC1 and SLC26A2) were inside the CpG islands (defined as CpG observed/expected>60%); interestingly, however, those of the remaining four genes were outside the CpG islands (Supplementary Figure 6). It is plausible that gene methylation at sites other than CpG islands are also involved in gene silencing in SSA/P tissues.
Of the six genes identified, PQLC1 and HDHD3 are not yet well characterised; their tissue distribution and function are unknown. The significance of epigenetic silencing of these genes in SSA/P as well as in colon tumours should be elucidated in detail in the future. RASL10B protein is a small monomeric GTPase with tumour suppressor potential, and epigenetic silencing of RASL10B in human HCC and breast cancer has been reported (Zou et al, 2006; Lin and Chuang, 2012). Although reduced expression of RASL10B in colonic tumours has not yet been reported, it is presumed to function in normal colon epithelial cells as an oncosuppressive factor, as in HCC and breast cancer. FLI1 is an ETS family member and EWS/FLI1 fusion gene (11;22 translocation) that is known to have oncogenic activity in Ewing's sarcoma (May et al, 1993). However, recently FLI1 expression was reportedly reduced by aberrant methylation in its promoter region in colorectal adenoma and cancer (Oster et al, 2011). Therefore, FLI1 epigenetic silencing may provide an advantage for cell growth in SSA/P cells. GJA3, same as connexin 46, is a gap junction protein (Hsieh et al, 1991). In general, expression of the connexin gene family is downregulated in cancer cells in association with the promotion of cell proliferation, or enhanced invasiveness. Although the expression of GJA3 in colorectal tumours has not yet been reported, silencing of GJA3 may contribute to promotion of cell proliferation and gap junction impairment resulting in crypt serration in SSA/P. Alternatively, it may be associated with mucin production because some connexins (e.g. connexin 30) are reportedly closely associated with mucin expression (Sentani et al, 2010). SLC26A2, also called diastrophic dysplasia sulphate transporter (DTDST), is an anion transporter and its epigenetic silencing in colon cancer cell lines has been reported recently by Yusa et al (2010). They also showed that knockdown of SLC26A2 in a colon cancer cell line increased proliferation. Therefore, silencing of SLC26A2 presumably promotes cell proliferation in SSA/P cells. However, there have been no reports to date indicating that patients with homozygous SLC26A2-inactivating germline mutation (diastrophic dysplasia) are predisposed to cancer. It is plausible that a single gene alteration without B-RAF mutation may not be sufficient for the development of SSA/P and subsequent cancer, although the possibility still remains that SLC26A2 silencing might be a simple consequence of the process of SSA/P formation. Thus, epigenetic silencing of these six genes may provide an advantage for cell growth, formation of serrated architecture, or mucin production, all of which are characteristic findings of SSA/P.
In this study, we employed the MIAMI method, which has lower resolution and sensitivity compared with the most recent comprehensive methods using next generation sequencing technology, and successfully identified several candidate genes. The use of such the newest technologies for genome-wide screening of methylated genes (sites) will be able to identify additional genes that are differentially methylated in SSA/P. Therefore, further experiments using these technologies will be needed to provide a more detailed analysis of the underlying mechanisms of carcinogenesis via SSA/P in the colon.
In conclusion, our results suggest that ACF are precursor lesions of the SSA/P-cancer sequence in patients with SSA/P. Our data also suggest that the B-RAF mutation and accumulated aberrant methylation of the six novel genes (PQLC1, HDHD3, RASL10B, FLI1, GJA3, or SLC26A2) are closely associated with development of SSA/P.
Change history
20 January 2015
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Ban S, Mitomi H, Horiguchi H, Sato H, Shimizu M (2014) Adenocarcinoma arising in small sessile serrated adenoma/polyp (SSA/P) of the colon: clinicopathological study of eight lesions. Pathology International 64: 123–132.
Beggs AD, Jones A, Shepherd N, Arnaout A, Finlayson C, Abulafi AM, Morton DG, Matthews GM, Hodgson SV, Tomlinson IP (2013) Loss of expression and promoter methylation of SLIT2 are associated with sessile serrated adenoma formation. PLoS Genet 9: e1003488.
Banerjee D, Gakhar G, Madgwick D, Hurt A, Takemoto D, Nguyen TA (2010) A novel role of gap junction connexin46 protein to protect breast tumors from hypoxia. Int J Cancer 127: 839–848.
Bettington M, Walker N, Clouston A, Brown I, Leggett B, Whitehall V (2013) The serrated pathway to colorectal carcinoma: current concepts and challenges. Histopathology 62: 367–386.
