Abstract
Dynamic DNA nanotechnology belongs to a larger umbrella of DNA nanotechnology that primarily uses DNA as a nanoscopic material to build mobile structures and cascaded reaction networks powered by DNA oligonucleotides. A widely used mechanism to construct a dynamic DNA system is toehold-mediated strand displacement reactions (TMSDRs). TMSDRs are easy to engineer because of the known base-pairing rules that follow the Watson–Crick model of DNA, sequence-dependent binding rates, and energies of DNAs, whose secondary structure is predictable. Due to these attributes, TMSDRs have been used to develop enzyme-free isothermal reaction networks with remarkable applications in diagnostics, therapeutics and DNA computing. In this review, we briefly introduce the working principle of TMSDRs, in silico design considerations, and diverse input and output signals that can be processed through TMSDRs. We then summarize recent applications where TMSDRs are successfully employed in detecting clinically relevant targets such as single nucleotide polymorphisms and variants, microRNAs and whole cells and to develop programmable drug delivery vehicles and regulation therapies including transcriptional and protein regulations. We also discuss TMSDRs driven biomedical applications of DNA hydrogels and DNA computing. Finally, we discuss the challenges in each of these applications and the prospects of TMSDRs in biomedical engineering.
Highlights
• Dynamic DNA nanotechnology can provide a viable solution to healthcare needs.
• Programmability and adaptability make dynamic DNA systems an excellent substrate for designing smart materials.
• This review discusses the most recent biomedical applications of toehold-mediated DNA strand displacement reactions.
AbstractSection Graphical Abstract
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Deoxyribonucleic acid (DNA) is commonly known as a genetic material that contains the necessary biological information for the development and growth of an organism. DNA was considered a read-only component until Seeman replicated the Holliday junction using short DNA strands called DNA oligonucleotides [1]. Using the complementary base pairing of DNA oligonucleotides, a thermodynamically stable immobile structure with nanoscale features was formed. This pioneering work where DNA was used as a nanoscopic material to assemble an artificial structure has led to the inception of an interdisciplinary emerging research area called DNA nanotechnology.
Among the major subtypes of DNA nanotechnology, static DNA technology uses DNA as a nanoscopic biomaterial to construct self-assembled 2- and 3-dimensional nanostructures such as DNA origami, nanotubes, lattices, nanopores, and cages [2,3,4,5]. On the other hand, dynamic DNA nanotechnology involves interaction between complementary DNA oligonucleotides to build active nanostructures and dynamic biochemical reaction networks. For example, DNA walkers, motors, and oscillators use various environmental and molecular stimuli to reconfigure due to changes in the thermodynamics and kinetics of DNA oligonucleotides [6,7,8].
The highly specific and predictable nature of complementary DNA oligonucleotides allow modular components to be rationally designed and integrated into more complex systems. The kinetics of DNA hybridization and dissociation can be accurately predicted, enabling in silico analysis [9, 10]. This makes DNA systems more predictive, modular, and programmable than genetic or other molecular platforms [11,12,13]. These attributes have been used to build extended nanostructures and programable cascades of binding and unbinding reactions that can perform operations similar to digital and analog electronic circuits. The complementary DNA sequences interact with each other through non-covalent and supramolecular interactions and reconfigure spontaneously in an aqueous solution [14], which enable realization of complex molecular interactions capable of performing computing functions.
There are different ways to bring dynamicity to a DNA system such as environmental stimuli, which involve changes in pH [15], temperature [16], and light [17]. However, this review focuses primarily on dynamic DNA systems that are driven by strand displacement reactions and use DNA oligonucleotides as fuel, which was first proposed by Simmel and Yurke [18]. Strand displacement reaction is a molecular process that brings motion in DNA systems, where a single-stranded (ss) DNA dissociates from a pre-hybridized strand to bind with another ssDNA oligonucleotide with a higher binding affinity. There are different mechanisms via which a strand displacement reaction can take place. This includes concentration disequilibria and concentration imbalances in a reaction that are driven by entropy differences [19, 20], catalytic reaction systems such as hybridization chain reaction (HCR) [21], and catalytic hairpin assembly [22]. Among all these methods, toehold-mediated strand displacement is one of the widely used mechanisms for bringing motion in DNA systems.
This review focuses on dynamic DNA systems driven by toehold-mediated strand displacement reactions (TMSDRs). In this paper, we introduce the working principle of TMSDRs, design strategies, computational tools, and diverse input and output signals that are employed by TMSDRs. We then discuss the recent versatile applications of TMSDRs in biosensing and therapeutics including those that use responsive DNA hydrogels and DNA computing. The biosensing section is divided into various subclasses based on the biological component that is being detected such as single nucleotide polymorphisms (SNPs), single nucleotide variants (SNVs), microRNA (miRNA) and whole cells. We then discuss in situ and DNA hydrogel based detection mechanisms. The therapeutics section is divided into subsections describing targeted drug delivery carriers and regulation therapies including DNA hydrogel based applications. We also discuss use of DNA computing to implement multi-input and output systems with practical applications. Finally, we discuss challenges involving TMSDRs and the potential future applications of dynamic DNA nanotechnology in biomedical engineering.
Toehold-mediated strand displacement reactions
Programmability, ease of modeling and conversion of diverse chemical and physical signals into ssDNA make TMSDRs an excellent substrate for designing and synthesizing arbitrary reaction networks and logic circuits with practical applications. In this section, we discuss some of these aspects of TMSDRs including the working principle.
Working principle of TMSDRs
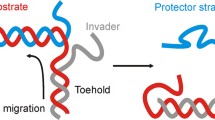
TMSDRs are fueled by DNA oligonucleotides and the strand displacement process is driven by thermodynamic and kinetic changes. Strand displacement initiates from ssDNA overhang region, called toehold. An invading strand can displace the partially complementary strand through branch migration without enzymatic assistance. This process is called toehold-mediated strand displacement reaction [7]. Referring to Fig. 1a, in a forward reaction, the complex ST has an exposed toehold domain (α*); when the U strand, which is fully complementary to the T strand, is present in the solution, it interacts with complex ST through the toehold domain (α) and displaces the shorter S strand. The coexistence of UT and S is thermodynamically more favorable than the coexistence of ST and U [23]. This is a forward-driven reaction as the complex UT is fully complementary and has no exposed toehold where strand S can bind. However, in a reversible reaction, the complex UT has an exposed toehold domain γ*, as shown in Fig. 1b, which is single-stranded compared to the earlier case where the UT complex was fully complementary (see Fig. 1a). In this case, the S strand with the exposed toehold domain γ interacts with the UT complex through the complementary domain γ* and might displace the U strand to bind to the T strand reversibly to form ST complex. As both strands U and S are partially complementary to the T strand, there is a competitive interaction of both the strands with the strand T, and the coexistence of complexes UT and ST depends on the sequence composition of domain γ with respect to domain α [9].
Schematic representation of toehold-mediated strand displacement reaction. DNA is drawn as directional lines where the sequence information is omitted, and the arrow indicates 5' to 3' direction of the backbone. The sequence is divided into different color domains that have specific functions, while * indicates a complementary domain. a A forward reaction takes place only in the forward direction such reactants get converted into products. The ssDNA oligonucleotide U initiates the displacement of the strand S from the ST complex (also known as a duplex) by binding with the complementary toehold domain (α*). In the subsequent steps, the strand S is displaced through branch migration, forming complex UT and free strand S [9]. b In a reversible reaction, the product can also be converted into a reactant simultaneously. When the complex UT has an exposed toehold domain γ*, the strand S can interact with it via complementary domain γ, leading to a reversible reaction where the complexes ST and UT can co-existence [6]
Design considerations for TMSDRs
For designing TMSDRs, the sequence of DNA oligonucleotide is generally divided into domains that simplify the task of sequence designing and provide an easier understanding of interactions between the complementary strands. Referring to Fig. 1b, domains γ and α are toehold domains that facilitate the strand displacement process, while domain β is a branch-migration domain that provides thermodynamic stability to the duplex complexes UT and ST.
Designing the toehold domain is one of the most important aspects of TMSDRs as it plays an important role in initiating and regulating the kinetics of the strand displacement process. Second-order reaction rate constants for TMSDRs are on the order of 10–106 M−1 s−1 for toeholds of 1–8 bases, respectively [9]. Referring to Fig. 1b, the incoming invading strand U might bind at the toehold domain and displace the incumbent strand S through branch migration. For a quicker branch migration, the binding of the invading strand S with the complementary toehold domain should be thermodynamically favorable, which depends on the sequence composition of the toehold domain. For example, a toehold domain with more Gs and Cs has higher binding strength than with more As and Ts. This is due to the reduction in free energy (ΔG°) for the sequence composed of Gs and Cs, which increases the thermodynamic stability of the duplex complex [10].
Computational and mathematical tools for analyzing TMSDRs
One immediate challenge in building TMSDRs based complex systems that use many DNA oligonucleotides is undesired crosstalk, meaning spurious hybridization between DNA strands [7]. Such undesired interactions can hinder the strand displacement process and the formation of correct complexes. One way to minimize this is by automating sequence design which can help to maximize the formation of desired complexes at the equilibrium. For that, based on the nearest-neighbor model, several computer-aided software packages have been developed to predict the thermodynamic parameters of DNA oligonucleotides, such as caDNAno [24], Cando [25], oxDNA [26], and MagicDNA [27]. Among these tools, Nucleic Acid Package (NUPACK) is preferred as it helps to identify the most probable crosstalk between DNA oligonucleotides and tune the reaction kinetics by analyzing the free energy of DNA complexes [28, 29].
