Abstract
Centromeres play a crucial role in ensuring the accurate separation of chromosomes during cell division. Despite the three rounds of genome sequencing technology undergone by Citrus sinensis (sweet orange), the presence of numerous repetitive DNA elements in its genome has led to substantial gaps in centromeric genomic mapping, leaving the composition of centromeric repeats unclear. To address this, we employed a combination of chromatin immunoprecipitation sequencing with the C. sinensis centromere-specific histone H3 variant antibody and centromere-specific bacterial artificial chromosome-3a sequencing to precisely locate the centromeres. This approach allowed us to identify a series of centromere-specific repeats, comprising five tandem repeats and nine long terminal repeat retrotransposons. Through comprehensive bioinformatics analysis, we gained valuable insights into potential centromeric evolution events and discovered the presence of DNA G-quadruplex structures of centromeric repeats in C. sinensis. Altogether, our study not only offers a valuable reference for centromeric genome assembly but also sheds light on the structural characteristics of C. sinensis centromeres.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Centromeres play a critical role in chromatid segregation during both mitosis and meiosis in eukaryotes (Hofstatter et al. 2022; Hou et al. 2021; Oliveira and Torres 2018). Morphologically, centromeres appear as the primary constriction on the metaphase chromosomes (Zhou et al. 2022), connecting with the spindle microtubules (Fernandes et al. 2019), and enabling chromosome segregation during cell division (Naish et al. 2021). The key distinguishing feature of centromeres is the presence of a centromere-specific histone H3 variant (CenH3) within the nucleosome (Liu et al. 2017), which epigenetically determines the position of the kinetochore (Oliveira and Torres 2018). Moreover, CenH3 serves as a marker for functional centromeres, aiding in the identification of true centromeric regions (Feng et al. 2020). Despite these advancements, defining centromeres has remained challenging due to the complexity and abundance of repetitive DNA sequences, particularly in important plant species.
Centromeric repeats demonstrate rapid evolutionary characteristics, contributing to the high degree of species specificity of centromeres (Naish et al. 2021). In plants, centromeres consist of a large number of repetitive DNA sequences, primarily including tandem repeats (TRs), long terminal repeat retrotransposons (LTR-RTs), and low-copy sequences (Comai et al. 2017; Plohl et al. 2014; Zhou et al. 2022). TRs represent the predominant component of centromeres in most plants. For instance, the Arabidopsis thaliana centromeres comprise TRs with a repeating unit of 180 bp (Naish et al. 2021; Wlodzimierz et al. 2023), while certain crops like rice (Oryza sativa L.), maize (Zea mays L.), and sugarcane (Saccharum officinarum L.) have TRs with repeating units as large as 140–156 bp (Hiatt et al. 2002; Huang et al. 2021; Zhang et al. 2017). LTR-RTs also play a vital role in the formation of centromeres. In Arabidopsis, the LTR-RTs ATHILA (Ty3/gypsy) and TR CEN180 are interspersed in centromeres, collectively constituting the central domain of the centromere (Naish et al. 2021). Similarly, rice centromeres harbor a significant number of LTR-RTs, such as centromere-specific retrotransposons of rice (CRR) and TR CentO (Song et al. 2021).
Notably, the aforementioned repetitive sequences, serving as primary structures of DNA, can both form the secondary structures of the canonical right-handed B-form double helix and atypical DNA structures like the G-quadruplex (G4) (Crespi and Ariel 2022; Liu et al. 2023). In addition to their involvement in gene expression (Fang et al. 2019; Gonzalo et al. 2022) and telomere maintenance (Miglietta et al. 2020), these non-B-form DNA secondary structures are associated with the deletion and inversion of fragments of centromeres (Liu et al. 2023). However, the connection between secondary and primary structures in centromeres remains largely unknown. Therefore, understanding TRs or LTR-RTs as the primary structural elements of centromeres is crucial for analyzing the composition, structure, and evolution of centromeres. To advance this knowledge, it is essential to develop more sophisticated methods to explore the repetitive sequences in plant centromeres.
Insights into centromeric repeats in most plants can now be revealed through various techniques, including chromatin immunoprecipitation sequencing (ChIP-seq) (Song et al. 2021; Su et al. 2019; Yang et al. 2018; Zhao et al. 2023), repetitive sequence analysis (Song et al. 2023), and new long-read DNA sequencing technologies (Deng et al. 2022; Hou et al. 2022; Naish et al. 2021; Nie et al. 2021). The utilization of anti-CenH3 antibodies in ChIP-seq has facilitated the determination of the sequence and localization of centromeres in wheat (Triticum aestivum L.) (Zhao et al. 2023), rice (Song et al. 2021), and oat (Avena sativa L.) (Liu et al. 2023). The advancement of longer and more accurate sequencing reads made it possible to construct complete genome assemblies in centromeres (Naish et al. 2021; Zhang et al. 2023). Although complete genomes have been achieved in a few species through gapless telomere-to-telomere (T2T) sequence assemblies (Belser et al. 2021; Giguere et al. 2022; Liu et al. 2020; Nurk et al. 2022; Zhang et al. 2022), completely assembling repetitive sequences containing numerous DNA elements remains challenging in most plants (Liu et al. 2022; Navratilova et al. 2022; Zhou et al. 2020).