Bird RP (1987) Observations and quantification of aberrant crypts in the murine colon treated with a colon carcinogen: preliminary findings. Cancer Lett 37: 147–151.
Brinkhuizen T, van den Hurk K, Winnepenninckx VJ, Hoon JP, van Marion AM, Veeck J, van Engeland M, van Steensel MA (2012) Epigenetic changes in Basal Cell Carcinoma affect SHH and WNT signaling components. PLoS One 7: e51710.
Cho NL, Redston M, Zauber AG, Carothers AM, Hornick J, Wilton A, Sontag S, Nishioka N, Giardiello FM, Saltzman JR, Gostout C, Eagle CJ, Hawk ET, Bertagnolli MM (2008) Aberrant crypt foci in the adenoma prevention with celecoxib trial. Cancer Prev Res (Phila Pa) 1: 21–31.
Cunningham JM, Christensen ER, Tester DJ, Kim CY, Roche PC, Burgart LJ, Thibodeau SN (1998) Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res 58: 3455–3460.
Dote H, Tsukuda K, Toyooka S, Yano M, Pass HI, Shimizu N (2004) Mutation analysis of the BRAF codon 599 in malignantpleural mesothelioma by enriched PCR-RFLP. Oncol Rep. 11: 361–363.
Drew DA, Devers TJ, O'Brien MJ, Horelik NA, Levine J, Rosenberg DW (2014) HD chromoendoscopy coupled with DNA mass spectrometry profiling identifies somatic mutations in microdissected human proximal aberrant crypt foci. Mol Cancer Res 12: 823–829.
Fujita K, Yamamoto H, Matsumoto T, Hirahashi M, Gushima M, Kishimoto J, Nishiyama K, Taguchi T, Yao T, Oda Y (2011) Sessile serrated adenoma with early neoplastic progression: a clinicopathologic and molecular study. Am J Surg Pathol 35: 295–304.
Gaiser T, Meinhardt S, Hirsch D, Killian JK, Gaedcke J, Jo P, Ponsa I, Miró R, Rüschoff J, Seitz G, Hu Y, Camps J, Ried T (2013) Molecular patterns in the evolution of serrated lesion of the colorectum. Int J Cancer 132: 1800–1810.
Haila S, Hästbacka J, Böhling T, Karjalainen-Lindsberg ML, Kere J, Saarialho-Kere U (2001) SLC26A2 (diastrophic dysplasia sulfate transporter) is expressed in developing and mature cartilage but also in other tissues and cell types. J Histochem Cytochem 49: 973–982.
Hatada I, Fukasawa M, Kimura M, Morita S, Yamada K, Yoshikawa T, Yamanaka S, Endo C, Sakurada A, Sano M, Kondo T, Horii A, Ushijima T, Sasaki H (2006) Genome-wide profiling of promoter methylation inhuman. Oncogene 25: 3059–3064.
Higuchi T, Sugihara K, Jass JR (2005) Demographic and pathological characteristics of serrated polyps of colorectum. Histopathology 47: 32–40.
Horii T, Morita S, Kimura M, Izuho H (2009) Epigenetic regulation of adipocyte differentiation by a Rho guanine nucleotide exchange factor, WGEF. PLoS One 4: e5809.
Hsieh CL, Kumar NM, Gilula NB, Francke U (1991) Distribution of genes for gap junction membrane channel proteins on human and mouse chromosomes. Somat Cell Mol Genet 17: 191–200.
Huang CS, Farraye FA, Yang S, O’Brien MJ (2011) The clinical significance of serrated polyps. Am J Gastroenterol 106: 229–240.
Hurlstone DP, Karajeh M, Sanders DS, Drew SK, Cross SS (2005) Rectal aberrant crypt foci identified using high-magnification-chromoscopic colonoscopy: biomarkers for flat and depressed neoplasia. Am J Gastroenterol 100: 1283–1289.
Jass JR, Baker K, Zlobec I, Higuchi T, Barker M, Buchanan D, Young J (2006) Advanced colorectal polyps with the molecular and morphological features of serrated polyps and adenomas: concept of a 'fusion' pathway to colorectal cancer. Histopathology 49: 121–131.
Kaji E, Uraoka T, Kato J, Hiraoka S, Suzuki H, Akita M, Saito S, Tanaka T, Ohara N, Yamamoto K (2012) Externalization of saw-tooth architecture in small serrated polyps implies the presence of methylation of IGFBP7. Dig Dis Sci. 57: 1261–1270.