Note that simple equilibrium analysis may not be accurate for reversible cascaded reactions where the output of one reaction serves as an input to another reaction. For that, using the law of mass action kinetics, ordinary differential equation based dynamic models can be built to understand the dynamics of cascaded TMSDRs [23]. These models require reaction rate constants for all interactions corresponding to individual TMSDRs, which can be calculated using the rate constant model proposed by Zhang et al. [9] and other similar models [30]. These tools generally enable the design-build-test cycle for TMSDRs and provide a starting basis for further optimization and building more complex systems.
TMSDRs with different input and output signals
TMSDRs are an excellent substrate for developing practical applications (Fig. 2a) that can respond to chemical and physical input signals (Fig. 2b). These input signals can be processed and transduced to produce different types of output signals in response to the input through TMSDRs (Fig. 2b) [31]. We now discuss recent developments where TMSDRs have been used to respond to and produce diverse input and output signals.
a TMSDRs have been used to develop highly sensitive biosensors to detect clinically relevant targets, programmable drug delivery carriers for targeted therapeutics, regulation strategies to control the activity of clinically relevant molecules, responsive soft biomaterials like smart DNA hydrogels and DNA computing based applications. b Processing different input and output signals through TMSDRs [31]. Various chemical and physical signals such as adenosine triphosphate (ATP), pH, light, temperature, and electric signals can be converted to ssDNA while this information can be processed and produced in the form of diverse output signals such as change in fluorescence using fluorescence quenching mechanism and fluorescence resonance energy transfer (FRET), colorimetric method, gel-electrophoresis and atomic force microscopy (AFM)
Processing chemical and physical input signals
One of the most used methods to process different types of inputs is via DNA aptamers that are designed to bind to various non-nucleic acid molecules such as small molecules, peptides, proteins, viruses, and cells with high affinity and specificity [32,33,34]. These DNA aptamers can be a part of TMSDRs such that any change in the input signal such as the concentration of ATP molecule, drives TMSDRs through its DNA aptamer that can bind to ATP or a complementary strand [35]. Building upon this, a more complex operation is achieved by Zhang et al. where a hairpin based DNA circuit is kinetically regulated via ATP and other molecules (see Fig. 3a) [36].
Examples of diverse input and output signals used by TMSDRs. a ATP as a signaling input. Here, a TMSDRs is initiated by ATP to produce a fluorescence signal through a ssDNA output. Reprinted with permission from Ref. [36], used under Creative Commons CC BY 4.0 license. b Temperature as a signaling input. Here, an increase in temperature above the melting point (Tm) opens the hairpin loop structure and releases a ssDNA output that can be used downstream in further cascaded reactions. Reprinted with permission from Ref. [37], used under Creative Commons CC BY license. c Colorimetric based output signal. Here, only in the presence of an input signal, a reporter-output complex can form that allows gold nanoparticles (AuNPs) to agglomerate, resulting in a detectable color change compared to dispersed AuNPs. Reprinted with permission from Ref. [38], used under Creative Commons CC BY 3.0 license. d Gel-electrophoresis based output signal. Here, the binding of HIV gene with a substrate probe (SP) results in an SP + HIV complex that has a higher molecular weight than SP alone. With the help of a fuel strand (FS), signal amplification can lead to higher fluorescence intensity. Reprinted with permission from Ref. [39], Copyright 2019, Royal Society of chemistry
TMSDRs can also respond to physical signals such as pH, light, temperature and electrical signals through chemical modifications in DNA oligonucleotides. For example, pH-responsive DNA logic circuits have been developed to regulate a DNA microgel [40] and reconfigurable DNA triplex structures [41]. Similarly, voltage-inducible oxidation based DNA nanoswitchs and nanodevices have been developed that employ TMSDRs to respond to electrical signals [42]. Additionally, temperature dependency on thermodynamic properties of DNA can be used to create conformational changes in DNA structure such as a fluorophore-tagged hairpin loop that melts at high temperatures (see Fig. 3b) [37]. Similarly, light can also act as a stimulus to trigger TMSDR using a photocleavable modified DNA linker that is sensitive to UV light [43].
Producing diverse output signals
In recent times, diverse output signals have been used to record the activity of TMSDRs. Some of these methods are fluorescence quenching, fFRET, electrochemical, colorimetric, gel-electrophoresis, and atomic force microscopy (AFM) as shown in Fig. 2b. Fluorescence quenching is one of the most common strategies employed in TMSDRs to record the dynamicity. In this approach, the physical spacing between the fluorophore and quencher molecules determines the fluorescence intensity in the output signal. On the other hand, in FRET, the physical spacing between acceptor and donor fluorophore molecules attached to the same or different DNA oligonucleotides determines the output fluorescence intensity. These approaches are widely used in detection mechanisms that use TMSDRs [44,45,46,47,48,49].
Additionally, a biochemical interaction such as between complementary DNA strands can be processed into a measurable electrical signal using a solid electrode surface that employs a redox reaction [50,51,52] while colorimetric readout strategies mainly use measurable color change upon reaction such as when gold nanoparticles (AuNPs) agglomerate (see Fig. 3c) [38] or using the peroxidase oxidation method, which provides a distinct colorimetric signal [53]. Finally, gel-electrophoresis along with fluorescence output signals have also been used for determining the presence of the HIV gene (see Fig. 3d) [39], and an atomic force microscopy (AFM) to study the conformational change in the structure of a DNA origami triggered by TMSDRs [54].
Biomedical applications of toehold-mediated strand displacement reactions
TMSDRs have been vastly explored in biomedical engineering due to their biocompatibility, modularity, tuneability, and enzyme-free functioning. This includes applications in biosensing such as detecting SNPs, SNVs, miRNA, and whole cells, and applications in therapeutics such as targeted drug delivery and regulation therapies. TMSDRs have also shown significant potential to develop responsive DNA hydrogels and to perform molecular computations. In the following section, we discuss each of these applications in detail.
Applications of TMSDRs in biosensing
An important goal of biological science and biomedical engineering is the detection of disease markers, bacteria, pathogens, and virus microorganisms to improve disease diagnosis and clinical care. Because of this, TMSDRs based detection mechanism is one of the most researched applications. Typically, in a biosensor, a bioreceptor binds to a target (input) molecule with high affinity via biochemical interactions. This is followed by a signal transducer that converts the biochemical interaction into a measurable signal via biophysical processes. In the final stage, the readout mechanism reads the measurable signal that quantitatively reflects the abundance of the target.
As mentioned earlier, a diverse range of target signals can be transduced via TMSDRs that has resulted in the development of several solution and surface based assays that exploit the programable nature of the TMSDRs. For example, in conventional methods, amplification of the detected signal is achieved by polymerase chain reaction (PCR) [55] and by isothermal methods such as rolling circle amplification (RCA) [56], and loop-mediated isothermal amplification (LAMP) [57] methods. However, these methods require probe labeling, heavily rely on enzyme manipulation and are time-consuming. TMSDRs based enzyme-free, one-pot reaction amplification system offers a simple yet sensitive way to detect up to the attomolar concentration and has been used to detect SNPs, miRNA, DNA, and whole cells.
Detection of single nucleotide polymorphisms and variants
SNPs and SNVs are essential biomarkers for diagnosing rare genetic diseases, which often involve point mutation in the genome. In the case of SNPs, the change is found in at least 1% of the population and serves as an essential biomarker in disease related genes and population based drug studies. On the other hand, in the case of SNVs, a single nucleotide change occurs without any frequency limitation and can be used to study gene variations related to disease diagnosis [58]. These mutations can be detected using rationally designed DNA nanostructures that employ TMSDRs, where a single base mutation in the DNA sequence may change the hybridization efficiency and thermodynamics of the reaction. For example, Ke et al. have demonstrated that RNA induced structural variations in DNA origami can be used for SNP genotyping [59]. Following this, Zhang et al. have developed a highly specific, labeling-free SNP detection system by combining TMSDRs and DNA origami [60]. Similarly, Dingzhong et al. have demonstrated the detection of SNPs using a hairpin based capture probe that is immobilized onto a gold electrode [61]. In the presence of the target strand, the hairpin loop unfolds via strand displacement reaction and hybridizes with a reporter probe.
In recent times, several TMSDRs based SNP detection methods have been proposed since small variations in the toehold sequence can significantly alter the dynamics of TMSDRs. In most studies, TMSDRs are used for signal amplification by recycling and disassociating the target strand. For instance, Gao et al. have proposed a locked nucleic acid (LNA) based switch where a capture DNA probe with an LNA base is used to detect SNP [62]. This capture probe binds to a residual probe on an electrode, and upon binding with the target strand, the capture probe displaces itself from the residual probe, turning the sensor OFF from the ON condition. Similarly, Wu et al. have developed an X-shaped LNA probe for SNV discrimination as shown in Fig. 4a [63]. The mutated target strand binds with the toehold domain of the X-probe and undergoes branch migration, and displaces the quencher strand, resulting in an enhanced fluorophore signal. In both these systems, the use of LNA has improved the system’s stability by increasing the melting temperature by 10 ºC. However, higher sensitivity was achieved in the latter case as each strand of the LNA probe can bind to multiple mutated strands. In an alternative approach, peptide nucleic acid-DNA hybrid probes have been used to increase the probe stability and affinity towards the target SNP [64].
