Abstract
Background:
This study evaluated soluble serum proteins as biomarkers to subset patients with metastatic colorectal cancer (mCRC) treated with chemotherapy±cediranib, a vascular endothelial growth factor (VEGF) signalling inhibitor (VEGFi). Exploring biomarkers at pre- and on-treatment may identify patient subgroups showing clinical benefit on cediranib combination.
Methods:
Two hundred and seven serum proteins were analysed in 588 mCRC patients at pre- and on-treatment with chemotherapy (FOLFOX/CAPOX)±cediranib 20 mg. Patients were enrolled in the phase III trial HORIZON II. We correlated baseline biomarker signatures and pharmacodynamic (PD) biomarkers with PFS and OS.
Results:
We identified a baseline signature (BS) of 47 biomarkers that included VEGFA, VEGFD, VEGFR2, VEGFR3 and TIE-2, which defined two distinct subgroups of patients. Patients treated with chemotherapy plus cediranib who had ‘high’ BS had shorter PFS (HR=1.82, P=0.003) than patients with ‘low’ BS. This BS did not correlate with PFS of the patients treated with chemotherapy plus placebo. In addition, we identified a profile of 16 PD proteins on treatment associated with PFS (HR=0.58, P<0.001) and OS (HR=0.52, P<0.001) in patients treated with chemotherapy plus cediranib. This PD profile did not correlate with PFS and OS in patients treated with chemotherapy plus placebo.
Conclusions:
Serum proteins may represent relevant biomarkers to predict the outcome of patients treated with VEGFi-based therapies. We report a BS and PD biomarkers that may identify mCRC patients showing increased benefit of combining cediranib with chemotherapy. These exploratory findings need to be validated in future prospective studies.
Similar content being viewed by others
Main
Colorectal cancer (CRC) is the third most diagnosed cancer in men and the second in women worldwide (Jemal et al, 2011). There has been a number of clinical trials investigating whether agents targeting vascular endothelial growth factor (VEGF) signalling (VEGF-signalling inhibitor (VEGFi)), provide benefit in treating a wide variety of distinct tumours, including CRC (Ferrara and Kerbel, 2005). Bevacizumab, an anti-VEGFA monoclonal antibody was the first drug targeting the VEGF-signalling pathway approved by the FDA in combination with 5-FU-based chemotherapy, and is presently standard of care in mCRC in many countries. Combining bevacizumab with IFL chemotherapy regimens has demonstrated clinical improvement in overall survival (OS) or progression-free survival (PFS) in CRC (Hurwitz et al, 2004); however, subsequent studies using next-generation chemotherapy regimens such as FOLFOX, while maintaining a PFS benefit, failed to show an OS benefit by the addition of bevacizumab (Saltz et al, 2008).
During the last decade, several other VEGF receptor tyrosine kinase inhibitors have also been developed (Abdullah and Perez-Soler, 2012). One of these agents is cediranib, a once-daily oral tyrosine kinase inhibitor with potent activity against all three VEGF receptors, and c-Kit (Wedge et al, 2005; Brave et al, 2011). Efficacy of cediranib plus FOLFOX/CAPOX (chemotherapy) vs placebo plus FOLFOX/CAPOX in patients with previously untreated mCRC has been assessed in the phase III HORIZON II trial (Hoff et al, 2012). This study met the co-primary end point of PFS prolongation with cediranib plus FOLFOX/CAPOX treatment compared with FOLFOX/CAPOX alone (HR=0.84; P=0.012). However, the OS end point was not met (HR=0.94; P=0.57). This result was consistent with other trials performed combining VEGFi’s with newer chemotherapy regimens in mCRC. Indeed, the PFS and OS results observed in the randomised controlled double blind phase III trial, HORIZON II were similar to those reported in a phase III trial assessing the efficacy of bevacizumab plus FOLFOX vs chemotherapy alone as first-line treatment for patients with mCRC (Saltz et al, 2008).
One of the main challenges for VEGFi’s is to identify the patient subgroups that receive most benefit from chemotherapy±VEGFi. Analyses of protein biomarkers in patient serum or plasma have been suggested as a feasible opportunity to investigate patient response to therapy (Tran et al, 2012), as blood sampling is readily accessible. However, pharmacodynamic (PD) changes in multiple markers induced by treatment with chemotherapy±VEGFi have not been widely studied, mainly due to the lack of appropriate sample collection in large controlled studies.
Understanding the changes in serum factors on treatment may help to predict groups of patients that may benefit more from certain treatments (Jain et al, 2006; Kopetz et al, 2010) and to discover additional signalling pathways that are regulated in response to treatment in mCRC. We have assessed samples from a large phase III trial asking three questions: (1) can serum biomarkers facilitate the segmentation of patient populations with differential response based on a baseline signature (BS), (2) among 207 soluble proteins, which changes are induced at 6/7 weeks and 13 weeks by chemotherapy and chemotherapy plus cediranib and (3) can PD biomarkers be associated with clinical benefit in patients treated with cediranib-based therapy?
Patients and methods
Patients and samples
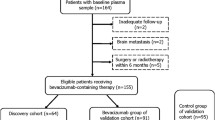
Eligible patients enrolled in the phase III double-blind HORIZON II study (ClinicalTrials.gov identifier NCT00399035) were ⩾18 years old with histologic/cytologic confirmation of metastatic (stage IV) CRC; had a World Health Organization (WHO) performance status of 0/1; and a life expectancy of ⩾12 weeks (Hoff et al, 2012). Patients must not have received prior systemic therapy for mCRC; any adjuvant (or neoadjuvant) therapy with oxaliplatin or 5-FU must have been received >12 months or >6 months, respectively, before study entry. Patients were initially randomly assigned 1 : 1 : 1 to receive once-per-day cediranib 30 mg, cediranib 20 mg, or placebo in combination with FOLFOX/CAPOX. Because recruitment to the cediranib 30 mg arm was discontinued (Hoff et al, 2012), we analysed protein biomarker levels in serum collected at baseline and on-treatment (6/7 and 13 weeks) from patients treated with cediranib 20 mg or placebo in combination with FOLFOX/CAPOX. In all, 582 serum samples were available at baseline (before treatment, T0), 587 after 6/7 weeks (T1) and 575 samples at 13 weeks (T2).