Citrus sinensis (sweet orange) is the most widely cultivated citrus species globally and ranks among the top-selling fresh fruits (Xu et al. 2013). Despite three generations of C. sinensis sequencing efforts (Wang et al. 2021; Xu et al. 2013), gaps in the genome still persist, particularly in regions rich in repetitive sequences (Song et al. 2023). To address this, our study focused on determining the centromeric regions of C. sinensis using a combination of ChIP-seq with a C. sinensis CenH3 antibody (CsCenH3-ChIP-seq) and centromere-specific bacterial artificial chromosome-3a sequencing (BAC-3a-seq). Through genome-wide BLAST analysis, we identified 23 centromeric candidate repeats, among which five TRs and nine LTR-RTs were found to be specific to C. sinensis centromeres. Additionally, colocalization analysis of CsCenH3-ChIP-seq and the centromeric repeat (named CL) from the published reference (Song et al. 2023) provided insights into the centromeric evolution event in C. sinensis. Moreover, we analyzed the DNA G4 structures of the centromeric repeats. Our findings contribute valuable information regarding the centromeric composition and the identification of centromere-specific molecular markers in C. sinensis.
Materials and methods
Chromosome preparation
Root tips from C. sinensis were cultivated in the dark for 10 days to facilitate chromosome preparation. The process of chromosome preparation followed established protocols as described in the literature (Song et al. 2023).
Cytogenetic analysis
For cytogenetic analysis, the probes CsCen1 (C. sinensis centromere 1) and BAC-3a were labeled using the BioPrime® DNA Labeling System (18094-011) kit. Fluorescence in situ hybridization (FISH) of CsCen1 and BAC-3a was performed according to previously reported procedures (Song et al. 2023). Immunofluorescence (IF) staining was conducted using published protocols (Xia et al. 2020). Subsequently, the FISH and IF staining slides were examined using a fluorescence microscope (ZEISS, Axio Imager M2, Germany). The appropriate filters for different fluorescence detections were applied, as specified in the published protocols (Song et al. 2023). Finally, the FISH and IF staining images were processed using Adobe Photoshop 2022.
CsCenH3-ChIP-seq and BAC-3a-seq
Anti-CsCenH3 antibodies were prepared, and libraries were constructed for CsCenH3-ChIP-seq following the previously published protocols (Song et al. 2023; Xia et al. 2020). Similarly, BAC-3a was sequenced using the Miseq sequencing platform (Illumina, USA). Before the assembly process, we estimated the genome size using a K-mer statistics analysis method (https://bioinformatics.uconn.edu/genome-size-estimation-tutorial/#). The reads were then assembled using SOAPdenovo software (https://sourceforge.net/projects/soapdenovo2/files/SOAPdenovo2/). Ultimately, we obtained a total of 12 scaffolds from the assembly process.
Bioinformatic analysis
RepeatExplorer2 (https://www.repeatexplorer.org/), RepeatMasker (https://www.repeatmasker.org/), and TRs Finder software (Benson 1999) (https://tandem.bu.edu/trf/trf.html) were utilized to screen centromeric candidate repeats. The distribution of repetitive sequences in genome version 3 (v3) (Wang et al. 2021) was determined through Blastn analysis in BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple sequence alignment was performed in ClusterW (https://www.genome.jp/tools-bin/clustalw) using Execute Multiple Alignment for alignment, and the results were plotted using ENDscript/ESPrint (https://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). RIdeogram (Hao et al. 2020) and Circos (https://github.com/zhangtaolab/Chorus2) were used to visualize CsCenH3-ChIP-seq, BAC-3a-seq, and centromeric candidate repeats on the genome. Lastly, DNA G4 structures in the centromeric candidate repeats were identified using pqsfinder (accessible at https://bioconductor.org/packages/release/bioc/vignettes/pqsfinder/inst/doc/pqsfinder.html).
Results
Centromere localization and identification of ten centromeric candidate repeats in C. sinensis using CsCenH3-ChIP-seq
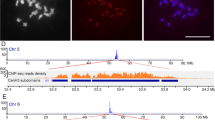
The specificity of anti-CenH3 antibodies for labeling centromeres in plants has been previously demonstrated (Liu et al. 2023; Zhao et al. 2023). To pinpoint the centromeric regions and determine the centromeric repeats of C. sinensis, we conducted ChIP using the CsCenH3 antibody. Initially, we performed IF staining on the nucleus of C. sinensis root tips using the CsCenH3 antibody (Fig. 1a, b, c), revealing 18 IF signals on the nucleus, which corresponded to the centromeric positions on 18 chromosomes (Fig. 1b, c). Subsequently, ChIP-seq with the CsCenH3 antibody was performed on nuclei isolated from C. sinensis leaf tissue. Significant peaks were detected at the ends of chromosomes 3 and 7 (Fig. 1d), while significant peaks were observed in the centromeric regions of C. sinensis chromosomes 8 and 9 (Fig. 1d). These findings indicate the presence of a low-quality assembly of C. sinensis centromeres.