Kambara T, Simms LA, Whitehall VL, Spring KJ, Wynter CV, Walsh MD, Baeker MA, Arnold S, McGivern A, Matsubara N, Tanaka N, Higuchi T, Young J, Jass JR, Leggett BA (2004) BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut 53: 1137–1144.
Kim J, Ng J, Arozulllah A, Ewing R, Llor X, Carroll RE, Benya RV (2008) Aberrant crypt focus size predicts distal polyp histopathology. Cancer Epidemiol Biomarkers Prev 17: 1155–1162.
Kim KM, Lee EJ, Ha S, Kang SY, Jang KT, Park CK, Kim JY, Kim YH, Chang DK, Odze RD (2011) Molecular features of colorectal hyperplastic polyps and sessile serrated adenoma/polyps from Korea. Am J Surg Pathol 35: 1274–1286.
Kobayashi Y, Aizawa A, Takizawa T, Yoshizawa C, Horiguchi H, Ikeuchi Y, Kakegawa S, Watanabe T, Maruyama K, Morikawa A, Hatada I, Arakawa H (2012) DNA methylation changes between relapse and remission of minimal change nephrotic syndrome. Pediatr Nephrol 27: 2233–2241.
Kubo M, Czuwara-Ladykowska J, Moussa O, Markiewicz M, Smith E, Silver RM, Jablonska S, Blaszczyk M, Watson DK, Trojanowska M (2003) Persistent down-regulation of Fli1, a suppressor of collagen transcription, in fibrotic scleroderma skin. Am J Pathol 163: 571–581.
Lin ZY, Chuang WL (2012) Genes responsible for the characteristics of primary cultured invasive phenotype hepatocellular carcinoma cells. Biomed Pharmacother 66: 454–458.
Maeda T, Suzuki K, Togashi K, Nokubi M, Saito M, Tsujinaka S, Kamiyama H, Konishi F (2011) Sessile serrated adenoma shares similar genetic and epigenetic features with microsatellite unstable colon cancer in a location-dependent manner. Exp Ther Med 2: 695–700.
May WA, Lessnick SL, Braun BS, Klemsz M, Lewis BC, Lunsford LB, Hromas R, Denny CT (1993) The Ewing's sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol Cell Biol 13: 7393–7398.
Miyanishi K, Takayama T, Ohi M, Hayashi T, Nobuoka A, Nakajima T, Takimoto R, Kogawa K, Kato J, Sakamaki S, Niitsu Y (2001) Glutathione S-transferase-pi overexpression is closely associated with K-ras mutation during human colon carcinogenesis. Gastroenterology 121: 865–874.
Mutch MG, Schoen RE, Fleshman JW, Rall CJ, Dry S, Seligson D, Charabaty A, Chia D, Umar A, Viner J, Hawk E, Pinsky PF (2009) A multicenter study of prevalence and risk factors for aberrant crypt foci. Clin Gastroenterol Hepatol 7: 568–574.
Nagasaka T, Sasamoto H, Notohara K, Cullings HM, Takeda M, Kimura K, Kambara T, MacPhee DG, Young J, Leggett BA, Jass JR, Tanaka N, Matsubara N (2004) Colorectal cancer with mutation in BRAF, KRAS, and wild-type with respect to both oncogenes showing different patterns of DNA methylation. J Clin Oncol 22: 4584–4594.
O'Brien MJ, Yang S, Mack C, Xu H, Huang CS, Mulcahy E, Amorosino M, Farraye FA (2006) Comparison of microsatellite instability, CpG island methylation phenotype, BRAF and KRAS status in serrated polyps and traditional adenomas indicates separate pathways to distinct colorectal carcinoma end points. Am J Surg Pathol 30: 1491–1501.
Oster B, Thorsen K, Lamy P, Wojdacz TK, Hansen LL, Birkenkamp-Demtröder K, Sørensen KD, Laurberg S, Orntoft TF, Andersen CL (2011) Identification and validation of highly frequent CpG island hypermethylation in colorectal adenomas and carcinomas. Int J Cancer 129: 2855–2866.
Roncucci L, Stamp D, Medline A, Cullen JB, Bruce WR (1991) Identification and quantification of aberrant crypt foci and microadenomas in the human colon. Hum Pathol 22: 287–294.
Seike K, Koda K, Oda K, Kosugi C, Shimizu K, Nishimura M, Shioiri M, Takano S, Ishikura H, Miyazaki M (2006) Assessment of rectal aberrant crypt foci by standard chromoscopy and its predictive value for colonic advanced neoplasms. Am J Gastroenterol 101: 1362–1369.