As samples were not available for all patients, the baseline characteristics of patients within the available data set (biomarker data set (n=582); BDS) were compared with patients in full data set (full data set; FDS) from the HORIZON II study to ensure that demography and treatment outcome were comparable between data sets. Age, sex and race (Black, Caucasian, Oriental or other) were compared along with the stratification covariates from the HORIZON II trial, namely, WHO performance status (0 vs 1 or 2), chemotherapy type (FOLFOX4, FOLFOX6 or CAPOX), study phase (i.e., whether patients contributed to the end-of-Phase II analysis from the HORIZON programme) and liver function (ALP ⩽320 U l−1 and albumin ⩾35 g l−1 vs other). The FDS and the BDS showed comparable demographics. The efficacy analyses for the reduced data sets were comparable with the primary trial results, indicating that there were no concerns of bias with the BDS and that, where comparisons were made with the hazard ratio (HR) and confidence intervals (CIs), they were reflective of the overall effect (Spencer et al, 2013).
Biomarker analysis
Collection of blood samples from consenting patients was prescribed (but not monitored) as follow: sampling into serum separated tubes and centrifuged within 1 h for 15 min at 3000 g, aliquoted into vials and stored immediately at –80 °C. Frozen serum samples were shipped and analysed centrally at Rules-Based Medicine (Myriad RBM, Austin, TX, USA). Analysed proteins were selected based on their relevance to angiogenesis and linked to tumour progression. Additional analytes were included if they were multiplexed with the requested markers. Each aliquot was thawed to measure 207 proteins that were quantified by using a Luminex bead-based multiplex immunodetection methodology. Myriad RBM’s multi-analyte profiles (MAPs) have been validated to Clinical Laboratory Standards Institute (formerly NCCLS) guidelines based upon the principles of immunoassay. Each assay is developed as a single test to establish the sensitivity and dynamic range necessary for that analyte. Key performance parameters such as lower limit of quantification, precision, cross-reactivity, linearity, spike-recovery, dynamic range, matrix interference, freeze-thaw stability and short-term sample stability are established for every assay (http://www.myriadrbm.com/). In all, 588 eligible patients were analysed as described in the CONSORT diagram (Supplementary Figure 1).
Statistical methods
Due to the reduced sample size, we chose to omit the primary covariates (described above) from our analyses. A comparison of the full HORIZON II analysis with and without the covariates showed very comparable HRs and CIs and these were consistent with the same analyses in the reduced biomarker data set (Spencer et al, 2013).
The BS was obtained by hierarchical clustering analyses performed using TIBCO Spotfire 3.1.1 (Boston, MA, USA) with the following parameters: Ward’s clustering method, half square Euclidean for the distance measure, average value for the ordering weight and Z-score calculation for the normalisation. The correlation with clinical end points was estimated using a Cox proportional hazards model.
Statistical analyses on the fold change from baseline to T1 or T2 were performed by paired t-test on the Log(T1/T0) and Log (T2/T0) in each treatment arm. For the differential changes between chemotherapy plus placebo (Chemo-placebo) vs chemotherapy plus cediranib (Chemo-cediranib) treatment, an unpaired t-test was performed on the difference in Log2(ratio) at T1 and T2. For example at T1, Log2(ratio)=(Log2(T1/T0) Chemo-cediranib arm) – (Log2(T1/T0) in Chemo-placebo arm). The proteins, including their mean baseline levels, standard deviation (s.d.) and standard error (s.e.) are shown in Table 1. A false discovery rate (FDR) analysis on the t-tests/paired t-tests using Storey’s method was carried out. Biomarkers described in this study showed P<0.05 and FDR<0.20.
For the analysis of the correlation between PD changes and clinical outcome, the patients were dichotomised into two groups for each biomarker based on increased vs decreased serum concentration at T1 relative to the baseline concentration. The impact of biomarker changes on OS and PFS in patients treated with chemotherapy plus placebo and in patients treated with chemotherapy plus cediranib was assessed. The association with clinical end points was estimated using a Cox proportional hazards model and P<0.05 was considered as significant. Nevertheless, given the number of proteins analysed, up to 5% of the PD biomarkers found associated with PFS or OS may have been found significant by chance. To minimise the impact of random findings, we focused the hierarchical clustering analysis on the proteins significantly associated with both PFS and OS to generate the PD signature. Hierarchical clustering analyses of patients and biomarkers was performed based on Log2(T1/T0) value using TIBCO Spofire 3.1.1 with the following parameters: Ward’s clustering method, half square Euclidean for the distance measure, average value for the ordering weight and Z-score calculation for the normalisation.
Results
Serum biomarker signature defines subgroups of mCRC patients associated with clinical outcomes
The possibility of defining subgroups of mCRC patients that may respond differentially to therapy has been explored in a number of small, often single arm, studies. Here, we explored samples from the HORIZON II phase III study with FOLFOX/CAPOX±cediranib to gain insight into how serum biomarkers may define response to therapy in mCRC. We analysed 207 circulating proteins by multiplex assays in serum obtained from patients diagnosed with mCRC and enrolled in the HORIZON II study just before treatment commenced (baseline; T0). Biomarkers were selected using two criteria. Specific proteins associated with angiogenesis and/or linked to tumour progression were prioritise, with additional exploratory analytes included by selecting specific multiplexed panels. The analysed biomarkers, mean, s.d. and s.e. are listed in Table 1.
Hierarchical clustering analysis identified 47 correlated proteins (Cluster 1) able to segregate mCRC patients into three groups (A, B and C) based on baseline pre-treatment serum concentrations (Figure 1A and B). This BS included angiogenic factors such as VEGFA, VEGFD, VEGFR2, VEGFR3, TIE-2 and NRP1. We next assessed the effect of chemotherapy±cediranib in the two most different patients groups with low (A) and high (C) BS (Figure 1C and D). Patients treated with chemotherapy plus cediranib who had high BS had a shorter PFS than those with low BS (HR=1.82, CI: 1.22–2.72, P=0.003). However, the BS did not predict PFS benefit in patients treated with chemotherapy plus placebo (HR=1.39, CI: 0.92–2.09, P=0.12). For OS, high BS was associated with shorter survival compared with low BS, regardless of the treatment received (HR=2.61, CI: 1.62–4.19, P<0.001 in chemo-cediranib group and HR=2.55, CI: 1.63–3.99, P<0.001 in chemo placebo group).