IF staining and genome-wide mapping of CsCenH3-ChIP-seq reads to the C. sinensis genome v3. (a) DAPI-stained interphase nucleus (blue) of C. sinensis. (b) Immunostaining for CsCenH3 (green) in the interphase nucleus. (c) DAPI-stained interphase nucleus (blue) showing immunofluorescence signals of CsCenH3 (green). Scale bars = 5 µm. (d) Circos plot representing the distribution of CsCenH3-ChIP-seq reads on the C. sinensis genome v3. The outer circle shows the nine chromosomes of C. sinensis, while the inner circle indicates the CsCenH3-ChIP-seq read peaks. The numbers around the circle indicate the length of the nine chromosomes in megabases (Mb). IF, immunofluorescence; CsCen, C. sinensis centromere; CsCenH3-ChIP-seq, chromatin -immunoprecipitation sequencing using CsCenH3 antibody; DAPI, 4’,6-diamidino-2-phenylindole; v3, version 3; chr, chromosomes
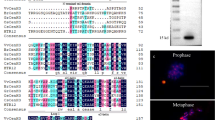
Additionally, we used the RepeatExplorer2 software to screen centromeric repeats from ChIP-seq, resulting in the identification of 10 centromeric candidate repeats (Table 1). Among these, the centromeric candidate repeat CsCen7 had previously been confirmed as a centromeric repeat in C. sinensis through FISH (Song et al. 2023). Notably, we made the intriguing observation that CsCen1, CsCen3, CsCen9, and the centromeric repeat CL predicted through bioinformatics analysis (Melters et al. 2013), all exhibited a length of 181 bp. Multiple sequence alignment analysis revealed a high similarity among CsCen1, CsCen3, CsCen9, and CL (Fig. 2a), and these four repeats exhibited an identical distribution in the C. sinensis genome v3 (Fig. S1). Furthermore, the FISH results showed a consistent distribution pattern of CsCen1 on the chromosomes (Fig. 2b, c, d, e) with CL (Song et al. 2023). Based on these findings, we propose that these four sequences are indeed the same repeats in C. sinensis.
CsCen1, CsCen3, and CsCen9 were identified as the same TR as the predicted centromeric repeat CL (a) Multiple sequence alignment of CL, CsCen1, CsCen3, and CsCen9. (b) FISH signals of CL (red) on metaphase chromosomes of C. sinensis. (c) DAPI-stained chromosomes (blue) showing FISH signals of CL (red). (d) CMA staining (yellow) on the same chromosomes as shown in Fig. 1c. (e) DAPI-stained chromosomes (blue) with CMA staining (yellow). Scale bars = 5 µm. CMA, Chromoycin A3; DAPI, 4’,6-diamidino-2-phenylindole; FISH, fluorescence in situ hybridization; CsCen, C. sinensis centromere
FISH mapping and sequencing of centromere-specific BAC-3a
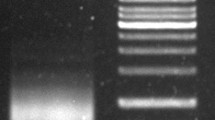
FISH signals of a single low-copy BAC-3a were observed to be distributed in the centromeres and near centromeric regions of 18 chromosomes in C. sinensis (Fig. 3a, b). This finding raised the possibility of the presence of centromere-specific repeats within BAC-3a, prompting us to undertake the sequencing and assembly of BAC-3a. Initially, we employed the Miseq sequencing platform to sequence the samples (Fig. 2c, d, e). The analysis of the base content and mass distribution of the 500-bp library indicated high sequencing quality of the repeats and a low error rate. After the assembly process, we ultimately obtained a total of 12 scaffolds. To visualize the distribution of BAC-3a in the centromeric region, we simultaneously located the reads of BAC-3a-seq and CsCenH3-ChIP-seq on the C. sinensis genome v3 (Fig. 4). The results clearly demonstrated that the regions enriched with BAC-3a all exhibited CsCenH3-ChIP-seq read peaks, providing strong evidence that BAC-3a indeed contains centromeric candidate repeats.
Sequencing and assembly of BAC-3a. (a), (b) FISH signals of BAC-3a (red) at the DAPI-stained chromosomal centromeres (blue) in C. sinensis. FISH signals of BAC-3a on different DAPI-stained metaphase chromosomes are shown. Scale bars = 5 µm. (c) Base content map of the 500-bp library. (d) Base quality distribution of the 500-bp library. (e) The 15-mer statistical map based on K-mer statistics. BAC, bacterial artificial chromosome; DAPI, 4’,6-diamidino-2-phenylindole; FISH, fluorescence in situ hybridization
CsCenH3-ChIP-seq peaks are present in all regions enriched with BAC-3a. The distribution of BAC-3a-seq reads and CsCenH3-ChIP-seq peaks on the C. sinensis genome v3. The density distribution of BAC-3a-seq is shown on the nine chromosomes, while the CsCenH3-ChIP-seq peaks are shown as the line labels below the nine chromosomes. BAC, bacterial artificial chromosome; BAC-3a-seq, bacterial artificial chromosome-3a sequencing; CsCen, C. sinensis centromere; CsCenH3-ChIP-seq, chromatin immunoprecipitation sequencing using CsCenH3 antibody; v3, version 3; Chr, chromosomes
Characterization of centromere-specific repeats in BAC-3a-seq: five TRs and eight LTR-RTs identified
To analyze potential centromeric candidate repeats in BAC-3a-seq reads, we adopted a strategy that combined RepeatMasker and TRs Finder to identify patterns within the 12 scaffolds. Initially, 48 TRs and 84 LTR-RTs were identified as preliminary candidates. Through a subsequent genome-wide BLAST of these 132 repeats, we selected five TRs and eight LTR-RTs that demonstrated higher quality and centromere-specific characteristics (Table 2). Among these, five TRs and eight LTR-RTs were found to be located at the centromere or in close proximity to the centromere within the C. sinensis genome v3 (Fig. 5). Furthermore, the distribution pattern of these eight LTR-RTs closely resembled that of CsCen7 (Fig. 5b), which has been validated as an excellent molecular marker for C. sinensis centromeres (Song et al. 2023). This finding suggests that these eight LTR-RTs may also serve as molecular markers for centromeres.
