Abstract
The Pyrenean desman (Galemys pyrenaicus) is a small endangered semi-aquatic mammal endemic to the Pyrenean Mountains and to the northern half of the Iberian Peninsula whose ecology and biology are still poorly known. The aim of this study was to identify Pyrenean desman faeces and to analyze its diet from this material using next-generation sequencing methods. We amplified and sequenced a small DNA minibarcode (133 bp) of the COI gene in twenty-four putative faeces samples of Pyrenean desman and successfully identified the species in 16 samples. Other identified species were mammals, birds and amphibians, evidencing the potential application of our methods to a larger panel of taxa.
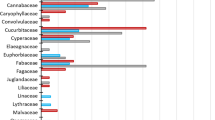
In the Pyrenean desman faeces, we were able to identify nineteen prey species with a positive match (more than 98% of identity with a reference sequence) and eleven putative prey species with lower identity scores (90–96%). The nineteen species belonged to four orders and eleven families among which Trichoptera and Hydropsychidae were the most frequent, respectively. Future improvements could be obtained by extending the reference DNA sequence collection to reach precise identifications over the Desman’s range and by increasing the sampling to gain a better knowledge of the local diet of this endangered species. Such information is of great importance to propose the best management measures for its conservation.
Similar content being viewed by others
References
Bertrand, A., 1993. Découvrir le Desman des Pyrénées. Ed. A.N.A., pp. 32.
Bertrand, A., 1994. Répartition géographique et écologie alimentaire de desman des pyrénées G. pyrenaicus dans les Pyrénées francaises. Diplôme Universitaire de Recherche, Toulouse, pp. 217.
Castién, E., Gosálbez, J., 1995. Diet of Galemyspyrenaicus (Geoffroy, 1811) inthe North of the Iberian peninsula. Netherlands J. Zool. 45 (3–4), 422–430.
Charbonnel, A., (Thèse de doctorat) 2015. Influence multi-échelle des facteurs environnementaux dans la répartition du Desman des Pyrénées (Galemys pyrenaicus) en France. l’Université de Toulouse, pp. 245.
Fernandes, M., et al., 2008. Galemys pyrenaicus. IUCN Red List of Threatened Species. Version 2013.2, Available at: https://doi.org/www.iucnredlist.org
Fernández-Salvador, R., et al., 1998. Feeding habits of the Iberian desman, Galemys pyrenaicus. Euro-American Mammal Congress, Santiago de Compostela (Spain).
Galan, M., Pagès, M., Cosson, J.F., 2012. Next-generation sequencing for rodent barcoding: species identification from fresh, degraded and environmental samples. PLoS ONE 7 (11).
Gillet, F., et al., 2015. PCR-RFLP identification of the endangered Pyrenean desman, Galemys pyrenaicus (Soricomorpha, Talpidae), based on faecal DNA. Mammalia, Available at: https://doi.org/www.degruyter.com/view/j/mamm.ahead-of-print/mammalia-2014-0093/mammalia-2014-0093.xml (accessed 01.11.14).
Gisbert, J., Garciá-Perea, R., 2014. Historiade la regresióndel desmán ibérico Galemys pyrenaicus (É. Geoffroy Saint-Hilaire, 1811) en el Sistema Central (Península Ibérica). Munibe Monographs Nature Series.
Hajibabaei, M., et al., 2011. Environmental barcoding: a next-generation sequencing approach forbiomonitoring applications using river benthos. PLoS ONE 6 (4), e17497, Available at: https://doi.org/www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3076369&tool=pmcentrez&rendertype=abstract (accessed 07.11.13).
Hebert, P.D.N., et al., 2004. Ten species in one: DNA barcoding reveals cryptic species inthe neotropical skipper butterfly Astraptes fulgerator. PNAS 101 (41).
Latinne, A., et al., 2014. Diet analysis of Leopoldamys neilli, a cave-dwelling rodent in southeast asia, using next-generation sequencing from feces. J. Cave Karst Stud. 76 (2), 139–145, Available at: https://doi.org/caves.org/pub/journal/PDF/v76/cave-76-02-139.pdf (accessed 25.02.15).
Lees, D.C., et al., 2010. DNA mini-barcodes in taxonomic assignment: a morphologically unique new homoneurous moth clade from the Indian Himalayas described in Micropterix (Lepidoptera, Micropterigidae). Zool. Scr. 39 (6), 642–661, Available at: https://doi.org/doi.wiley.com/10.1111/j.1463-6409.2010.00447.x (accessed 14.11.13).
Meusnier, I., et al., 2008. A universal DNA mini-barcode for biodiversity analysis. BMC Genomics 9, 214, Available at: https://doi.org/www.ncbi.nlm.nih.gov/pubmed/18474098 (accessed 07.11.13).