Hierarchical clustering analysis describes two subgroups of patient defined by a signature of 47 soluble biomarkers at baseline that correlates with clinical outcomes. (A) Heat map representing a hierarchical clustering analysis of the patients and soluble proteins. This analysis identified two groups of patients (A vs C) with distinct baseline concentration of 47 correlated serum proteins (Cluster 1). (B) Detailed representation of the marker in Cluster 1. Kaplan–Meier curves and Cox regression analyses show the progression-free survival (PFS) time (C) and overall survival (OS) time (D) of the patient groups (A vs C) treated with chemotherapy plus placebo (chemo-plac) and chemotherapy plus cediranib (chemo-cediranib). Abbreviations: CI, confidence interval; HR, hazard ratio.
These data suggest that the BS of 47 biomarkers may be able to segregate mCRC patient populations with regard to PFS and OS.
Chemotherapy plus placebo and chemotherapy plus cediranib induce broad PD biomarker changes
Pharmacodynamic changes in serum biomarker levels following treatment with chemotherapy±cediranib may differentiate patient responses and give initial insight into physiological response to therapy. To determine the changes induced on treatment in this study, the 207 biomarkers were quantified at two time points on treatment at 6/7 weeks (T1) and 13 weeks (T2) and compared with their baseline levels in 251 (T1) and 247 (T2) patients receiving Chemo-placebo (Figure 2; Supplementary Tables 1 and 2) and 330 (T1) and 323 (T2) patients on Chemo-cediranib (Figure 3; Supplementary Tables 3 and 4).
Pharmacodynamic (PD) biomarker changes after chemotherapy plus placebo (CP) treatment. (A) Scatter plot showing biomarkers displayed according to their fold change and P-value after T1 (week 6/7) and T2 (week 13) on CP. Each dot represents a biomarker. Fold changes for each biomarker are represented by the Log2(T1 or T2/T0) from baseline level and distributed according to the P-value. The percentage change from baseline corresponding to the Log2(T1or T2/T0) scale is shown. Significance was determined by paired t-test on Log2(T1 or T2/T0) and a P-value of <0.01. 119 (T1) and 132 (T2) markers were found changed on treatment with chemotherapy. The most significant PD biomarkers (P<1E−12) are listed. Changes for all the biomarkers are available in Supplementary Tables 1 and 2. Venn diagrams show the number of biomarkers exclusively and commonly changed (B), of which downregulated (C), or upregulated (D) at T1 or T2 after CP treatment. For full biomarker names, see Table 1.
Pharmacodynamic biomarker changes after chemotherapy plus cediranib (CC) treatment. (A) Scatter plot showing biomarkers ordered by fold change and P-value on CC. Each dot represents a biomarker. Fold changes for each biomarker are represented by the Log2(T1or T2/T0) from baseline level (T0) to T1 (week 6/7) or T2 (week 13) and distributed according to the P-value of the fold change. The percentage of change from baseline corresponding to the Log2(T1or T2/T0) scale is shown. Significance was determined by paired t-test on Log2(T1or T2/T0) and a P-value of <0.01. The most significant PD biomarkers (P<1E−13) are listed. Changes for all biomarkers are available in Supplementary Tables 3 and 4. Venn diagrams show the number of biomarkers exclusively and commonly changed (B), of which downregulated (C), or upregulated (D) at T1 or T2. For full biomarker names, see Table 1.
Analysis of the biomarker changes induced by Chemo-placebo revealed a large number of modulated proteins, 119 markers at T1 and 132 at T2 (Figure 2A). In all, 107 (74%) of these markers were changed both at T1 and at T2. In all, 59 (84%) of the down-egulated and 48 (65%) of the upregulated proteins were changed at both time points suggesting that most of the PD changes were durable for at least 13 weeks (Figure 3B–D). Among the most consistent and significant changes shown over time, we observed an increase in COL4, FB1-1C, VCAM1, TBG and AFP and a decrease in VEGFC, TGFβ1, PDGFbb, PAI-1 and S100-A12 on chemotherapy.
On Chemo-cediranib, 125 (T1) and 126 (T2) markers changed, representing over 50% of the markers analysed (Figure 3A). In all, 106 (73%) of these markers changed both at T1 and at T2. In all, 64 (77%) of the downregulated and 41 (65%) of the upregulated proteins changed at both time points (Figure 3B–D). This indicated that most of the PD changes observed at T1 were maintained at least until T2. Some pro-angiogenic markers were reduced by combination treatment. For example, decreases in VEGFR-2 and -3, VEGFC, PDGFbb and TIE-2 levels on treatment were observed. c-Kit, another target of cediranib, however, showed only slight changes that were inconsistent between time points (3.5% upregulation at T1 and 3.4% downregulation at T2) and VEGFR-1 did not demonstrate a change at either time point. Many factors involved in cell migration such as FN, CXCL5, CXCL1, TIMP, AXL-RTK, MRC2, MMP9, CCL24 and CRP were downregulated on Chemo-cediranib treatment.
PD changes induced by addition of cediranib to chemotherapy
Treatment-related changes in circulating factors may reflect physiological biomarkers or adaptive changes of the tumour following therapy. Identification of serum factors modulated by VEGFi may help define novel PD markers that characterise the patient response to chemotherapy and VEGF-signalling inhibitors and have potential to identify acquired resistance to therapy.
Most biomarkers modulated by the Chemo-cediranib combination were also significantly affected by the chemotherapy treatment alone. For example, the level of angiogenic factors such as VEGFC, PDGFbb and VEGFR-3 and factors involved in cell migration (FN, CXCL5, CXCL1, TIMP1, CRP, CCL23, CCL24 and MMP9) decreased on Chemo-placebo (Figure 2). However, a small number of proteins such as PlGF (PGF) or VEGFA were reduced on Chemo-placebo whereas they were maintained or upregulated in patients treated with Chemo-cediranib. This suggests a specific effect of cediranib addition on the PD biomarker profile.
To further investigate the effect of cediranib addition to chemotherapy on serum biomarker levels compared with chemotherapy plus placebo, we analysed the differential changes induced between Chemo-placebo and Chemo-cediranib at 6/7 weeks (T1) and 13 weeks (T2) (Figure 4). The change from baseline of individual patients was averaged for patients treated with chemo-placebo and patients treated with chemo-cediranib and compared between treatment groups. Addition of cediranib to chemotherapy led to a significant inhibition of TIE-2, VEGFR-2 and -3, NRP1 and to an upregulation of PlGF and VEGFA indicating an effect on VEGF-signalling pathways. A modest downregulation of other angiogenic factors and targets of cediranib was observed in cediranib-treated patients for VEGFR-1 only at T2, c-Kit only at T1 and VEGFD at T1 and T2. No difference between the two treatment arms was observed for VEGFB and VEGFC at any time. The addition of cediranib decreased TIE-2 and COL4 and increased A-FABP concentrations compared with chemotherapy alone.