Specific distribution of five TRs and eight LTR-RTs at the centromere in the C. sinensis genome v3. (a) The distribution of five TRs in the C. sinensis genome v3. (b) The distribution of CsCen7 and eight LTR-RTs in the C. sinensis genome v3. The numbers around the circle indicate the length of the nine chromosomes in megabases (Mb). TR, tandem repeat; LTR-RT, long terminal repeat retrotransposon; CsCen, C. sinensis centromere; v3, version 3; chr, chromosomes
Involvement of CL in the evolution of C. sinensis centromeres
The FISH results revealed that the predicted centromeric repeats CL (Song et al. 2023) and CsCen1 (Fig. 2b, c, d, e) were indeed localized at the ends of the C. sinensis chromosome, with no FISH signal observed at the centromeres (Fig. S2). To further investigate this, we positioned CsCenH3-ChIP-seq reads and CL on the C. sinensis genome v3 (Fig. 6a) and found that CsCenH3-ChIP-seq reads and CL co-localized at the ends of chromosomes 2 and 7 (Fig. 6a, black arrows). This co-localization suggests that CsCenH3 is highly enriched in regions where CL is abundantly distributed. Based on these observations, it is possible that during the evolution of C. sinensis, CL and CsCenH3 might have translocated to the ends of the chromosomes, possibly due to the breakage of the centromeric region of the chromosome (Fig. 6b). Alternatively, they may have directly migrated from the centromeres to the ends through chromosomal rearrangement (Fig. 6b).
The distribution of CL at the end of chromosomes was related to the evolution of C. sinensis centromeres. (a) The distribution of CL and CsCenH3-ChIP-seq reads on the C. sinensis genome v3. The density distribution of CL (green) is shown on the nine chromosomes, while the CsCenH3-ChIP-seq read peaks (red) are shown as the line labels below the nine chromosomes. The black arrows indicate two overlaps in the distribution of CL and CsCenH3-ChIP-seq reads. (b) A model diagram illustrating the involvement of CL in the evolution of the C. sinensis centromere. CsCen, C. sinensis centromere; CsCenH3-ChIP-seq, chromatin immunoprecipitation sequencing using CsCenH3 antibody. CL, the centromeric repeat from the published reference (Song et al. 2023); v3, version 3; Chr, chromosomes; H3, Histone H3
Abundance of DNA G4 structures: distribution in centromeric LTR-RTs and extensive enrichment in centromeric TRs
Previous studies have suggested that CenH3 can identify centromeres by recognizing the secondary structure of DNA (Liu et al. 2023). In plants, non-B-form DNA tends to form in the regions where CenH3 binds (Liu et al. 2023), and DNA G4 structures have been observed in LTR-RTs (Lexa et al. 2014). In light of this, we used pqsfinder to detect the distribution of DNA G4 structures in the 24 centromeric candidate repeats mentioned earlier. Screening criteria were set with scores greater than 25, leading to the selection of 12 centromeric repeats with DNA G4 structures. Notably, the analysis revealed that DNA G4 structures were distributed in the centromeric LTR-RTs and showed significant enrichment in the centromeric TRs (Table 3). Notably, among the identified repeats, CsCen141 showed the highest percentage of G4 sequences (74.84%), the largest number of G4 structures (6), the highest score (115), and the most G-tetrads (6).
Discussion
Facilitating gap-free genome assembly in C. sinensis with centromeric repeats
Centromeric repeats play a crucial role in achieving a gap-free genome assembly for C. sinensis. However, due to the high genomic heterozygosity of C. sinensis (Song et al. 2023; Wang et al. 2021), the analysis of centromeres lags behind that of other major cash crops. Additionally, the sequencing of centromeric repeats is challenging due to the presence of numerous repeating DNA elements (Naish et al. 2021), which hinder gap-free genome assembly for C. sinensis. Therefore, it is essential to identify more centromeric candidate repeats to enhance our understanding of C. sinensis centromeres and improve genome assembly.