Applications of TMSDRs in biosensing. a Use of TMSDRs for SNVs detection. Here, an X-shaped DNA probe with a quencher and fluorophore molecules disassociates when the complementary target strand binds with the DNA probe [63]. Further addition of two ssDNA probes, AP1 and AP2, releases the fluorescence tagged strand and the target strand to undergo multiple cycles of TMSDRs to amplify the fluorescence signal. Reprinted with permission from Ref. [63], copyright 2017, Elsevier B.V. b Use of TMSDRs for miRNA detection. Here, the target miRNA binds to the toehold and causes the displacement of the DNA walker probes, which further hybridize with a ferrocene-tagged signal probes [65]. The latter is released from the system using endonuclease, resulting in a detectable fluctuation in the oxidation current of the ferrocene. Reprinted with permission from Ref. [65], copyright 2020, American Chemical Society. c Use of TMSDRs for whole cell detection. Here, a hydrogel is functionalized with a DNA aptamer that can attach to specific cells during a dynamic flow [66]. The attached cells are programmed to be released at certain physiological conditions using strand displacement for further assay. Reprinted with permission from Ref. [66], copyright 2015, American Chemical Society. d Use of TMSDRs for real-time in situ detection. Here, a modular exchange network can detect dynamic changes in the target protein using a cascaded TMSDRs [23]. Reprinted with permission from Ref. [23], copyright 2020, Oxford University Press. e Use of TMSDRs driven DNA hydrogel for biosensing. Here, DNA oligonucleotide with aptamer-toehold sequence assists in specific binding to the cancer cells [67]. Two hairpin loops bind to the toehold and undergo a chain reaction to form hydrogel around the cell. Reprinted with permission from Ref. [67], copyright 2017, American Chemical Society
TMSDRs have also been used for signal decoding instead of amplification [68]. For example, Choi et al. have developed an encoding system comprising gold nanoparticles (AuNPs) immobilized with invader strands and a decoding system that decodes the invader strands using TMSDRs [69]. In this work, AuNPs encode the incoming target strands into invaders, which are diffused into DNA hydrogel microparticles. In the decoding step, the invader strands displace the quencher strands and emit a specific fluorescent signal. One important advantage of this method is that multiple invader strands can be immobilized on the surface, leading to a multiplex SNP detection system. Other studies combined the TMSDRs with other amplification methods to develop highly sensitive SNP detection systems. For example, Hu et al. have discriminated single base variations by employing cooperativity with TMSDRs [70] while Gu et al. have used HCR with TMSDRs for SNP genotyping with a detection limit of 20 aM [71].
With the increased use of TMSDRs for detecting SNPs and SNVs, new design considerations and strategies have also been reported [72, 73]. For example, Zhang et al. have studied the effects of mismatch location on TMSDRs kinetics using the oxDNA molecular dynamics simulation tool [74]. Their study concludes that mismatches near the toehold or the branch migration may lead to an improved specificity and discrimination of SNPs via TMSDRs.
Detection of MicroRNAs
MicroRNAs (miRNAs) are essential in regulating gene expressions in different immunological pathways. Quantifying the up-regulated miRNAs can play a significant role in early disease detection such as cancers [75], cardiovascular diseases [76], and neurological conditions [77]. Typically, miRNAs are detected by blotting techniques and quantitative reverse-transcription polymerase chain reaction (qRT-PCR). Over the last decade, isothermal amplification methods such as HCR, hairpin assembly [78], and rolling circle amplification [79] have been used for detecting miRNA. However, point-of-care and nanocomposite miRNA detection systems that employ TMSDRs have recently gained much attention as these systems mostly use linear DNA probes, which are comparatively easier to develop than traditional methods. For example, Lee et al. have developed a paper based colorimetric miRNA biosensor where the target miRNA initiates TMSDRs that amplify the DNA enzyme (DNAzyme) probes [80]. The free DNAzyme probes are adsorbed onto graphene oxide based paper, creating a color change perceptible to the naked eye. Similarly, Yang et al. have used the paper spray mass spectroscopy method for miRNA detection in blood where photocleavable compounds are functionalized onto AuNPs, which are released through TMSDRs in the presence of the target miRNA [81]. On exposure to light, photocleavable compounds are cleaved and quantified using spectroscopy with a detection limit of 9.72 fM.
TMSDRs have also been employed to determine the target miRNA using a personal glucometer [82]. Here, glucoamylase-encapsulated liposomes are functionalized onto magnetic beads, which are coupled with cascaded amplification reactions to produce glucose signals in response to the target miRNA. Similarly, Wang et al. have developed a portable glucometer based biosensor to detect miRNA using magnetic beads, which are immobilized with DNA probes [83].
Many authors have used DNA probes modified with nanoparticles to develop nanocomposite miRNA detection systems. For example, Lu et al. have used AuNPs as a substrate to immobilize a large number of DNA walkers and signal probes where the target miRNA triggers TMSDRs to facilitate the cleavage of the signal probes, resulting in the detection of the signal probes as shown in Fig. 4b [65]. Quantum dots-based miRNA detection systems have been developed by several authors [84,85,86]. Different geometric shapes have also been used to increase structural stability and avoid enzymatic degradation [87, 88]. In addition, the exosomal miRNA [89] and small RNAs [90] have also been detected using TMSDRs. Even though many authors have used various techniques to increase the stability and specificity of TMSDRs based miRNA detection systems, the signal may be distorted in human samples due to interference from other biomolecules. Further design improvements are required for more viable use of these systems in clinical settings.
Detection of whole cell
Transmembrane receptors on the cell surface can be an important indicator of specific cell populations. However, isolating cells in a viable condition from a huge heterogeneous population can be challenging. To address this, many authors have used the specificity of DNA aptamers and the programmability of TMSDRs to develop cell detection and separation assays. For example, Gaddes et al. have developed aptamer functionalized hydrogels for cell attachment as shown in Fig. 4c [66]. In this mechanism, polyvalent aptamers are synthesized using DNA toehold backbones that can bind to multiple cells via respective aptamers. To release the captured cells, the toehold backbone is disassembled by hybridizing with a complementary sequence. Although this method is non-invasive, the activity of the cells is reduced compared to that of the natural cells. To increase accuracy and sensitivity, Chang et al. have used multiple DNA aptamers integrated with TMSDRs to perform AND logic gate operation to detect multiple surface markers present on the tumor cells [91]. Similarly, Guo et al. have used tumor specific DNA aptamers functionalized on a zinc based microchip to retrieve the captured cells via a complementary toehold [92].
To increase the specificity, another type of whole cell biosensors have used antibodies instead of DNA aptamers as a bioreceptor. For example, Li et al. have used antibody crosslinked DNA probes coated with immuno-magnetic beads to develop a highly specific whole cell biosensor [93]. Here, antibody crosslinked DNA probes bind to the over-expressed antigens of the cancer cells, and immuno-magnetic beads help in the magnetic separation of the cells with high specificity.
TMSDRs driven whole cell detection and separation methods can replace the invasive biopsy technique as these methods often do not require any chemical modifications on the cell surface and are highly specific due to the utilization of aptamers and antibodies based DNA hybrids. However, maintaining cell activity and viability after the cell release could be challenging, and further advancements are needed for effective cell separation.
Real-time in situ detection mechanisms
A real-time in situ detection mechanism, which can be easily adapted to detect clinically relevant molecules modularly, can be used to develop multi-layered, multi-input biosensors with complex network connectivity. For example, Agrawal et al. have developed an exchange network to detect biologically relevant proteins such as human α-thrombin [23] —a serine protease that controls the activation of the coagulation cascade [94]. The exchange network uses DNA aptamers as bioreceptors to detect specific proteins and a cascade of TMSDRs to produce a programmable DNA oligonucleotide output (see Fig. 4d). Using this strategy, four orthogonal biosensors are constructed that can detect the rise and fall in protein concertation in real-time. Building on this, as described earlier, Zhang et al. engineered a hairpin based ligand binding sequence that employs DNA aptamer where binding of the ligand with the aptamer causes a conformational change in the hairpin stem to produce a detectable change in the fluorescence signal (Fig. 3a) [36]. This strategy might provide the necessary flexibility in designing sensitive modular detection mechanisms. Such in situ mechanisms can play an essential role in pharmacokinetics to understand the interaction of a drug with other biomolecules in real-time.
A critical challenge in developing real-time in situ detection mechanisms is the limited reaction lifetime due to the limited concentration of fuel strands. To address this, Deng et al. have used ATP-fueled TMSDRs to fabricate a programmable network where toehold length is altered to reset the reaction network using restriction endonuclease [95]. Similarly, Liu et al. have developed an advanced dissipative network by combining multiple fuels such as deoxyribonucleotide triphosphates and ATP [96].
Toehold-triggered DNA hydrogels for biosensing
Hydrogels are entangled hydrophilic polymer network structures that can swell and retain water in the swollen state [97]. DNA hydrogels are composed of DNA oligonucleotides that polymerize when complementary strands are hybridized to form networks. Similarly, TMSDRs can be used to direct the formation of DNA hydrogels via strand displacement that can trigger polymerization. In one of the earliest works, to create thermoresponsive DNA hydrogels, complementary base-pairing of DNA oligonucleotides is used along with N-dimethylacrylamide and N-acryloyloxysuccinimide as copolymers [98]. Since then, many DNA hydrogels have been synthesized using different isothermal amplification methods like DNA-walker [99], RCA [100], and hairpin based amplification methods [101].