Differential pharmacodynamic (PD) changes induced by cediranib addition to chemotherapy. (A) Scatter plot showing biomarkers displayed according to their differential fold change and P-value between chemotherapy plus placebo (CP; n=252) and chemotherapy plus cediranib (CC; n=330) treatment arms at T1 and T2. The differential fold changes are expressed by the difference in Log2 ratio. For example at T1, Log(T1/T0)=(Log2 (T1/T0) in CC arm) – (Log2 (T1/T0) in CP arm). Significances were determined by unpaired t-test on Log2(T1 or T2/T0) in each arm and P-values <0.01 were considered as significant. 50 (T1) and 57 (T2) markers were found differentially changed by addition of cediranib. The most significantly changed biomarkers (P<1E−7) are shown. (B) Comparison of the percentage of change between CP and CC arms for the 10 biomarkers most significantly downregulated or upregulated markers by addition of cediranib treatment at T1 or T2.
These data indicate that serum concentrations of multiple proteins are modulated on treatments with chemotherapy±cediranib. The differential PD changes observed in patients on cediranib result from the combination effect with chemotherapy and gives insight into the effect of cediranib addition to chemotherapy.
PD biomarkers signature’s association with clinical response in patients treated with chemotherapy plus cediranib
Pharmacodynamic changes of serum proteins on treatment may be associated with tumour response and disease progression. As PD modulation on treatment may be different in each patient, it is important to classify patients according to changes in each biomarker. To gain insight into how PD changes may influence response to therapy, we have dichotomised patients into two groups for each protein based on whether the biomarker was increased or decreased (relative to baseline) on treatment. To identify biomarkers associated with cediranib benefit, we analysed whether the PD changes (increased vs decreased at T1) were linked with PFS and OS in patients treated with chemo-cediranib (Supplementary Figure 2A and D) and chemo-placebo (Supplementary Figure 2B and E). The HRs and P-values for all the proteins are represented as volcano plots. To identify the PD proteins only associated with outcomes on chemo-cediranib, we excluded the proteins significantly associated with outcome in the chemo-placebo group (likely to be prognostic biomarkers). Scatter plots (Supplementary Figure 2C and F) and forest plots (Figure 5A and B) illustrate the individual proteins associated with PFS and OS benefit on chemo-cediranib using this approach. In all, 25 PD proteins correlated with PFS. Of these most notable were MMP7, vWF, IL-8, MIF, TIE-2, KLK7, A-FABP, TNC or VEGFA. Patients who had increased concentrations of these proteins had improved PFS compared with patients showing decreases. Similarly, in the 40 PD proteins associated with OS, the patients on chemo-cediranib who showed an increase in MMP7, IL-8, CRP, A-FABP, TIMP1, VEGFD, IL-1RA, CTSD or COL-4 had a longer OS time than those with decreased concentrations.
Pharmacodynamic biomarkers can predict PFS and OS benefit of patients treated with chemotherapy plus cediranib compared with chemotherapy plus placebo. Effect of the significant (P<0.05) PD biomarkers on PFS (A) and OS (B) in patients treated with chemotherapy±cediranib at T1. Biomarkers are ordered according to the P-value of the HR in patients treated with chemo-cediranib. A pink star indicates biomarkers associated with OS and PFS in patients treated with chemotherapy plus cediranib. (C) Heat map representing the hierarchical clustering analysis of the patients based on the pharmacodynamic changes from baseline to T1. The 16 biomarkers found significantly associated with cediranib benefit on PFS and OS in our previous univariate analysis were include in this signature. This analysis revealed two main clusters of patient showing an overall decrease vs increase in serum concentration of the 16 biomarkers. Kaplan–Meier curves and Cox regression analyses show the PFS (D) and OS (E) of the patient groups (increased vs decreased) treated with chemotherapy plus placebo (chemo-plac) and chemotherapy plus cediranib (chemo-cediranib). Abbreviations: CI, confidence interval; HR, hazard ratio; OS, overall survival; PFS, progression-free survival.
Hierarchical clustering analysis identified two distinct groups of patients based on the PD changes in 16 serum biomarkers found associated with PFS and OS benefit (pink stars annotation in Figure 5A and B) in chemo-cediranib-treated patients (Figure 5C–E). Patients in the increased PD signature (PDS) had a longer PFS (Figure 5D) and OS (Figure 5D) than those in the decreased PDS on chemo-cediranib (PFS: HR=0.58, CI: 0.46–0.73, P<0.001/OS: HR=0.52, CI: 0.40–0.66, P<0.001). The PDS did not correlate with PFS (P=0.46) or OS (P=0.44) in patients treated with chemo-placebo (Figure 5D and E). There was no significant difference in age, gender, race, WHO status, k-ras status or the number of metastatic sites between the two groups. Previous analysis of Horizon II showed there was a number of general prognostic biomarker at baseline, none were specifically associated with cediranib benefit (Jürgensmeier et al, 2013; Spencer et al, 2013). We assessed whether a subset of these prognostic biomarkers showed differential association with the either subset. There was a slight difference (maximum of two-fold) with a large variance in the level of the baseline prognostic markers ICAM, VCAM, TIMP, CEA and CRP between the two groups (Supplementary Table 5).
Discussion
Vascular endothelial growth factor signaling inhibitors, including cediranib, have shown PFS benefits in mCRC, when added to FOLFOX/CAPOX, but limited OS benefit (Saltz et al, 2008; Hoff et al, 2012; Schmoll et al, 2012). Nevertheless, FOLFOX and CAPOX, as well as FOLFIRI are widely used either alone or in combination with bevacizumab to treat patients with mCRC but it is presently unclear, which patients respond best to either treatment regime and serum protein profile on treatments have not been widely characterised to date. This study provides insight into the influence of commonly used therapies on serum proteins present in first-line mCRC patients, and how levels of these proteins identifies groups of patients that potentially respond better to these treatments.
Circulating biomarkers provide a feasible, minimal-invasive opportunity to study physio-pathological processes in cancer patients. They can be applied to disease diagnostic, prognosis/predictive assessment before treatment (Hanrahan et al, 2010; Nikolinakos et al, 2010; Abajo et al, 2012a, 2012b) and to study PD information post-treatment (Jain et al, 2006; Kopetz et al, 2010). Here, we analysed 207 serum proteins in ∼580 patients with mCRC at baseline and on-treatment with FOLFOX/CAPOX±cediranib enrolled in the HORIZON II phase III trial. There were three major findings in our exploratory analyses. First, we identified a signature of 47 markers at baseline that defines patient subgroups associated with PFS and OS. Second, we characterised the PD effects of chemotherapy and chemotherapy plus cediranib treatments identifying factors differentially modulated by cediranib addition to chemotherapy. Finally, we identified a signature of 16 PD biomarkers associated with greater potential clinical benefit in response to chemotherapy plus cediranib.