In this study, we used CsCenH3-ChIP-seq and centromere-specific BAC-3a-seq in C. sinensis, which allowed us to screen and identify 23 centromeric candidate repeats using bioinformatics methods. Among these repeats, CL exhibited high homology with CsCen1, CsCen3, and CsCen9 (Fig. 2a), and the FISH signals of CL and CsCen1 on the chromosomes were consistent (Fig. 2c), indicating that these four TRs belong to the same repeat. Moreover, the distribution pattern of the LTR-RTs CsCenL9, CsCenL27, CsCenL31, CsCenL49, CsCenL50, CsCenL53, CsCenL63, and CsCenL68 in the C. sinensis genome v3 resembled that of CsCen7 (Fig. 5b). Previous FISH mapping showed 18 CsCen7 signals at the centromeric region of each chromosome in C. sinensis (Song et al. 2023), suggesting that these eight LTR-RTs would also generate FISH signals at the centromeres. Additionally, we identified five RTs with nine centromere-specific LTR-RTs through genome-wide BLAST analysis, indicating that these nine LTR-RTs can serve as molecular markers to locate C. sinensis centromeres. With the potential use of T2T sequence assemblies and 14 centromere-specific repeats, a complete genome of C. sinensis may be achieved in the future.
Centromeric evolution provides clues for constructing ancestral chromosome karyotypes of C. sinensis
Over the past 12,000 years, more than 2,500 plant species have been domesticated from their wild ancestors to yield crops suitable for human consumption and economic development (Fernie and Yan 2019; Zhao et al. 2021). However, the complexities and variations in chromosomal evolutionary events have posed challenges in inferring ancestral karyotypes. With advancements in genome sequencing and analysis, studying the genomes of species can offer valuable insights for constructing ancestral karyotypes. In this study, we observed that the CsCenH3-ChIP-seq read peaks were not solely concentrated at the centromere of chromosomes; there were evident peaks at the ends and proximal parts of some chromosomes (Fig. 1d). Additionally, we observed overlaps between the peaks and the centromeric repeat CL at the ends of chromosomes 2 and 7. Since CenH3 is indicative of true centromeric regions, we infer that chromosomes 2 and 7 have been involved in the evolutionary process of C. sinensis (Fig. 6b). It is plausible that chromosomes 2 and 7 may have originated from a shared ancestor chromosome that underwent breakage at the centromere, and subsequent chromosomal rearrangement may have occurred between the centromeric regions of chromosomes 2 and 7 and their respective ends. Recently, a Python-based command-line tool named Whole-Genome Duplication Integrated (WGDI) analysis has demonstrated the capability to perform polyploid inference, genome homology hierarchical inference, and ancestral chromosome karyotype construction (Sun et al. 2022). Combining our findings with the application of WGDI in citrus may offer promising avenues for constructing ancestral karyotypes and exploring potential chromosomal rearrangement events in the near future.
DNA G4 structures in the CsCenH3 binding regions prefer to form in TRs
Plant DNA G4 structures have been implicated in various physiological processes, including the regulation of gene expression and translation, as well as plant growth, development, and stress responses (Cagirici and Sen 2020; Cho et al. 2018; Feng et al. 2022; Garg et al. 2016; Griffin and Bass 2018; Kwok et al. 2015; Yadav et al. 2017). Given that centromeric regions are known to be enriched with non-B-form DNA structures in oat (Liu et al. 2023), it is reasonable to infer that DNA G4 structures are also present in C. sinensis centromeres. Using pqsfinder, we identified 24 centromeric candidate repeats of C. sinensis, including CL. As anticipated, DNA G4 structures were detected in both TRs and LTR-RTs. Notably, the DNA G4 proportion in TRs was found to be significantly higher than that in LTR-RTs, with CsCen141 exhibiting the highest DNA G4 proportion. These results suggest that DNA G4 structures are not only distributed in the CsCenH3 binding regions in C. sinensis but also tend to form more frequently in TRs compared to LTR-RTs. Currently, blood group 4 (BG4) is utilized as an in vitro G4 binding protein for visualizing DNA G4 structures in human and plant cells (Biffi et al. 2013; Fang et al. 2019; Feng et al. 2022; Zhang et al. 2018). The application of BG4-based ChIP-seq or IP-seq, in combination with TR analysis, holds promise for further exploration of the relationship between DNA G4 structures and centromeres.
Conclusions
In summary, our study successfully identified the centromeric regions of C. sinensis through CsCenH3-ChIP-seq and BAC-3a-seq, resulting in the selection of 23 centromeric candidate repeats. Subsequently, we screened and identified five centromere-specific TRs and nine LTR-RTs from these centromeric candidate repeats by conducting genome-wide BLAST analysis against the C. sinensis genome v3. Furthermore, we analyzed the evolution of C. sinensis centromeres and the distribution of DNA G4 structures. These findings offer novel insights into the composition and evolution of C. sinensis centromeres, laying a solid theoretical foundation for further exploring the relationship between the primary and secondary structures of centromeres.
Availability of data and materials
Data and materials will be made available on request. The sequences of all centromeric candidate repeats are included in Dataset S1.