DNA-responsive hydrogels driven by TMSDRs offer many advantages as a biosensor. They can function in aqueous media and mimic cellular response by size expansion, even when the target is at a low concentration. For example, Song et al. have developed a liquid non-invasive biopsy technique for live cell analysis by employing the sol–gel phase transition of a hydrogel to detect the circulating tumor cells as shown in Fig. 4e [67]. This method achieved high specificity using a DNA aptamer trigger that binds to epithelial cell adhesion molecule (EpCAM) on the cell surface. In the presence of the target EpCAM, TMSDRs induce hydrogel encapsulation of the viable cells for further testing. Another group of researchers has developed a hydrogel microneedle patch that can detect miRNA in the dermal interstitial fluid [102]. Here, the target miRNA acts as an input trigger to break the crosslinks of the hydrogel using strand displacement and produce an amplified fluorescence signal.
TMSDRs triggered DNA hydrogels have also been utilized to enhance fluorescence signal. For example, Liu et al. have used a biotinylated DNA hairpin probe as a fluorescence amplifier [103]. In the presence of the target catalyst and streptavidin, the DNA probes aggregate with streptavidin using TMSDRs to form a submicron hydrogel, resulting in enhanced fluorescence. Such mechanisms could be used to develop highly sensitive biosensors to detect clinically relevant targets relatively easily.
Employing other techniques to enhance sensitivity and stability of TMSDRs based biosensors
Detecting the lowest amount of relevant disease biomarkers could be key in early diagnosis. This led to an effort to develop detection mechanisms that employ different amplification methods to enhance the detection limits. Some of the methods include HCR, catalytic hairpin assembly (CHA), entropy-driven catalysis (EDC), and DNA walkers. For example, Zhang et al. have developed a highly sensitive TMSDRs based aptasensor that employs HCR and DNA aptamer to detect a highly toxic mycotoxin with a detection limit of 0.01 pg mL−1 [104]. CHA is also a commonly used amplifying mechanism to develop ultrasensitive biosensors [105]. For example, Khajouei et al. have fabricated a colorimetric biosensor that undergoes CHA and this led to the formation of DNA hydrogel via TMSDRs in the presence of the initiator strands [106].
Combinational methods have also been used to increase the stability and sensitivity of the biosensors. For instance, Xing et al. have developed a highly sensitive dual amplification method that combines EDC and HCR to detect nucleic acids and small molecules accurately [107]. An ultrasensitive biosensor that uses DNA walkers has been developed to detect DNA in human serum with a detection limit of 36 fM [108]. DNAzymes are also used with TMSDRs to improve the reaction stability and affinity towards the target. Using this, Zhao et al. have developed an electrochemical biosensor to detect lead with a detection limit of 0.32 pM [51]. Alternatively, Song et al., used multiplex blocker displacement amplification to sense SNVs, with variant allele frequency below 1%, which can go as low as 0.019% in human cell lines [109]. Other traditional amplification methods like PCR [110], LAMP [111], and RCA [112], have also been used along with TMSDRs to enhance the sensitivity of the biosensors. Further details of the strategies mentioned above for signal amplification have been reviewed extensively elsewhere [113, 114].
Applications of TMSDRs in therapeutics
TMSDRs can be designed to perform logical operations and signal transduction by converting the target signal into a DNA output signal, which can then be used to exhibit kinetic control over the drug release. We now discuss such mechanisms that employ TMSDRs for targeted drug delivery and regulation therapies.
Targeted drug delivery
Targeted therapy is one of the most significant applications in biomedical engineering. Detection mechanisms can be coupled to drug delivery carriers to form a range of applications termed theranostics, where the output signal from a detection mechanism can induce the targeted drug release. Such a mechanism can reduce drug toxicity by increasing the specificity and minimizing mistarget bindings. For example, Zhang et al. have developed a programmable DNA nanomachine that uses extracellular and intracellular inputs to control photodynamic therapy precisely through transmembrane TMSDRs as shown in Fig. 5a [115]. Similarly, Mirzaiebadizi et al. have developed DNA nanorobots that are immobilized in the pores of mesoporous silica nanoparticle drug carriers [116] and the targeted drug release is programmed when the two target strands interact with the nanorobots via an AND gate operation.
Applications of TMSDRs in targeted drug delivery and regulation therapies. a Use of TMSDRs driven DNA nanomachine for transmembrane drug delivery. Here, DNA strands on the nanoparticles bind to input 1, a transmembrane receptor, which is over-expressed in the cancer cells, leading to endocytosis of the DNA nanomachine [115]. Hybridization of the toehold with intracellular miRNA, which is input 2, leads to the activation of phototherapy inside the cell. Reprinted with permission from Ref. [115], copyright 2021, American Chemical Society. b Use of TMSDRs driven DNA nanostructures for targeted drug delivery. Here, interaction with the target strand disassembles the DNA tetrahedron, which can be used for controlled drug release [117]. Reprinted with permission from Ref. [117], used under Creative Commons CC BY-NC 3.0 license. c Use of TMSDRs for regulating gene transcription. Here, complementary ssDNA and ssRNA reassociate on mutual interaction with another hybrid and release NF-kB decoys that block NF-kB proteins, thereby inhibiting gene expression [54]. Reprinted with permission from Ref. [54], used under Crown copyright. d Use of TMSDRs for regulating protein activity. Here, a regulatory circuit inhibits excess thrombin using a threshold controller network, which is designed to release inhibitor strands through TMSDRs that suppress thrombin activity when it is above a threshold [118]. Reprinted with permission from Ref. [118], copyright 2012, American Chemical Society. e Use of TMSDRs driven DNA hydrogel for targeted drug release. Here, AuNPs are functionalized with thiol groups to bind with DNA strands, which is designed to hybridize with other initiator strands and polymerize to form a DNA network hydrogel [119]. The target ligand binds to the toehold switch and disassembles the DNA shell in a controlled manner to release the drug. Reprinted with permission from Ref. [119], copyright 2019, American Chemical Society
Additionally, DNA nanostructure can be coupled with TMSDRs to develop drug carriers, which are capable of sensing and responding. For example, Chandrasekaran et al. have developed a DNA tetrahedron structure that has the potential to carry the relevant drug in the internal cavity and can be disassembled through TMSDRs in a targeted manner [117] (see Fig. 5b). In general, one of the major challenges in targeted drug delivery is designing drug-binding short DNA sequences. Many studies shed light on drug-binding DNA domains such as repeats of GC sequence that bind to a particular group of anti-cancer antibiotic drugs [120]. Hence, intricate sequence designing is necessary for functionalizing dynamic DNA systems for therapeutics.
Regulating clinically relevant molecules
TMSDRs have also been applied to regulate gene transcription and protein translocation synthetically, using DNA oligonucleotides as a therapeutic material. For example, Ke et al. have developed an RNA–DNA hybrid nanostructure to regulate NF-κB transcription factor [54]—an essential regulator in several autoimmune disorders like rheumatoid arthritis, systemic lupus erythematosus, and vasculitis [121]. In this mechanism, on recognizing complementary sequences, the hybrid nanostructure is reassembled using TMSDRs to form a functional structure. Subsequently, NF-κB decoys are released upon binding of RNA-RNA and DNA-DNA sequences (see Fig. 5c). These decoys can bind to the DNA-binding domain of the NF-κB protein complex and block the translocation of NF-κB into the nucleus, thereby blocking the gene transcription and reducing hyper-immune reactions. Similarly, Zhang et al. have used DNA tetrahedron and TMSDRs to regulate the activity of apyrimidinic endonuclease 1 enzyme, which is involved in DNA repair pathways [122]. An allosteric enzyme regulation is achieved by transitioning the target strand of the tetrahedron from the external side to the internal side of the DNA tetrahedron using TMSDRs.
Relying more on TMSDRs based logic circuits, Han et al. have developed a regulatory circuit that uses DNA aptamers to manipulate the activity of the human α-thrombin as shown in Fig. 5d [118]. Excess amounts of thrombin can lead to life-threatening conditions such as a variety of functional disorders [94]. This regulatory circuit can suppress thrombin activity dynamically using specific DNA aptamers in an enzyme-free environment. Such approaches might be useful in developing targeted controlled therapies for diseases with dysfunctional pathways.
Toehold-triggered DNA hydrogels for therapeutics
Self-assembled DNA boxes and cages have been used as drug carriers in many studies. However, designing these structures is a complex process, and they have poor stability and control over drug encapsulation and targeted delivery, respectively. In contrast, DNA hydrogels have a high drug-loading capacity and are made stable using crosslinking of DNA. Moreover, TMSDRs can be used in DNA hydrogels to enhance the control over drug release. For example, Bi et al. have developed nanohydrogels with high loading capacity and precise control over drug release using a DNA four-way junction (DNA-4WJ) probe [123]. In the presence of target miRNA, the DNA-4WJ probe undergoes a cascaded reaction and crosslinks into a nanohydrogel, which can be further functionalized with drug-loading sides and aptamers for specific drug release. Similarly, Xiao et al. have developed a DNA hydrogel where DNA probes are immobilized onto AuNPs that serve as a scaffold for hydrogel polymerization triggered by specific DNA aptamers (Fig. 5e) [119]. Highly targeted drug delivery in tumor cells is achieved using cell surface receptors and a toehold switch where dynamic drug release is accomplished by varying the toehold length to provide a release time range from 5 to 60 min.