A signature of 47 soluble biomarkers was associated with clinical outcomes in mCRC. This BS was predictive of PFS benefit in chemo-cediranib-treated patients. In this signature, high baseline concentration of angiogenic markers (VEGFA, VEGFD, VEGFR2, VEGFR3, NRP1 and TIE-2) was associated with shorter PFS. Lower baseline plasma VEGFA correlates with longer time to progression in patients treated with bevacizumab (Burstein et al, 2008). In response to sunitinib, lower baseline levels of plasma VEGFA and VEGFR3 were associated with prolonged PFS (Rini et al, 2008). Low levels of ICAM1, another marker associated with our signature, have also been associated with improved PFS in patients treated with chemotherapy plus bevacizumab (Dowlati et al, 2008). The BS had a strong prognostic effect (independent of treatment arms) on OS. Factors such as CEA, VEGFA, CRP and TIMP1 have already been described as prognostic biomarkers in mCRC (Aldulaymi et al, 2010; Frederiksen et al, 2011; Bystrom et al, 2012; Jürgensmeier et al, 2013). The fact that our analysis also identified previously reported biomarkers supports the robustness of this multiplex approach and increases confidence in the potential of the other predictive/prognostic markers present in our signature. However, because all the patients received chemotherapy, it was not possible to determine the predictive vs prognostic value of these biomarkers in HORIZON II serum samples because of the lack of a placebo only arm in the trial design.
There were a large number of factors modulated by treatment with chemotherapy. It was striking that many of the proteins known to play a role in angiogenesis are modified by chemotherapy alone. The VEGF-signalling pathway (VEGFA, VEGFC and VEGFR-3) was also downregulated whereas Ang and TIE-2 were increased by chemotherapy. The changes observed in combination with cediranib will need to be interpreted in this context of a high impact of chemotherapy on the PD changes. As observed in previous studies with cediranib in monotherapy and combination or bevacizumab in combination with docetaxel (Baar et al, 2009; Drevs et al, 2007; Willett et al, 2009; Batchelor et al, 2010, 2013; van Cruijsen et al, 2010; Cunningham et al, 2013), VEGFA and PlGF increased in response to cediranib addition. The difference in VEGFA levels between the two arms in our study was mainly due to a decrease in the chemotherapy alone arm. Indeed, VEGFA levels did not change at T1 on chemo-cediranib but decreased on chemo-placebo. At T2, the increase in VEGFA was modest (∼10%) compared with PlGF (∼100%) indicating that PlGF may represent a better PD marker for cediranib, at least when combined with chemotherapy. The only 3-arm study with cediranib that evaluated biomarkers with chemotherapy (lomustine) vs cediranib vs the combination of both in patients with rGBM (Batchelor et al, 2013) showed decreases in VEGFA in the lomustine arm, increases in the cediranib monotherapy arm with the combination resulted in an increase. As previously described on cediranib monotherapy and combination therapy (Drevs et al, 2007; van Cruijsen et al, 2010; Cunningham et al, 2013), we observed a decrease in VEGFR-2 in cediranib-treated patients. Interestingly, we found other angiogenic factors such as COL4, VEGFR3, NRP1, TIE-2, ANG-2 and ENG downregulated by cediranib addition, perhaps indicating effects on vasculature. One of the most significant changes between the two treatment arms was the increase in TSH in chemo-cediranib-treated patients. The TSH elevation has been a consistent observation in all clinical studies with cediranib however in general, changes in the TSH levels were reversible following removal of cediranib and did not require treatment (Drevs et al, 2007; Hoff et al, 2012). In line with a previous study in patients with rectal carcinoma treated with bevacizumab (Xu et al, 2009), addition of cediranib increased CXCL12 levels compared with chemotherapy alone. CXCL12 is a potent chemo-attractant for myeloid cells (Jin et al, 2006; Sugiyama et al, 2006) and was associated with acquired resistance to an antibody to VEGFA in preclinical models (Shojaei et al, 2007). However, PD changes in CXCL12 did not correlate with outcome in patients treated with chemo-cediranib in our study.
Our study reports a number of PD changes in serum proteins associated with PFS and/or OS in patients treated with chemo-cediranib but not in patients treated with chemo-placebo. However, given the number of proteins analysed, some of the PD biomarkers associated with PFS or OS may have been found significant by chance. Therefore, these associations have to be interpreted with caution until further validation. To increase our confidence in some potential predictive biomarkers, we focused the hierarchical clustering analysis on the proteins significantly associated with both PFS and OS. Among the proteins identified in this PD signature, increased concentration of inflammatory biomarkers such as CRP, IL-6 and IL-8 was associated with longer PFS and/or OS. Similarly, increased concentrations of MMP1, MMP7, MMP10 and TIMP-1, all involved in extracellular matrix remodelling, were associated with clinical benefit in patients receiving chemo-cediranib. These observations may suggest that induction of an inflammatory response on treatment may be associated with improved efficacy. Consistent with our findings, a previous study (Tran et al, 2012) has shown that patients with high baseline concentrations of inflammatory or immunomodulatory factors (including IL-6, IL-8 and TIMP1) had significantly worst prognosis, but derived greater relative OS benefit from pazopanib in renal cancer. Interestingly, we found that increased concentration on treatment of angiogenic markers such as VEGFA, VEGFD, Ang and TIE-2 was associated with prolonged PFS and OS in patients receiving chemo-cediranib. However, previous studies with cediranib did not show association between VEGFA modulation on treatment and clinical response (unpublished data and Batchelor et al, 2013), indicating that further work is required to determine whether this finding may be applicable in other disease settings. This suggests that the PD changes in serum proteins may segment patients with mCRC who respond to cediranib by inducing feedback on the VEGF-signalling axis, as a result of intrinsic sensitivity to inhibitor. Therefore, increased serum VEGFA concentration on treatment may be an indicator of patients bearing tumours that may benefit from further VEGFi treatment.