References
Belser C, Baurens F-C, Noel B, Martin G, Cruaud C, Istace B, et al. Telomere-to-telomere gapless chromosomes of banana using nanopore sequencing. Commun Biol. 2021;4:1047–58. https://doi.org/10.1038/s42003-021-02559-3.
Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–80. https://doi.org/10.1093/nar/27.2.573.
Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem. 2013;5:182–6. https://doi.org/10.1038/nchem.1548.
Cagirici HB, Sen TZ. Genome-wide discovery of G-quadruplexes in wheat: distribution and putative functional roles. G3-Genes Genom Genet. 2020;10:2021–32. https://doi.org/10.1534/g3.120.401288.
Cho H, Cho HS, Nam H, Jo H, Yoon J, Park C, et al. Translational control of phloem development by RNA G-quadruplex-JULGI determines plant sink strength. Nat Plants. 2018;4:376–90. https://doi.org/10.1038/s41477-018-0157-2.
Comai L, Maheshwari S, Marimuthu MPA. Plant centromeres. Curr Opin Plant Biol. 2017;36:158–67. https://doi.org/10.1016/j.pbi.2017.03.003.
Crespi M, Ariel F. Non-B DNA structures emerging from plant genomes. Trends Plant Sci. 2022;27:624–6. https://doi.org/10.1016/j.tplants.2022.03.004.
Deng Y, Liu S, Zhang Y, Tan J, Li X, Chu X, et al. A telomere-to-telomere gap-free reference genome of watermelon and its mutation library provide important resources for gene discovery and breeding. Mol Plant. 2022;15:1268–84. https://doi.org/10.1016/j.molp.2022.06.010.
Fang Y, Chen L, Lin K, Feng Y, Zhang P, Pan X, et al. Characterization of functional relationships of R-loops with gene transcription and epigenetic modifications in rice. Genome Res. 2019;29:1287–97. https://doi.org/10.1101/gr.246009.118.
Feng C, Yuan J, Bai H, Liu Y, Su H, Liu Y, et al. The deposition of CENH3 in maize is stringently regulated. Plant J. 2020;102:6–17. https://doi.org/10.1111/tpj.14606.
Feng Y, Tao S, Zhang P, Sperti FR, Liu G, Cheng X, et al. Epigenomic features of DNA G-quadruplexes and their roles in regulating rice gene transcription. Plant Physiol. 2022;188:1632–48. https://doi.org/10.1093/plphys/kiab566.
Fernandes JB, Wlodzimierz P, Henderson IR. Meiotic recombination within plant centromeres. Curr Opin Plant Biol. 2019;48:26–35. https://doi.org/10.1016/j.pbi.2019.02.008.
Fernie AR, Yan J. De novo domestication: an alternative route toward new crops for the future. Mol Plant. 2019;12:615–31. https://doi.org/10.1016/j.molp.2019.03.016.
Garg R, Aggarwal J, Thakkar B. Genome-wide discovery of G-quadruplex forming sequences and their functional relevance in plants. Sci Rep. 2016;6:28211. https://doi.org/10.1038/srep28211.
Giguere DJ, Bahcheli AT, Slattery SS, Patel RR, Browne TS, Flatley M, et al. Telomere-to-telomere genome assembly of Phaeodactylum tricornutum. PeerJ. 2022. https://doi.org/10.7717/peerj.13607.
Gonzalo L, Tossolini I, Gulanicz T, Cambiagno DA, Kasprowicz-Maluski A, Smolinski DJ, et al. R-loops at microRNA encoding loci promote co-transcriptional processing of pri-miRNAs in plants. Nat Plants. 2022;8:402–18. https://doi.org/10.1038/s41477-022-01125-x.
Griffin BD, Bass HW. Review: Plant G-quadruplex (G4) motifs in DNA and RNA; abundant, intriguing sequences of unknown function. Plant Sci. 2018;269:143–7. https://doi.org/10.1016/j.plantsci.2018.01.011.
Hao Z, Lv D, Ge Y, Shi J, Weijers D, Yu G, et al. RIdeogram: drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ Comput Sci. 2020. https://doi.org/10.7717/peerj-cs.251.
Hiatt EN, Kentner EK, Dawe RK. Independently regulated neocentromere activity of two classes of tandem repeat arrays. Plant Cell. 2002;14:407–20. https://doi.org/10.1105/tpc.010373.
Hofstatter PG, Thangavel G, Lux T, Neumann P, Vondrak T, Novak P, et al. Repeat-based holocentromeres influence genome architecture and karyotype evolution. Cell. 2022;185:3153–68.e18. https://doi.org/10.1016/j.cell.2022.06.045.
Hou H, Kyriacou E, Thadani R, Klutstein M, Chapman JH, Cooper JP. Centromeres are dismantled by foundational meiotic proteins Spo11 and Rec8. Nature. 2021;591:671–6. https://doi.org/10.1038/s41586-021-03279-8.
Hou X, Wang D, Cheng Z, Wang Y, Jiao Y. A near-complete assembly of an Arabidopsis thaliana genome. Mol Plant. 2022;15:1247–50. https://doi.org/10.1016/j.molp.2022.05.014.