For more practical applications of DNA hydrogels, further work is needed to control their mechanical, physical, and kinetic properties and swelling capacity. For example, the mechanical properties and stability of DNA hydrogels can be modified using hydrophilic polymers and cross-linkers [124], while the kinetics of polymerization can be controlled via toehold that regulates the crosslinking ability of DNA hydrogel as studied by Jonášová et al. [125]. Their study showed that increasing the toehold length increases the crosslinking rate. By exploiting these features, Cangialosi et al. have used specific DNA toeholds to trigger a 100-fold expansion of a DNA hydrogel [126]. These synthetic systems can produce controlled shape-changing effects in the presence of specific DNA sequences. Such systems can be used as logical circuits for therapeutics using the regulated phase switching of hydrogels for drug delivery.
TMSDRs driven DNA computing for biomedical applications
Programmability and tunability make TMSDRs an excellent tool for performing complex mathematical, biological and computational operations. These operations have been used to develop advanced biosensing and targeted drug delivery systems. We have already described some of these applications in the previous sections. Here, we discuss additional applications that specifically use DNA computing to process multiple inputs and outputs simultaneously.
Brown et al. have developed a TMSDRs based multi-layer diagnostic logic circuit that can detect 4 dengue serotypes (Fig. 6a) [127]. In this mechanism, the output sequence is released by cleavage of chimeric sequences by DNAzymes, which are triggered by TMSDRs. On the other hand, Lopez et al. have used TMSDRs to translate the outcome of a machine learning (ML) based multi-gene classifier into molecular interactions that can differentiate viral and bacterial infections [128]. This approach presents an opportunity for cost-effective gene expression analysis. Similarly, Zhang et al. have implemented a TMSDRs based classifier to phenotype healthy and cancerous cells from patient serum samples [129]. The classifier is first trained in silico by feeding the up and down regulated miRNA profiles from publicly available omics data. This information is then translated into TMSDRs that operate the classifier at the molecular level with an accuracy of 86.4%. Further applications have been developed where three-way DNA junctions are used to sense small molecules like ATP [130]. Similarly, Chang et al. have fabricated a point-of-care device that uses a BRET system for a bioluminescent readout on a smartphone [131]. The device can sense three different miRNAs using an AND gate operation implemented through TMSDRs.
Applications of TMSDRs based DNA computing. a DNA computing for biosensing. Here, a multi-input cascaded network can detect 4 serotypes of dengue. When Dengue A and DEN- k (k = 1,2,3,4) are present, the chimeric substrate is cleaved by the DNAzyme, producing an activator strand for cascaded signaling. Reprinted with permission from Ref [127], copyright 2014, Wiley–VCH. b DNA computing for targeted drug delivery. Here, multiple cancer cell surface markers are tagged with DNA aptamers, enabling TMSDRs to determine cellular viability and build a targeted photodynamic therapeutic device. The AND operation is activated by the first input aptamer, which displaces the incumbent strand and begins the cascaded layer reaction by exposing the second toehold. Reprinted with permission from Ref [132], copyright 2015, American Chemical Society. c-d DNA computing for programmable networks; c Here, the digital display works inside a well-plate and the logical circuit uses DNAzyme to function. The Boolean gate is operated in such a way that in the presence of an input strand, the FRET is cleaved by DNAzyme, releasing the FRET labelled DNA strand. Reprinted with permission from Ref [133], copyright 2014, Wiley–VCH; d Here, the fuel strand hybridizes with the target strand, and the resulting complex forms a waste product of lower energy. The consumption of the fuel strand causes the release of the target strand, which spontaneously hybridizes with the high-energy output strand. Reprinted with permission from Ref. [134] under creative common By-NC 4.0 license
TMSDRs based DNA computing is also used for targeted drug delivery. For example, You et al. have realized a Boolean operation to detect multiple cancer cell-surface markers, and in response, a photoinduced therapy is triggered as shown in Fig. 6b [132]. Using the specificity of the DNA aptamer, false positives by the surface markers of the neighboring cells are avoided and the release of photoinduced therapy is triggered by a photosensitizer that produces reactive oxygen species. Similarly, Wu et al. have developed a logical circuit with a 3-bit binary code that can diagnose live cancer cells and eliminate them using phototherapy [135]. This mechanism achieved a targeted and reliable operation using a novel error correction method function that uses redundant modules. These modules trace the logic gate results and target only cancer cells for phototherapy while avoiding normal cells. On the other hand, Morihiro et al. have developed a three-input logic gate that releases small molecules in response to miRNA inputs using the Staudinger reduction method [136].
TMSDRs based DNA computing has also been used to implement programmable networks with exciting applications. For example, Poje et al. have developed a graphical processing unit that translates the molecular information into a 7-segment display that uses fluorescent and can perform mathematical operations to diagnose Ebola virus sequences as shown in Fig. 6c [133]. On the other hand, Grosso et al. have developed dissipative TMSDRs where temporal control over reaction dynamics can be achieved through a shift in the free energy landscape of the reaction as shown in Fig. 6d [134]. This approach can be used for a transient fluorescent recording of DNA assemblies and temporal control over cascaded TMSDRs. Other exciting applications have also been reported such as storage devices [137], and neural networks [138]. For instance, weak and reversible interaction of the non-complementary DNA and RNA strands can be used for information storage and processing based on the concentration of these short-lived complexes [139]. However, with more complex operations, the system's modularity and efficient computing time need to be maintained [140, 141].
Conclusion
Dynamic DNA nanotechnology is a relatively new field with different applications emerging in biomedical engineering due to its unique properties, such as enzyme-free isothermal amplification, tunable reaction kinetics and biocompatibility. As discussed in this review, TMSDRs have been widely used to develop programmable biosensors and targeted drug delivery carriers. TMSDRs driven smart hydrogels and DNA computing have also gained significant attention for biomedical applications. These approaches can be used in the applications of responsive biomaterial, for regulated assembly of nanostructures, and dissipative DNA networks for drug release. More focused research intend to make TMSDRs based diagnostic and therapeutic systems more robust, specific, and sensitive.
Among all these emerging applications, the dynamic regulation of biological pathways needs to be studied more. Specifically, a system for regulating hypersensitive immune response using an all-DNA system can be a breakthrough in treating immunological diseases. Although transcription regulatory systems like synthetic riboregulators are widely used for regulating gene expression, such approaches have limited practical use due to the slow kinetics and instability of the RNA strands compared to DNA systems [142]. Real-time regulatory systems mimicking immunological disease pathways for theranostics applications are highly desirable in biomedicine.
Significant challenges need to be addressed in using TMSDRs to implement accurate and reliable systems with clinical use. First, a complex system requires many DNA oligonucleotides and any error in the sequence can hinder the desired operation [143]. For example, the chemical synthesis of synthetic DNA oligonucleotides such as using phosphoramidite chemistry, generally has 98–99% coupling efficiency and the remaining side products contain several types of errors such as insertions, base substitutions and deletions [144]. A change in a single nucleotide can alter the binding energy and kinetics of TMSDRs [145]. This may lead to crosstalk, signal leaks and incorrect complex formations due to undesired interactions between DNA strands. However, different purification methods can help in reducing the side products [146]. Second, TMSDRs generally employ short DNA strands, which are typically of ~ 20–50 base pairs, and are highly susceptible to degradation by cellular nucleases like exonucleases or endonucleases. Because of that, TMSDRs are highly unstable in blood serum and other serum-supplemented cell cultures and this limits their use in in vitro and in vivo gene based reaction platforms. [147, 148]. However, new mechanisms have been developed such as specific design considerations [148] and chemically modified DNA strands [149] to improve the functionality of TMSDRs in intracellular environments. Third, unlike biological pathways, which are fueled by ATP, enzymes, and other biological catalysts, TMSDRs are fueled by DNA strands. The reaction reaches an equilibrium once the initial amount of fuel strands is consumed. Therefore, the absence of powerful cellular machinery in TMSDRs causes the depletion of energy resources and that limits the total output yield and reaction run time. However, as mentioned in the previous section, new approaches have been developed to address this limitation [95, 96]. Finally, in silico designing tools for TMSDRs have limited predictability as loop sequence and tertiary interaction are often not accounted while predicting the free energy and cross interactions of DNA oligonucleotides [150].
To conclude, TMSDRs have given many exciting applications in biomedical engineering. Many new applications such as the polymerization of soft materials, whole cell biosensing, polyvalent ligands and targeted drug delivery, are emerging that are pushing the field of nanotechnology ahead. Dynamic DNA nanotechnology could provide a promising toolkit for diagnosis and therapy of diseases.
Availability of data and materials
Not applicable.
References
Seeman NC. Nucleic acid junctions and lattices. J Theor Bio. 1982;99(2):237–47. https://doi.org/10.1016/0022-5193(82)90002-9.
Agrawal DK, Jiang R, Reinhart S, Mohammed AM, Jorgenson TD, Schulman R. Terminating DNA tile assembly with nanostructured caps. ACS Nano. 2017;11(10):9770–9. https://doi.org/10.1021/acsnano.7b02256.
Jorgenson TD, Mohammed AM, Agrawal DK, Schulman R. Self-assembly of hierarchical DNA nanotube architectures with well-defined geometries. ACS Nano. 2017;11(2):1927–36. https://doi.org/10.1021/acsnano.6b08008.
Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440(7082):297–302. https://doi.org/10.1038/nature04586.