The challenge in interpreting soluble serum biomarker data to monitor response to therapies is that they are not necessarily tumour derived. For example, A-FABP was one of the most significant markers differentially upregulated on cediranib addition to chemotherapy. A-FABP is mainly expressed in adipocytes and is involved in glucose and lipid systemic metabolism (Hotamisligil et al, 1996; Cao et al, 2008). Increased A-FABP concentration on treatment was strongly associated with improved PFS and OS in patients treated with chemo-cediranib. Interestingly, PD changes in other adipokines such as CRP, IL-6 and IL-10 were also associated with benefit on cediranib in our study. Adipose tissue is well known to play a role in inflammatory processes as reported by the links between obesity, macrophages and inflammation (Weisberg et al, 2003; Tilg and Moschen, 2006). This suggests that some PD changes may be associated with subgroups of patients with endocrine activity related to adipose tissue. The presence of visceral fat tissue has been correlated sensitivity to VEGFi’s in the clinic (Guiu et al, 2010; Steffens et al, 2011), suggesting that these associations warrant further investigation.
With regard to the PD signature, it does not appear that the difference in benefit is merely due to different baseline prognostic features of the induced biomarker and reduced biomarker groups. In this analysis, we included a number of independent prognostic markers. A small difference in mean CEA, TIMP-1 and CRP was observed but the difference was only two-fold or less. There were no differences in these markers comparing the chemo-placebo and chemo-cediranib groups. In Horizon II, patients with a baseline CEA level in the range of 0–50 ng ml−1 showed better prognosis than those exhibiting a baseline value of above 50 ng ml−1 (Jürgensmeier et al, 2013). Moreover, baseline CEA, TIMP or CRP levels are not associated with differences in benefit from addition of cediranib in Horizon II (Spencer et al, 2013). There are limitations to the analysis we have performed, and areas where the findings can be built on. For example, it would be informative to exploit the data set further by performing a multivariate analysis on baseline markers in each patient group to learn more about other factors associated with potential benefit on treatment. While we identify a number of interesting candidate biomarkers that may be associated with benefit from cediranib, these require validation using independent data sets.
Monitoring soluble biomarkers can be a powerful tool to gain additional insight into patient subgroups responding differently to drug in the context of a clinical trial. To deploy the approach more broadly further validation work would be required to assemble a minimal set of markers which delivers optimal predictivity in a simple test, and then subjected to prospective validation. For PD biomarkers, the changes from baseline may be investigated after 6–7 weeks based on these observations, but shorter time points may be appropriate improving selection of treatment options. While the 16 potential biomarkers identified are correlated, in theory one protein, but more realistically a subset of markers could be used to assess response. MMP7 is the most significantly associated with PFS and OS in cediranib-treated patients. It will be interesting to assess the link to outcome in other mCRC sample sets from patients treated with chemotherapy and angiogenic therapy.
In conclusion, this work provides baseline and PD biomarkers associated with clinical outcomes in mCRC patients treated with chemotherapy±cediranib. In addition, we described a comprehensive data set on the serum PD biomarkers changed by chemotherapy±cediranib. Exploring a wide range of serum biomarkers has stimulated interesting insight into the effect of these different treatments. Clinical studies remain ongoing to assess the potential of cediranib in cancer. The hypotheses generated by our data using serum samples from mCRC patients treated with chemotherapy ±cediranib could potentially apply to patients with other tumour types and could therefore be tested in future studies.
Change history
14 October 2014
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Abajo A, Bitarte N, Zarate R, Boni V, Lopez I, Gonzalez-Huarriz M, Rodriguez J, Bandres E, Garcia-Foncillas J (2012a) Identification of colorectal cancer metastasis markers by an angiogenesis-related cytokine-antibody array. World J Gastroenterol 18: 637–645.
Abajo A, Boni V, Lopez I, Gonzalez-Huarriz M, Bitarte N, Rodriguez J, Zarate R, Bandres E, Garcia-Foncillas J (2012b) Identification of predictive circulating biomarkers of bevacizumab-containing regimen efficacy in pre-treated metastatic colorectal cancer patients. Br J Cancer 107: 287–290.
Abdullah SE, Perez-Soler R (2012) Mechanisms of resistance to vascular endothelial growth factor blockade. Cancer 118: 3455–3467.
Aldulaymi B, Bystrom P, Berglund A, Christensen IJ, Brunner N, Nielsen HJ, Glimelius B (2010) High plasma TIMP-1 and serum CEA levels during combination chemotherapy for metastatic colorectal cancer are significantly associated with poor outcome. Oncology 79: 144–149.
Baar J, Silverman P, Lyons J, Fu P, Abdul-Karim F, Ziats N, Wasman J, Hartman P, Jesberger J, Dumadag L, Hohler E, Leeming R, Shenk R, Chen H, McCrae K, Dowlati A, Remick SC, Overmoyer B (2009) A vasculature-targeting regimen of preoperative docetaxel with or without bevacizumab for locally advanced breast cancer: impact on angiogenic biomarkers. Clin Cancer Res 15: 3583–3590.
Batchelor T, Mulholland P, Neyns B, Nabors L, Campone M, Wick A, Mason W, Mikkelsen T, Phuphanich S, Ashby L, DeGroot J, Gattamaneni R, Cher L, Rosenthal M, Payer F, Jürgensmeier J, Jain R, Sorensen A, Xu J, Liu Q, van den Bent M (2013) Phase III randomized trial comparing the efficacy of cediranib as monotherapy, and in combination with lomustine, versus lomustine alone in patients with recurrent glioblastoma. J Clin Oncol 31 (26): 3212–3218.
Batchelor TT, Duda DG, di Tomaso E, Ancukiewicz M, Plotkin SR, Gerstner E, Eichler AF, Drappatz J, Hochberg FH, Benner T, Louis DN, Cohen KS, Chea H, Exarhopoulos A, Loeffler JS, Moses MA, Ivy P, Sorensen AG, Wen PY, Jain RK (2010) Phase II study of cediranib, an oral pan-vascular endothelial growth factor receptor tyrosine kinase inhibitor, in patients with recurrent glioblastoma. J Clin Oncol 28: 2817–2823.
Brave SR, Ratcliffe K, Wilson Z, James NH, Ashton S, Wainwright A, Kendrew J, Dudley P, Broadbent N, Sproat G, Taylor S, Barnes C, Silva JC, Farnsworth CL, Hennequin L, Ogilvie DJ, Jürgensmeier JM, Shibuya M, Wedge SR, Barry ST (2011) Assessing the activity of cediranib, a VEGFR-2/3 tyrosine kinase inhibitor, against VEGFR-1 and members of the structurally related PDGFR family. Mol Cancer Ther 10: 861–873.