Huang Y, Ding W, Zhang M, Han J, Jing Y, Yao W, et al. The formation and evolution of centromeric satellite repeats in Saccharum species. Plant J. 2021;106:616–29. https://doi.org/10.1111/tpj.15186.
Kwok CK, Ding Y, Shahid S, Assmann SM, Bevilacqua PC. A stable RNA G-quadruplex within the 5’-UTR of Arabidopsis thaliana ATR mRNA inhibits translation. Biochem J. 2015;467:91–102. https://doi.org/10.1042/BJ20141063.
Lexa M, Kejnovský E, Šteflová P, Konvalinová H, Vorlíčková M, Vyskot B. Quadruplex-forming sequences occupy discrete regions inside plant LTR retrotransposons. Nucleic Acids Res. 2014;42:968–78. https://doi.org/10.1093/nar/gkt893.
Liu Y, Su H, Liu Y, Zhang J, Dong Q, Birchler J-A, et al. Cohesion and centromere activity are required for phosphorylation of histone H3 in maize. Plant J. 2017;92:1121–31. https://doi.org/10.1111/tpj.13748.
Liu J, Seetharam A-S, Chougule K, Ou S, Swentowsky K-W, Gent J-I, et al. Gapless assembly of maize chromosomes using long-read technologies. Genome Biol. 2020;21:121. https://doi.org/10.1186/s13059-020-02029-9.
Liu H, Wang X, Liu S, Huang Y, Guo Y-X, Xie W-Z, et al. Citrus Pan-genome to Breeding Database (CPBD): a comprehensive genome database for citrus breeding. Mol Plant. 2022;15:1503–5. https://doi.org/10.1016/j.molp.2022.08.006.
Liu Q, Yi C, Zhang Z, Su H, Liu C, Huang Y, et al. Non-B-form DNA tends to form in centromeric regions and has undergone changes in polyploid oat subgenomes. Proc Natl Acad Sci U S A. 2022;120:e2211683120. https://doi.org/10.1073/pnas.2211683120.
Melters D-P, Bradnam K-R, Young H-A, Telis N, May M-R, Ruby J-G, et al. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 2013;14:R10. https://doi.org/10.1186/gb-2013-14-1-r10.
Miglietta G, Russo M, Capranico G. G-quadruplex-R-loop interactions and the mechanism of anticancer G-quadruplex binders. Nucleic Acids Res. 2020;48:11942–57. https://doi.org/10.1093/nar/gkaa944.
Naish M, Alonge M, Wlodzimierz P, Tock A-J, Abramson B-W, Schmucker A, et al. The genetic and epigenetic landscape of the Arabidopsis centromeres. Science. 2021;374:eabi7489. https://doi.org/10.1126/science.abi7489.
Navratilova P, Toegelova H, Tulpova Z, Kuo YT, Stein N, Dolezel J, et al. Prospects of telomere-to-telomere assembly in barley: analysis of sequence gaps in the MorexV3 reference genome. Plant Biotechnol J. 2022;20:1373–86. https://doi.org/10.1111/pbi.13816.
Nie S, Wang B, Ding H, Lin H, Zhang L, Li Q, et al. Genome assembly of the Chinese maize elite inbred line RP125 and its EMS mutant collection provide new resources for maize genetics research and crop improvement. Plant J. 2021;108:40–54. https://doi.org/10.1111/tpj.15421.
Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze A-V, Mikheenko A, et al. The complete sequence of a human genome. Science. 2022;376:44–53. https://doi.org/10.1126/science.abj6987.
Oliveira L-C, Torres G-A. Plant centromeres: genetics, epigenetics and evolution. Mol Biol Rep. 2018;45:1491–7. https://doi.org/10.1007/s11033-018-4284-7.
Plohl M, Meštrović N, Mravinac B. Centromere identity from the DNA point of view. Chromosoma. 2014;123:313–25. https://doi.org/10.1007/s00412-014-0462-0.
Song J-M, Xie W-Z, Wang S, Guo Y-X, Koo D-H, Kudrna D, et al. Two gap-free reference genomes and a global view of the centromere architecture in rice. Mol Plant. 2021;14:1757–67. https://doi.org/10.1016/j.molp.2021.06.018.
Song S, Liu H, Miao L, He L, Xie W, Lan H, et al. Molecular cytogenetic map visualizes the heterozygotic genome and identifies translocation chromosomes in Citrus sinensis. J Genet Genomics. 2023;50:410–21. https://doi.org/10.1016/j.jgg.2022.12.003.
Su H, Liu Y, Liu C, Shi Q, Huang Y, Han F. Centromere satellite repeats have undergone rapid changes in polyploid wheat subgenomes. Plant Cell. 2019;31:2035–51. https://doi.org/10.1105/tpc.19.00133.
Sun P, Jiao B, Yang Y, Shan L, Li T, Li X, et al. WGDI: a user-friendly toolkit for evolutionary analyses of whole-genome duplications and ancestral karyotypes. Mol Plant. 2022;15:1841–51. https://doi.org/10.1016/j.molp.2022.10.018.
Wang L, Huang Y, Liu Z, He J, Jiang X, He F, et al. Somatic variations led to the selection of acidic and acidless orange cultivars. Nat Plants. 2021;7:954–65. https://doi.org/10.1038/s41477-021-00941-x.