Schaffter SW, Schneider J, Agrawal DK, Pacella MS, Rothchild E, Murphy T, Schulman R. Reconfiguring DNA nanotube architectures via selective regulation of terminating structures. ACS Nano. 2020;14(10):13451–62. https://doi.org/10.1021/acsnano.0c05340.
Zhang QL, Wang Y, Wang LL, Xie F, Wu RY, Ma XY, et al. Programming Non-Nucleic Acid Molecules into Computational Nucleic Acid Systems. Angew Chem Int Ed. 2023;62(2):e202214698. https://doi.org/10.1002/anie.202214698.
Zhang DY, Seelig G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat Chem. 2011;3(2):103–13.
Seeman NC, Sleiman HF. DNA nanotechnology. Nat Rev Mater. 2017;3(1):1–23.
Zhang DY, Winfree E. Control of DNA strand displacement kinetics using toehold exchange. J Am Chem Soc. 2009;131(47):17303–14. https://doi.org/10.1021/ja906987s.
SantaLucia J, Allawi HT, Seneviratne PA. Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry. 1996;35(11):3555–62.
Agrawal DK, Marshall R, Noireaux V, Sontag ED. In vitro implementation of robust gene regulation in a synthetic biomolecular integral controller. Nat Commun. 2019;10(1):5760. https://doi.org/10.1038/s41467-019-13626-z.
Agrawal DK, Dolan EM, Hernandez NE, Blacklock KM, Khare SD, Sontag ED. Mathematical models of protease-based enzymatic biosensors. ACS Synth Biol. 2020;9(2):198–208. https://doi.org/10.1021/acssynbio.9b00279.
Westbrook A, Tang X, Marshall R, Maxwell CS, Chappell J, Agrawal DK, et al. Distinct timescales of RNA regulators enable the construction of a genetic pulse generator. Biotechnol Bioeng. 2019;116(5):1139–51. https://doi.org/10.1002/bit.26918.
Aldaye FA, Palmer AL, Sleiman HF. Assembling materials with DNA as the guide. Science. 2008;321(5897):1795–9. https://doi.org/10.1126/science.1154533.
Wu N, Willner I. pH-stimulated reconfiguration and structural isomerization of origami dimer and trimer systems. Nano Lett. 2016;16(10):6650–5. https://doi.org/10.1021/acs.nanolett.6b03418.
Chen Z, Chen K, Xie C, Liao K, Xu F, Pan L. Cyclic transitions of DNA origami dimers driven by thermal cycling. Nanotechnology. 2022;34(6). https://doi.org/10.1088/1361-6528/aca02f.
Dai Z, Lo PK. Photo-switchable patterning of gold nanoparticles along 3D DNA nanotubes. Nanoscale. 2018;10(12):5431–5. https://doi.org/10.1039/c7nr09650j.
Yurke B, Turberfield AJ, Mills AP Jr, Simmel FC, Neumann JL. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406(6796):605–8. https://doi.org/10.1038/35020524.
Qian L, Winfree E, Bruck J. Neural network computation with DNA strand displacement cascades. Nature. 2011;475(7356):368–72. https://doi.org/10.1038/nature10262.
Zhang DY, Turberfield AJ, Yurke B, Winfree E. Engineering entropy-driven reactions and networks catalyzed by DNA. Science. 2007;318(5853):1121–5. https://doi.org/10.1126/science.1148532.
Dirks RM, Pierce NA. Triggered amplification by hybridization chain reaction. Proc Natl Acad Sci U S A. 2004;101(43):15275–8. https://doi.org/10.1073/pnas.0407024101.
Li B, Ellington AD, Chen X. Rational, modular adaptation of enzyme-free DNA circuits to multiple detection methods. Nucleic Acids Res. 2011;39(16):e110. https://doi.org/10.1093/nar/gkr504.
Agrawal DK, Schulman R. Modular protein-oligonucleotide signal exchange. Nucleic Acids Res. 2020;48(12):6431–44. https://doi.org/10.1093/nar/gkaa405.
Douglas SM, Marblestone AH, Teerapittayanon S, Vazquez A, Church GM, Shih WM. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009;37(15):5001–6. https://doi.org/10.1093/nar/gkp436.
Kim D-N, Kilchherr F, Dietz H, Bathe M. Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Res. 2011;40(7):2862–8. https://doi.org/10.1093/nar/gkr1173.
Snodin BEK, Randisi F, Mosayebi M, Šulc P, Schreck JS, Romano F, et al. Introducing improved structural properties and salt dependence into a coarse-grained model of DNA. J Chem Phys. 2015;142(23):234901. https://doi.org/10.1063/1.4921957.
Huang CM, Kucinic A, Johnson JA, Su HJ, Castro CE. Integrated computer-aided engineering and design for DNA assemblies. Nat Mater. 2021;20(9):1264–71. https://doi.org/10.1038/s41563-021-00978-5.
Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, et al. NUPACK: Analysis and design of nucleic acid systems. J Comput Chem. 2011;32(1):170–3. https://doi.org/10.1002/jcc.21596.
Fornace ME, Porubsky NJ, Pierce NA. A unified dynamic programming framework for the analysis of interacting nucleic acid strands: enhanced models, scalability, and speed. ACS Synth Biol. 2020;9(10):2665–78. https://doi.org/10.1021/acssynbio.9b00523.
Srinivas N, Ouldridge TE, Sulc P, Schaeffer JM, Yurke B, Louis AA, et al. On the biophysics and kinetics of toehold-mediated DNA strand displacement. Nucleic Acids Res. 2013;41(22):10641–58. https://doi.org/10.1093/nar/gkt801.
Scalise D, Schulman R. Controlling matter at the molecular scale with DNA circuits. Annu Rev Biomed Eng. 2019;21:469–93. https://doi.org/10.1146/annurev-bioeng-060418-052357.
Takahashi M. Aptamers targeting cell surface proteins. Biochimie. 2018;145:63–72. https://doi.org/10.1016/j.biochi.2017.11.019.
Chai C, Xie Z, Grotewold E. SELEX (Systematic Evolution of Ligands by EXponential Enrichment), as a Powerful Tool for Deciphering the Protein–DNA Interaction Space. In: Yuan L, Perry SE, editors. Plant transcription factors: methods and protocols. Totowa, NJ: Humana Press; 2011. p. 249–58.
Zou X, Wu J, Gu J, Shen L, Mao L. Application of Aptamers in Virus Detection and Antiviral Therapy. Front Microbiol. 2019;10:1462. https://doi.org/10.3389/fmicb.2019.01462.
Xing Y, Yang Z, Liu D. A responsive hidden toehold to enable controllable DNA strand displacement reactions. Angew Chem Int Ed Engls. 2011;50(50):11934–6. https://doi.org/10.1002/anie.201105923. PMID: 22012587.
Zhang QL, Wang LL, Liu Y, Lin J, Xu L. A kinetically controlled platform for ligand-oligonucleotide transduction. Nat Commun. 2021;12(1):4654. https://doi.org/10.1038/s41467-021-24962-4.
Jonstrup AT, Fredsøe J, Andersen AH. DNA hairpins as temperature switches thermometers and ionic detectors. Sensors. 2013;13(5):5937–44. https://doi.org/10.3390/s130505937. PMID.
Li X, Song T, Chen Z, Shi X, Chen C, Zhang Z. A universal fast colorimetric method for DNA signal detection with DNA strand displacement and gold nanoparticles. J Nanomater. 2015;2015:407184. https://doi.org/10.1155/2015/407184.
Li Q, Liu Z, Zhou D, Pan J, Liu C, Chen J. A cascade toehold-mediated strand displacement strategy for label-free and sensitive non-enzymatic recycling amplification detection of the HIV-1 gene. Analyst. 2019;144(6):2173–8. https://doi.org/10.1039/C8AN02340A.
Tang Q, Lai W, Wang P, Xiong X, Xiao M, Li L, et al. Multi-Mode Reconfigurable DNA-Based Chemical Reaction Circuits for Soft Matter Computing and Control. Angew Chem Int Ed Engl. 2021;60(27):15013–9. https://doi.org/10.1002/anie.202102169. PMID: 33893703.
Qi M, Shi P, Zhang X, Cui S, Liu Y, Zhou S, Zhang Q. Reconfigurable DNA triplex structure for pH responsive logic gates. RSC Adv. 2023;13(15):9864–70. https://doi.org/10.1039/D3RA00536D.
Ranallo S, Amodio A, Idili A, Porchetta A, Ricci F. Electronic control of DNA-based nanoswitches and nanodevices. Chem Sci. 2016;7(1):66–71. https://doi.org/10.1039/C5SC03694A.
Xing C, Chen Z, Dai J, Zhou J, Wang L, Zhang KL, et al. Light-controlled, toehold-mediated logic circuit for assembly of DNA tiles. ACS Appl Mater Interfaces. 2020;12(5):6336–42. https://doi.org/10.1021/acsami.9b21778.
Song X, Ding Q, Pu Y, Zhang J, Sun R, Yin L, et al. Application of the Dimeric G-Quadruplex and toehold-mediated strand displacement reaction for fluorescence biosensing of ochratoxin A. Biosens Bioelectron. 2021;192:113537. https://doi.org/10.1016/j.bios.2021.113537. PMID: 34339903.
Feng DQ, Liu G. Target-activating and toehold displacement Ag NCs/GO biosensor-mediating signal shift and enhancement for simultaneous multiple detection. Anal Chem. 2021;93(48):16025–34. https://doi.org/10.1021/acs.analchem.1c03570. PMID: 34817158.