Burstein HJ, Chen YH, Parker LM, Savoie J, Younger J, Kuter I, Ryan PD, Garber JE, Chen H, Campos SM, Shulman LN, Harris LN, Gelman R, Winer EP (2008) VEGF as a marker for outcome among advanced breast cancer patients receiving anti-VEGF therapy with bevacizumab and vinorelbine chemotherapy. Clin Cancer Res 14: 7871–7877.
Bystrom P, Berglund A, Nygren P, Wernroth L, Johansson B, Larsson A, Glimelius B (2012) Evaluation of predictive markers for patients with advanced colorectal cancer. Acta Oncol 51: 849–859.
Cao H, Gerhold K, Mayers JR, Wiest MM, Watkins SM, Hotamisligil GS (2008) Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell 134: 933–944.
Cunningham D, Wong RP, D'Haens G, Douillard JY, Robertson J, Stone AM, Van Cutsem E HORIZON I Study Group (2013) Cediranib with mFOLFOX6 vs bevacizumab with mFOLFOX6 in previously treated metastatic colorectal cancer. Br J Cancer 108: 493–502.
Dowlati A, Gray R, Sandler AB, Schiller JH, Johnson DH (2008) Cell adhesion molecules, vascular endothelial growth factor, and basic fibroblast growth factor in patients with non-small cell lung cancer treated with chemotherapy with or without bevacizumab—an Eastern Cooperative Oncology Group Study. Clin Cancer Res 14: 1407–1412.
Drevs J, Siegert P, Medinger M, Mross K, Strecker R, Zirrgiebel U, Harder J, Blum H, Robertson J, Jürgensmeier JM, Puchalski TA, Young H, Saunders O, Unger C (2007) Phase I clinical study of AZD2171, an oral vascular endothelial growth factor signaling inhibitor, in patients with advanced solid tumors. J Clin Oncol 25: 3045–3054.
Ferrara N, Kerbel RS (2005) Angiogenesis as a therapeutic target. Nature 438: 967–974.
Frederiksen C, Qvortrup C, Christensen IJ, Glimelius B, Berglund A, Jensen BV, Nielsen SE, Keldsen N, Nielsen HJ, Brunner N, Pfeiffer P (2011) Plasma TIMP-1 levels and treatment outcome in patients treated with XELOX for metastatic colorectal cancer. Ann Oncol 22: 369–375.
Guiu B, Petit JM, Bonnetain F, Ladoire S, Guiu S, Cercueil JP, Krause D, Hillon P, Borg C, Chauffert B, Ghiringhelli F (2010) Visceral fat area is an independent predictive biomarker of outcome after first-line bevacizumab-based treatment in metastatic colorectal cancer. Gut 59: 341–347.
Hanrahan EO, Lin HY, Kim ES, Yan S, Du DZ, McKee KS, Tran HT, Lee JJ, Ryan AJ, Langmuir P, Johnson BE, Heymach JV (2010) Distinct patterns of cytokine and angiogenic factor modulation and markers of benefit for vandetanib and/or chemotherapy in patients with non-small-cell lung cancer. J Clin Oncol 28: 193–201.
Hoff PM, Hochhaus A, Pestalozzi BC, Tebbutt NC, Li J, Kim TW, Koynov KD, Kurteva G, Pinter T, Cheng Y, van Eyll B, Pike L, Fielding A, Robertson JD, Saunders MP (2012) Cediranib plus FOLFOX/CAPOX versus placebo plus FOLFOX/CAPOX in patients with previously untreated metastatic colorectal cancer: a randomized, double-blind, phase III study (HORIZON II). J Clin Oncol 30: 3596–3603.
Hotamisligil GS, Johnson RS, Distel RJ, Ellis R, Papaioannou VE, Spiegelman BM (1996) Uncoupling of obesity from insulin resistance through a targeted mutation in aP2, the adipocyte fatty acid binding protein. Science 274: 1377–1379.
Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, Berlin J, Baron A, Griffing S, Holmgren E, Ferrara N, Fyfe G, Rogers B, Ross R, Kabbinavar F (2004) Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. New Engl J Med 350: 2335–2342.
Jain RK, Duda DG, Clark JW, Loeffler JS (2006) Lessons from phase III clinical trials on anti-VEGF therapy for cancer. Nat Clin Pract Oncol 3: 24–40.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61: 69–90.
Jin DK, Shido K, Kopp HG, Petit I, Shmelkov SV, Young LM, Hooper AT, Amano H, Avecilla ST, Heissig B, Hattori K, Zhang F, Hicklin DJ, Wu Y, Zhu Z, Dunn A, Salari H, Werb Z, Hackett NR, Crystal RG, Lyden D, Rafii S (2006) Cytokine-mediated deployment of SDF-1 induces revascularization through recruitment of CXCR4+ hemangiocytes. Nat Med 12: 557–567.
Jürgensmeier JM, Schmoll HJ, Robertson JD, Brooks L, Taboada M, Morgan SR, Wilson D, Hoff PM (2013) Prognostic and predictive value of VEGF, sVEGFR-2 and CEA in mCRC studies comparing cediranib, bevacizumab and chemotherapy. Br J Cancer 108: 1316–1323.
Kopetz S, Hoff PM, Morris JS, Wolff RA, Eng C, Glover KY, Adinin R, Overman MJ, Valero V, Wen S, Lieu C, Yan S, Tran HT, Ellis LM, Abbruzzese JL, Heymach JV (2010) Phase II trial of infusional fluorouracil, irinotecan, and bevacizumab for metastatic colorectal cancer: efficacy and circulating angiogenic biomarkers associated with therapeutic resistance. J Clin Oncol 28: 453–459.
Nikolinakos PG, Altorki N, Yankelevitz D, Tran HT, Yan S, Rajagopalan D, Bordogna W, Ottesen LH, Heymach JV (2010) Plasma cytokine and angiogenic factor profiling identifies markers associated with tumor shrinkage in early-stage non-small cell lung cancer patients treated with pazopanib. Cancer Res 70: 2171–2179.
Rini BI, Michaelson MD, Rosenberg JE, Bukowski RM, Sosman JA, Stadler WM, Hutson TE, Margolin K, Harmon CS, DePrimo SE, Kim ST, Chen I, George DJ (2008) Antitumor activity and biomarker analysis of sunitinib in patients with bevacizumab-refractory metastatic renal cell carcinoma. J Clin Oncol 26: 3743–3748.