Sentani K, Oue N, Sakamoto N, Anami K, Naitou Y, Aoyagi K, Sasaki H, Yasui W (2010) Upregulation of connexin 30 in intestinal phenotype gastric cancer and its reduction during tumor progression. Pathobiology 77: 241–248.
Snover DC, Ahnen DJ, Burt RW (2010) Serrated polyps of the colon and rectum and serrated polyposis. In Bosman FT, Carneiro F, Hruban RH, Theise ND, (eds). WHO classification of tumours of the digestive system. IARC P: Lyon, France, pp 160–165.
Shpitz B, Bomstein Y, Mekori Y, Cohen R, Kaufman Z, Neufeld D, Galkin M, Bernheim J (1998) Aberrant crypt foci in human colons: distribution and histomorphologic characteristics. Hum Pathol 29: 469–475.
Spring KJ, Zhao ZZ, Karamatic R, Walsh MD, Whitehall VL, Pike T, Simms LA, Young J, James M, Montgomery GW, Appleyard M, Hewett D, Togashi K, Jass JR, Leggett BA (2006) High prevalence of sessile serrated adenomas with BRAF mutations: a prospective study of patients undergoing colonoscopy. Gastroenterology 131: 1400–1407.
Takayama T, Katsuki S, Takahashi Y, Ohi M, Nojiri S, Sakamaki S, Kato J, Kogawa K, Miyake H, Niitsu Y (1998) Aberrant crypt foci of the colon as precursors of adenoma and cancer. N Engl J Med 339: 1277–1284.
Takayama T, Nagashima H, Maeda M, Nojiri S, Hirayama M, Nakano Y, Takahashi Y, Sato Y, Sekikawa H, Mori M, Sonoda T, Kimura T, Kato J, Niitsu Y (2011) Randomized double-blind trial of sulindac and etodolac to eradicate aberrant crypt foci and to prevent sporadic colorectal polyps. Clin Cancer Res 17: 3803–3811.
Takayama T, Ohi M, Hayashi T, Miyanishi K, Nobuoka A, Nakajima T, Satou T, Takimoto R, Kato J, Sakamaki S, Niitsu Y (2001) Analysis of K-ras, APC, and beta-catenin in aberrant crypt foci in sporadic adenoma, cancer, and familial adenomatous polyposis. Gastroenterology 121: 599–611.
Takayama T, Miyanishi K, Hayashi T, Kukitsu T, Takanashi K, Ishiwatari H, Kogawa T, Abe T, Niitsu Y (2005) Aberrant crypt foci: detection, gene abnormalities, and clinical usefulness. Clin Gastroenterol Hepatol 3 (7 Suppl 1): S42–S45.
You JF, Buhard O, Ligtenberg MJ, Kets CM, Niessen RC, Hofstra RM, Wagner A, Dinjens WN, Colas C, Lascols O, Collura A, Flejou JF, Duval A, Hamelin R (2010) Tumours with loss of MSH6 expression are MSI-H when screened with a pentaplex of five mononucleotide repeats. Br J Cancer 103: 1840–1845.
Yusa A, Miyazaki K, Kimura N, Izawa M, Kannagi R (2010) Epigenetic silencing of the sulfate transporter gene DTDST induces sialyl Lewisx expression and accelerates proliferation of colon cancer cells. Cancer Res 70: 4064–4073.
Zou H, Hu L, Li J, Zhan S, Cao K (2006) Cloning and characterization of a novel small monomeric GTPase, RasL10B, with tumor suppressor potential. Biotechnol Lett 28: 1901–1908.
Acknowledgements
We are grateful to Drs Shunto J and Kataoka K for referring SSA/P patients to our hospital, and Kajimoto M and Nakanishi Y for their expert technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Inoue, A., Okamoto, K., Fujino, Y. et al. B-RAF mutation and accumulated gene methylation in aberrant crypt foci (ACF), sessile serrated adenoma/polyp (SSA/P) and cancer in SSA/P. Br J Cancer 112, 403–412 (2015). https://doi.org/10.1038/bjc.2014.545
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2014.545
- Springer Nature Limited
Keywords
This article is cited by
-
Cryopreservation effect on DNA methylation profile in rainbow trout spermatozoa
Scientific Reports (2023)
-
DNA methylation changes that precede onset of dysplasia in advanced sessile serrated adenomas
Clinical Epigenetics (2019)
-
Associations of dietary fat with risk of early neoplasia in the proximal colon in a population-based case–control study
Cancer Causes & Control (2018)
-
Genome-wide DNA methylation profiling reveals cancer-associated changes within early colonic neoplasia
Oncogene (2017)