Wlodzimierz P, Rabanal F-A, Burns R, Naish M, Primetis E, Scott A, et al. Cycles of satellite and transposon evolution in Arabidopsis centromeres. Nature. 2023;618:557–65. https://doi.org/10.1038/s41586-023-06062-z.
Xia Q-M, Miao L-K, Xie K-D, Yin Z-P, Wu X-M, Chen C-L, et al. Localization and characterization of Citrus centromeres by combining half-tetrad analysis and CenH3-associated sequence profiling. Plant Cell Rep. 2020;39:1609–22. https://doi.org/10.1007/s00299-020-02587-z.
Xu Q, Chen L-L, Ruan X, Chen D, Zhu A, Chen C, et al. The draft genome of sweet orange (Citrus sinensis). Nat Genet. 2013;45:59–66. https://doi.org/10.1038/ng.2472.
Yadav V, Hemansi N, Kim N, Tuteja N, Yadav P. G quadruplex in plants: a ubiquitous regulatory element and its biological relevance. Front Plant Sci. 2017;8:1163. https://doi.org/10.3389/fpls.2017.01163.
Yang X, Zhao H, Zhang T, Zeng Z, Zhang P, Zhu B, et al. Amplification and adaptation of centromeric repeats in polyploid switchgrass species. New Phytol. 2018;218:1645–57. https://doi.org/10.1111/nph.15098.
Zhang W, Zuo S, Li Z, Meng Z, Han J, Song J, et al. Isolation and characterization of centromeric repetitive DNA sequences in Saccharum spontaneum. Sci Rep. 2017;7:41659. https://doi.org/10.1038/srep41659.
Zhang S, Sun H, Wang L, Liu Y, Chen H, Li Q, et al. Real-time monitoring of DNA G-quadruplexes in living cells with a small-molecule fluorescent probe. Nucleic Acids Res. 2018;46:7522–32. https://doi.org/10.1093/nar/gky665.
Zhang Y, Fu J, Wang K, Han X, Yan T, Su Y, et al. The telomere-to-telomere gap-free genome of four rice parents reveals SV and PAV patterns in hybrid rice breeding. Plant Biotechnol J. 2022;20:1642–4. https://doi.org/10.1111/pbi.13880.
Zhang L, Liang J, Chen H, Zhang Z, Wu J, Wang X. A near-complete genome assembly of Brassica rapa provides new insights into the evolution of centromeres. Plant Biotechnol J. 2023;21:1022–32. https://doi.org/10.1111/pbi.14015.
Zhao Q, Meng Y, Wang P, Qin X, Cheng C, Zhou J, et al. Reconstruction of ancestral karyotype illuminates chromosome evolution in the genus Cucumis. Plant J. 2021;107:1243–59. https://doi.org/10.1111/tpj.15381.
Zhao J, Xie Y, Kong C, Lu Z, Jia H, Ma Z, et al. Centromere repositioning and shifts in wheat evolution. Plant Commun. 2023;4:100556. https://doi.org/10.1016/j.xplc.2023.100556.
Zhou C, Olukolu B, Gemenet D-C, Wu S, Gruneberg W, Cao M-D, et al. Assembly of whole-chromosome pseudomolecules for polyploid plant genomes using outbred mapping populations. Nat Genet. 2020;52:1256–64. https://doi.org/10.1038/s41588-020-00717-7.
Zhou J, Liu Y, Guo X, Birchler JA, Han F, Su H. Centromeres: from chromosome biology to biotechnology applications and synthetic genomes in plants. Plant Biotechnol J. 2022;20:2051–63. https://doi.org/10.1111/pbi.13875.
Acknowledgements
We appreciate Dr. Simon Moore and Mr. Dengyue Zheng for English language editing.
Funding
This work was supported by the National Natural Science Foundation of China (31970525), the National Key Research and Development Project of China (2019YFD1001401-GJ03), and the Cultivating Fund Project of Hubei Hongshan Laboratory (2022hspy002).
Author information
Authors and Affiliations
Contributions
Shipeng Song: investigation, methodology, validation, visualization, formal analysis, original draft (writing), review and editing (writing). Hui Liu: investigation, methodology, validation, visualization, formal analysis. Luke Miao: investigation, methodology, validation, visualization. Hong Lan: investigation, methodology, validation, visualization. Chunli Chen: conceptualization, funding acquisition, project administration, supervision.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests. The authors declare that they have no financial or nonfinancial interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Fig. S1.
Circos plot of ten centromeric candidate repeats on sweet orange genome v3. Fig. S2. The distribution of FISH signals of CL and CsCen1 on nine pseudo chromosomes of C. sinensis.
Additional file 2: Dataset S1.
Sequences of centromeric candidate repeats.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Song, S., Liu, H., Miao, L. et al. Centromeric repeats in Citrus sinensis provide new insights into centromeric evolution and the distribution of G-quadruplex structures. HORTIC. ADV. 1, 7 (2023). https://doi.org/10.1007/s44281-023-00010-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s44281-023-00010-7