Saltz LB, Clarke S, Diaz-Rubio E, Scheithauer W, Figer A, Wong R, Koski S, Lichinitser M, Yang TS, Rivera F, Couture F, Sirzen F, Cassidy J (2008) Bevacizumab in combination with oxaliplatin-based chemotherapy as first-line therapy in metastatic colorectal cancer: a randomized phase III study. J Clin Oncol 26: 2013–2019.
Schmoll HJ, Cunningham D, Sobrero A, Karapetis CS, Rougier P, Koski SL, Kocakova I, Bondarenko I, Bodoky G, Mainwaring P, Salazar R, Barker P, Mookerjee B, Robertson J, Van Cutsem E (2012) Cediranib with mFOLFOX6 versus bevacizumab with mFOLFOX6 as first-line treatment for patients with advanced colorectal cancer: a double-blind, randomized phase III study (HORIZON III). J Clin Oncol 30: 3588–3595.
Shojaei F, Wu X, Malik AK, Zhong C, Baldwin ME, Schanz S, Fuh G, Gerber HP, Ferrara N (2007) Tumor refractoriness to anti-VEGF treatment is mediated by CD11b+Gr1+ myeloid cells. Nat Biotechnol 25: 911–920.
Spencer S, Pommier A, Morgan S, Barry S, Robertson J, Hoff P, Jürgensmeier J (2013) Prognostic/predictive value of 207 serum factors in colorectal cancer treated with cediranib and/or chemotherapy. Br J Cancer 109: 2765–2773.
Steffens S, Grunwald V, Ringe KI, Seidel C, Eggers H, Schrader M, Wacker F, Kuczyk MA, Schrader AJ (2011) Does obesity influence the prognosis of metastatic renal cell carcinoma in patients treated with vascular endothelial growth factor-targeted therapy? Oncologist 16: 1565–1571.
Sugiyama T, Kohara H, Noda M, Nagasawa T (2006) Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity 25: 977–988.
Tilg H, Moschen AR (2006) Adipocytokines: mediators linking adipose tissue, inflammation and immunity. Nat Rev Immunol 6: 772–783.
Tran HT, Liu Y, Zurita AJ, Lin Y, Baker-Neblett KL, Martin AM, Figlin RA, Hutson TE, Sternberg CN, Amado RG, Pandite LN, Heymach JV (2012) Prognostic or predictive plasma cytokines and angiogenic factors for patients treated with pazopanib for metastatic renal-cell cancer: a retrospective analysis of phase 2 and phase 3 trials. Lancet Oncol 13: 827–837.
van Cruijsen H, Voest EE, Punt CJ, Hoekman K, Witteveen PO, Meijerink MR, Puchalski TA, Robertson J, Saunders O, Jürgensmeier JM, van Herpen CM, Giaccone G (2010) Phase I evaluation of cediranib, a selective VEGFR signalling inhibitor, in combination with gefitinib in patients with advanced tumours. Eur J Cancer 46: 901–911.
Wedge SR, Kendrew J, Hennequin LF, Valentine PJ, Barry ST, Brave SR, Smith NR, James NH, Dukes M, Curwen JO, Chester R, Jackson JA, Boffey SJ, Kilburn LL, Barnett S, Richmond GH, Wadsworth PF, Walker M, Bigley AL, Taylor ST, Cooper L, Beck S, Jürgensmeier JM, Ogilvie DJ (2005) AZD2171: a highly potent, orally bioavailable, vascular endothelial growth factor receptor-2 tyrosine kinase inhibitor for the treatment of cancer. Cancer Res 65: 4389–4400.
Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW Jr (2003) Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest 112: 1796–1808.
Willett CG, Duda DG, di Tomaso E, Boucher Y, Ancukiewicz M, Sahani DV, Lahdenranta J, Chung DC, Fischman AJ, Lauwers GY, Shellito P, Czito BG, Wong TZ, Paulson E, Poleski M, Vujaskovic Z, Bentley R, Chen HX, Clark JW, Jain RK (2009) Efficacy, safety, and biomarkers of neoadjuvant bevacizumab, radiation therapy, and fluorouracil in rectal cancer: a multidisciplinary phase II study. J Clin Oncol 27: 3020–3026.
Xu L, Duda DG, di Tomaso E, Ancukiewicz M, Chung DC, Lauwers GY, Samuel R, Shellito P, Czito BG, Lin PC, Poleski M, Bentley R, Clark JW, Willett CG, Jain RK (2009) Direct evidence that bevacizumab, an anti-VEGF antibody, up-regulates SDF1alpha, CXCR4, CXCL6, and neuropilin 1 in tumors from patients with rectal cancer. Cancer Res 69: 7905–7910.
Acknowledgements
This work was supported by a FP7-funded Initial Training Network grant to JMJ (TranSViR) by the EU that included the fellowship for AP. We thank Bhavika Patel for critical reading of the manuscript. Funding for this study was provided by AstraZeneca.
Author Contributions
AP, RS, SS, STB, JDR, SM, PMH and JMJ contributed to conception and design, analysis and interpretation of data, and writing, review and/or revision of manuscript. AP, RS and SS contributed to development of methodology. PMH, SS and SRM contributed to acquisition of data. SRM and PMH contributed to administrative, technical or material support. PMH, JDR and JMJ contributed to study supervision.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
RS, SKMS, STB, JDR SRM are AstraZeneca employees and own stock. AJCP and JMJ are former employees of AstraZeneca. PMH has received fees for consultancy and participation in advisory boards from AstraZeneca.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Pommier, A., Shaw, R., Spencer, S. et al. Serum protein profiling reveals baseline and pharmacodynamic biomarker signatures associated with clinical outcome in mCRC patients treated with chemotherapy ± cediranib. Br J Cancer 111, 1590–1604 (2014). https://doi.org/10.1038/bjc.2014.436
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2014.436
- Springer Nature Limited
Keywords
This article is cited by
-
EMT-mediated regulation of CXCL1/5 for resistance to anti-EGFR therapy in colorectal cancer
Oncogene (2022)
-
Circulating biomarkers during treatment in patients with advanced biliary tract cancer receiving cediranib in the UK ABC-03 trial
British Journal of Cancer (2018)
-
A functional bioassay to determine the activity of anti-VEGF antibody therapy in blood of patients with cancer
British Journal of Cancer (2016)
-
A job for quantum dots: use of a smartphone and 3D-printed accessory for all-in-one excitation and imaging of photoluminescence
Analytical and Bioanalytical Chemistry (2016)