Zhang Y, Li Z, Su W, Zhong G, Zhang X, Wu Y, et al. A highly sensitive and versatile fluorescent biosensor for pathogen nucleic acid detection based on toehold-mediated strand displacement initiated primer exchange reaction. Anal Chim Acta. 2022;1221:340125. https://doi.org/10.1016/j.aca.2022.340125. PMID: 35934404.
Khodakov DA, Khodakova AS, Linacre A, Ellis AV. Toehold-mediated nonenzymatic DNA strand displacement as a platform for DNA genotyping. J Am Chem Soc. 2013;135(15):5612–9. https://doi.org/10.1021/ja310991r.
Monserud JH, Macri KM, Schwartz DK. Toehold-mediated displacement of an adenosine-binding aptamer from a DNA duplex by its ligand. Angew Chem Int Ed Engl. 2016;55(44):13710–3. https://doi.org/10.1002/anie.201603458. PMID: 27689920.
Chen RP, Blackstock D, Sun Q, Chen W. Dynamic protein assembly by programmable DNA strand displacement. Nat Chem. 2018;10(4):474–81. https://doi.org/10.1038/s41557-018-0016-9. PMID: 29531373.
Hong F, Chen X, Cao Y, Dong Y, Wu D, Hu F, Gan N. Enzyme- and label-free electrochemical aptasensor for kanamycin detection based on double stir bar-assisted toehold-mediated strand displacement reaction for dual-signal amplification. Biosens Bioelectron. 2018;112:202–8. https://doi.org/10.1016/j.bios.2018.04.017. PMID: 29709830.
Zhao J, Zheng T, Gao J, Xu W. Toehold-mediated strand displacement reaction triggered by nicked DNAzymes substrate for amplified electrochemical detection of lead ion. Electrochimica Acta. 2018;274:16–22. https://doi.org/10.1016/j.electacta.2018.04.083.
Wang Y, Wang Y, Liu S, Sun W, Zhang M, Jiang L, et al. Toehold-mediated DNA strand displacement-driven super-fast tripedal DNA walker for ultrasensitive and label-free electrochemical detection of ochratoxin A. Anal Chim Acta. 2021;1143:21–30. https://doi.org/10.1016/j.aca.2020.11.013. PMID: 33384119.
Park Y, Lee CY, Park KS, Park HG. Enzyme-free colorimetric detection of Cu2+ by utilizing target-triggered DNAzymes and toehold-mediated DNA strand displacement events. Chemistry. 2017;23(68):17379–83. https://doi.org/10.1002/chem.201704346.
Ke W, Hong E, Saito RF, Rangel MC, Wang J, Viard M, et al. RNA–DNA fibers and polygons with controlled immunorecognition activate RNAi, FRET and transcriptional regulation of NF-κB in human cells. Nucleic Acids Res. 2018;47(3):1350–61. https://doi.org/10.1093/nar/gky1215.
Kralik P, Ricchi M. A basic guide to real time PCR in microbial diagnostics: definitions, parameters, and everything. Front Microbiol. 2017;8:108. https://doi.org/10.3389/fmicb.2017.00108.
Zhang C, Li D, Li D, Wen K, Yang X, Zhu Y. Rolling circle amplification-mediated in situ synthesis of palladium nanoparticles for the ultrasensitive electrochemical detection of microRNA. Analyst. 2019;144(12):3817–25. https://doi.org/10.1039/c9an00427k.
Becherer L, Borst N, Bakheit M, Frischmann S, Zengerle R, von Stetten F. Loop-mediated isothermal amplification (LAMP) – review and classification of methods for sequence-specific detection. Anal Methods. 2020;12(6):717–46. https://doi.org/10.1039/C9AY02246E.
He Q, He Q, Liu X, Wei Y, Shen S, Hu X, et al. Genome-wide prediction of cancer driver genes based on SNP and cancer SNV data. Am J Cancer Res. 2014;4(4):394.
Ke Y, Lindsay S, Chang Y, Liu Y, Yan H. Self-assembled water-soluble nucleic acid probe tiles for label-free RNA hybridization assays. Science. 2008;319(5860):180–3. https://doi.org/10.1126/science.1150082. PMID: 18187649.
Zhang Z, Zeng D, Ma H, Feng G, Hu J, He L, et al. A DNA-Origami chip platform for label-free SNP genotyping using toehold-mediated strand displacement. Small. 2010;6(17):1854–8. https://doi.org/10.1002/smll.201000908.
Wang D, Tang W, Wu X, Wang X, Chen G, Chen Q, et al. Highly selective detection of single-nucleotide polymorphisms using a quartz crystal microbalance biosensor based on the toehold-mediated strand displacement reaction. Anal Chem. 2012;84(16):7008–14. https://doi.org/10.1021/ac301064g.
Gao ZF, Ling Y, Lu L, Chen NY, Luo HQ, Li NB. Detection of single-nucleotide polymorphisms using an ON-OFF switching of regenerated biosensor based on a locked nucleic acid-integrated and toehold-mediated strand displacement reaction. Anal Chem. 2014;86(5):2543–8. https://doi.org/10.1021/ac500362z.
Wu F, Chen M, Lan J, Xia Y, Liu M, He W, et al. A universal locked nucleic acid-integrated X-shaped DNA probe design for amplified fluorescence detection of single-nucleotide variant. Sens Actuators B Chem. 2017;241:123–8. https://doi.org/10.1016/j.snb.2016.10.066.
Ding S, Yu X, Zhao Y, Zhao C. Identification of single nucleotide polymorphisms by a peptide nucleic acid-based sandwich hybridization assay coupled with toehold-mediated strand displacement reactions. Anal Chim Acta. 2023;1242:340810. https://doi.org/10.1016/j.aca.2023.340810.
Lu H, Hailin T, Yi X, Wang J. Three-dimensional DNA nanomachine combined with toehold-mediated strand displacement reaction for sensitive electrochemical detection of MiRNA. Langmuir. 2020;36(36):10708–14. https://doi.org/10.1021/acs.langmuir.0c01415.
Gaddes ER, Gydush G, Li S, Chen N, Dong C, Wang Y. Aptamer-based polyvalent ligands for regulated cell attachment on the hydrogel surface. Biomacromol. 2015;16(4):1382–9. https://doi.org/10.1021/acs.biomac.5b00165.
Song P, Ye D, Zuo X, Li J, Wang J, Liu H, et al. DNA hydrogel with aptamer-toehold-based recognition, cloaking, and decloaking of circulating tumor cells for live cell analysis. Nano Lett. 2017;17(9):5193–8. https://doi.org/10.1021/acs.nanolett.7b01006.
Kaur A, Sapkota K, Dhakal S. Multiplexed nucleic acid sensing with single-molecule FRET. ACS Sensors. 2019;4(3):623–33. https://doi.org/10.1021/acssensors.8b01373.
Choi W, Park E, Bae S, Choi K-H, Han S, Son K-H, et al. Multiplex SNP genotyping using SWITCH: sequence-specific nanoparticle with interpretative toehold-mediated sequence decoding in hydrogel. Small. 2022;18(8):e2105538. https://doi.org/10.1002/smll.202105538.
Hu S, Li N, Liu F. Combining cooperativity with sequestration: a novel strategy for discrimination of single nucleotide variants. Chem Commun. 2018;54(26):3223–6. https://doi.org/10.1039/c8cc00838h.
Gu C, Kong X, Liu X, Gai P, Li F. Enzymatic biofuel-cell-based self-powered biosensor integrated with DNA amplification strategy for ultrasensitive detection of single-nucleotide polymorphism. Anal Chem. 2019;91(13):8697–704. https://doi.org/10.1021/acs.analchem.9b02510.
Gao YM, Qiao HY, Pan V, Wang ZG, Li JJ, Wei YN, et al. Accurate genotyping of fragmented DNA using a toehold assisted padlock probe. Biosens Bioelectron. 2021;179:113079. https://doi.org/10.1016/j.bios.2021.113079. PMID: WOS:000632849900004.
Hao H, Li Y, Yang B, Lou S, Guo Z, Lu W. Simulation-guided rational design of DNA probe for accurate discrimination of single-nucleotide variants based on “Hill-Type” cooperativity. Anal Chem. 2023;95(5):2893–900. https://doi.org/10.1021/acs.analchem.2c04446.
Zhang L, Chen J, He M, Su X. Molecular dynamics simulation-guided toehold mediated strand displacement probe for single-nucleotide variants detection. Exploration (Beijing). 2022;2(1):20210265. https://doi.org/10.1002/exp.20210265.
Yi M, Xu L, Jiao Y, Luo S, Li A, Wu K. The role of cancer-derived microRNAs in cancer immune escape. J Hematol Oncol. 2020;13(1):25. https://doi.org/10.1186/s13045-020-00848-8.
Mirna M, Paar V, Rezar R, Topf A, Eber M, Hoppe UC, et al. MicroRNAs in inflammatory heart diseases and sepsis-induced cardiac dysfunction: a potential scope for the future? Cells. 2019;8(11):1352. https://doi.org/10.3390/cells8111352.
Rastegar-Moghaddam SH, Ebrahimzadeh-Bideskan A, Shahba S, Malvandi AM, Mohammadipour A. MicroRNA-22: a novel and potent biological therapeutics in neurological disorders. Mol Neurobiol. 2022;59(5):2694–701. https://doi.org/10.1007/s12035-022-02769-8.