Némoz, M., Bertrand, A., 2008. Plan National d’Actions en faveurdu Desman des Pyrénéés (Galemys pyrenaicus), 2009–2014. Société Francaise pour l’Etude et la Protection des Mammifères/Ministère de l’Ecologie, de l’Energie, du Développement Durable et de l’Aménagement du Territoire, 1.
Nores, C, Queiroz, A.I., Gisbert, J., 2007. Galemys pyrenaicus, (E. Geoffroy Saint-Hilaire, 1811). In: Palomo, L, Gisbert, J., Blanco, J. (Eds.), Atlas y libro rojo de los mamíferos terrestres de Espan˜a. Dirección General para la Biodiversidad – SECEM – SECEMU, Madrid, pp. 92–98.
Palmeirim, J.M., Hoffmann, S.H., 1983. Galemys pyrenaicus. Mamm. Species 207, 1–5.
Piñol, J., et al., 2014. A pragmatic approach to the analysis of diets of generalist predators: the use of next-generation sequencing with no blocking probes. Mol. Ecol. Resour. 14 (1), 18–26, Available at: https://doi.org/www.ncbi.nlm.nih.gov/pubmed/23957910 (accessed 10.02.15).
Piry, S., et al., 2012. |SE|S|AM|E| Barcode: NGS-oriented software foramplicon characterization – application to species and environmental barcoding. Mol. Ecol. Resour. 12 (6), 1151–1157, Available at: https://doi.org/www.ncbi.nlm.nih.gov/pubmed/22823139 (accessed 30.03.15).
Puisségur, C, 1935. Recherches sur le Desman des Pyrénées. Bull. Soc. Hist. Nat. Toulouse 67, 163–227.
Ratnasingham, S., Hebert, P.D.N., 2007. BOLD: The Barcode of Life Data System (https://doi.org/www.barcodinglife.org). Mol. Ecol. Notes 7, 355–364.
Richard, P.B., 1986. Les Desman des Pyrénées, un mammifère inconnu à découvrir. Science et Découvertes, Ed., Le Rocher, Monaco, 118 pp.
Richard, P.B., Vallette Vialard, A., 1969. Le Desman des Pyrénées (Galemys pyrenaicus): premières notes sursabiologie. LaTerre et la Vie 116 (3), 225–245.
Santamarina, J., 1993. Feeding ecology of a vertebrate assemblage inhabiting a stream of NW Spain (Riobo; Ulla basin). Hydrobiologia 252 (2), 175–191, Available at: https://doi.org/link.springer.com/10.1007/BF00008154
Santamarina, J., Guitian, J., 1988. Quelques données sur le régime alimentaire du desman (Galemys pyrenaicus) dans le nord-ouest de l’Espagne. Mammalia 52 (3).
Shehzad, W., et al., 2012. Carnivore diet analysis based on next-generation sequencing: application to the leopard cat (Prionailurus bengalensis) in Pakistan. Mol. Ecol. 21 (8), 1951–1965, Available at: https://doi.org/www.ncbi.nlm.nih.gov/pubmed/22250784 (accessed 28.03.14).
Soininen, E.M., et al., 2009. Analysing diet of small herbivores: the efficiency of DNA barcoding coupled with high-throughput pyrosequencing for deciphering the composition of complex plant mixtures. Front. Zool. 6,16, Available at: https://doi.org/www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2736939&tool=pmcentrez&rendertype=abstract (accessed 17.12.14).
Stone, R.D., 1987. The activity pattern of the Pyrenean desman (Galemys pyrenaicus) (Insectivora: Talpidae), as determined under natural condition. J. Zool. Lond. 213, 95–106.
Trutat, E., 1891. Essai sur l’histoire naturelle du Desman des Pyrénées. Douladoure-Privat, Toulouse, pp. 107.
Virgilio, M., et al., 2010. Comparative performances of DNA barcoding across insect orders. BMC Bioinformatics 11, 206, Available at: https://doi.org/www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2885370&tool=pmcentrez&rendertype=abstract
Zeale, M.R.K., et al., 2011. Taxon-specific PCR for DNA barcoding arthropod prey in bat faeces. Mol. Ecol. Resour. 11 (2), 236–244, Available at: https://doi.org/www.ncbi.nlm.nih.gov/pubmed/21429129 (accessed 09.03.15).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gillet, F., Tiouchichine, ML., Galan, M. et al. A new method to identify the endangered Pyrenean desman (Galemys pyrenaicus) and to study its diet, using next generation sequencing from faeces. Mamm Biol 80, 505–509 (2015). https://doi.org/10.1016/j.mambio.2015.08.002
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1016/j.mambio.2015.08.002