Wu Y, Fu C, Shi W, Chen J. Recent advances in catalytic hairpin assembly signal amplification-based sensing strategies for microRNA detection. Talanta. 2021;235:122735. https://doi.org/10.1016/j.talanta.2021.122735.
Deng R, Tang L, Tian Q, Wang Y, Lin L, Li J. Toehold-initiated rolling circle amplification for visualizing individual microRNAs in situ in single cells. Angew Chem Int Ed Engl. 2014;53(9):2389–93. https://doi.org/10.1002/anie.201309388.
Lee J, Na H-K, Lee S, Kim W-K. Advanced graphene oxide-based paper sensor for colorimetric detection of miRNA. Mikrochim Acta. 2021;189(1):35. https://doi.org/10.1007/s00604-021-05140-1.
Yang Y, Wang W, Liu H, Tong L, Mu X, Chen Z, Tang B. Sensitive quantification of MicroRNA in blood through multi-amplification toehold-mediated DNA-strand-displacement paper-spray mass spectrometry (TSD-PS MS). Angew Chem Int Ed Engl. 2022;61(9):e202113051. https://doi.org/10.1002/anie.202113051.
Kong Y, Liu X, Liu C, Xue Q, Li X, Wang H. A dandelion-like liposomes-encoded magnetic bead probe-based toehold-mediated DNA circuit for the amplification detection of MiRNA. Analyst. 2019;144(15):4694–701. https://doi.org/10.1039/c9an00887j.
Wang Q, He Y, He S, Yu S, Jiang Y, Wang F. An entropy-driven DNA nanomachine for microRNA detection using a personal glucose meter. Chem Commun. 2023;59(10):1345–8. https://doi.org/10.1039/d2cc06479k.
Miao P, Tang Y, Wang B, Meng F. Near-infrared Ag2S quantum dots-based DNA logic gate platform for miRNA diagnostics. Anal Chem. 2016;88(15):7567–73. https://doi.org/10.1021/acs.analchem.6b01044.
Chu Y, Wu R, Fan G-C, Deng A-P, Zhu J-J. Enzyme-free photoelectrochemical biosensor based on the co-sensitization effect coupled with dual cascade toehold-mediated strand displacement amplification for the sensitive detection of MicroRNA-21. ACS Sustain Chem Eng. 2018;6(9):11633–41. https://doi.org/10.1021/acssuschemeng.8b01857.
Ma F, Zhang Q, Zhang C-Y. Catalytic self-assembly of quantum-dot-based MicroRNA nanosensor directed by toehold-mediated strand displacement cascade. Nano Lett. 2019;19(9):6370–6. https://doi.org/10.1021/acs.nanolett.9b02544.
Li L, Meng Y, Li L, Wang S, Ding J, Zhou W. A tetrahedral DNA nanoflare for fluorometric determination of nucleic acids and imaging of microRNA using toehold strands. Mikrochim Acta. 2019;186(12):824. https://doi.org/10.1007/s00604-019-3931-6.
Li C-C, Hu J, Zou X, Luo X, Zhang C-Y. Construction of a structure-switchable toehold dumbbell probe for sensitive and label-free measurement of MicroRNA in cancer cells and tissues. Anal Chem. 2022;94(3):1882–9. https://doi.org/10.1021/acs.analchem.1c05066.
Wang Q, Liu J, Zeng J, Yang Z, Ran F, Wu L, et al. Determination of miRNA derived from exosomes of prostate cancer via toehold-aided cyclic amplification combined with HRP enzyme catalysis and magnetic nanoparticles. Anal Biochem. 2021;630:114336. https://doi.org/10.1016/j.ab.2021.114336.
Zhao B, Wang W, Li N, Garcia-Lezana T, Che C, Wang X, et al. Digital-resolution and highly sensitive detection of multiple exosomal small RNAs by DNA toehold probe-based photonic resonator absorption microscopy. Talanta. 2022;241:123256. https://doi.org/10.1016/j.talanta.2022.123256.
Chang X, Zhang C, Lv C, Sun Y, Zhang M, Zhao Y, et al. Construction of a multiple-aptamer-based DNA logic device on live cell membranes via associative toehold activation for accurate cancer cell identification. J Am Chem Soc. 2019;141(32):12738–43. https://doi.org/10.1021/jacs.9b05470.
Guo S, Huang H, Deng X, Chen Y, Jiang Z, Xie M, et al. Programmable DNA-responsive microchip for the capture and release of circulating tumor cells by nucleic acid hybridization. Nano Res. 2018;11(5):2592–604. https://doi.org/10.1007/s12274-017-1885-8.
Li Y, Chen H, Dai Y, Chen T, Cao Y, Zhang J. Cellular interface supported toehold strand displacement cascade for amplified dual-electrochemical signal and its application for tumor cell analysis. Anal Chim Acta. 2019;1064:25–32. https://doi.org/10.1016/j.aca.2019.03.021.
Wolberg AS, Campbell RA. Thrombin generation, fibrin clot formation and hemostasis. Transfus Apher Sci. 2008;38(1):15–23. https://doi.org/10.1016/j.transci.2007.12.005. PMID: 18282807.
Deng J, Walther A. Fuel-driven transient DNA strand displacement circuitry with self-resetting function. J Am Chem Soc. 2020;142(50):21102–9. https://doi.org/10.1021/jacs.0c09681.
Liu Y, Fu S, Liu J, Su X. A DNA-based dissipation system that synchronizes multiple fuels. Chemistry. 2023;29(39):e202301156. https://doi.org/10.1002/chem.202301156.
Ottenbrite RM, Park K, Okano T. Biomedical applications of hydrogels handbook: Springer Science & Business Media. 2010. p. 432 ISBN: 9781441959195.
Nagahara S, Matsuda T. Hydrogel formation via hybridization of oligonucleotides derivatized in water-soluble vinyl polymers. Polym Gels Networks. 1996;4(2):111–27. https://doi.org/10.1016/0966-7822(96)00001-9.
Li C, Li H, Ge J, Jie G. Versatile fluorescence detection of microRNA based on novel DNA hydrogel-amplified signal probes coupled with DNA walker amplification. Chem Commun. 2019;55(27):3919–22. https://doi.org/10.1039/c9cc00565j.
Kim H-S, Abbas N, Shin S. A rapid diagnosis of SARS-CoV-2 using DNA hydrogel formation on microfluidic pores. Biosens Bioelectron. 2021;177:113005. https://doi.org/10.1016/j.bios.2021.113005.
Liao W-C, Lilienthal S, Kahn JS, Riutin M, Sohn YS, Nechushtai R, Willner I. pH- and ligand-induced release of loads from DNA-acrylamide hydrogel microcapsules. Chem Sci. 2017;8(5):3362–73. https://doi.org/10.1039/c6sc04770j.
Yang Q, Wang Y, Liu T, Wu C, Li J, Cheng J, et al. Microneedle array encapsulated with programmed DNA hydrogels for rapidly sampling and sensitively sensing of specific MicroRNA in dermal interstitial fluid. ACS Nano. 2022;16(11):18366–75. https://doi.org/10.1021/acsnano.2c06261.
Liu YX, Xie TJ, Li CH, Ye QC, Tian LL, Li YF, et al. A crosslinked submicro-hydrogel formed by DNA circuit-driven protein aggregation amplified fluorescence anisotropy for biomolecules detection. Anal Chim Acta. 2021;1154:338319. https://doi.org/10.1016/j.aca.2021.338319.
Zhang H, Ye S, Huang L, Fan S, Mao W, Hu Y, et al. An electrochemical biosensor for the detection of aflatoxin B1 based on the specific aptamer and HCR biological magnification. Anal Methods. 2023;15(1):99–108. https://doi.org/10.1039/D2AY01682F.
Tian Z, Zhou C, Zhang C, Wu M, Duan Y, Li Y. Recent advances of catalytic hairpin assembly and its application in bioimaging and biomedicine. J Mater Chem B. 2022;10(28):5303–22. https://doi.org/10.1039/D2TB00815G.
Khajouei S, Ravan H, Ebrahimi A. Developing a colorimetric nucleic acid-responsive DNA hydrogel using DNA proximity circuit and catalytic hairpin assembly. Analytica Chimica Acta. 2020;1137:1–10. https://doi.org/10.1016/j.aca.2020.08.059.
Xing C, Chen Z, Zhang C, Wang J, Lu C. Target-directed enzyme-free dual-amplification DNA circuit for rapid signal amplification. J Mater Chem B. 2020;8(47):10770–5. https://doi.org/10.1039/D0TB02114H.
Wang K, He M-Q, Zhai F-H, Wang J, He R-H, Yu Y-L. Autonomous DNA nanomachine based on cascade amplification of strand displacement and DNA walker for detection of multiple DNAs. Biosens Bioelectron. 2018;105:159–65. https://doi.org/10.1016/j.bios.2018.01.044.
Song P, Chen SX, Yan YH, Pinto A, Cheng LY, Dai P, et al. Selective multiplexed enrichment for the detection and quantitation of low-fraction DNA variants via low-depth sequencing. Nat Biomed Eng. 2021;5(7):690–701. https://doi.org/10.1038/s41551-021-00713-0.
Zhang Z, Weng Z, Yao J, Liu D, Zhang L, Zhang L, Xie G. Toehold-mediated nonenzymatic DNAstrand displacement coupling UDG mediated PCR and multi-code magnetic beads for DNA genotyping. Microchem J. 2022;178:107340. https://doi.org/10.1016/j.microc.2022.107340.
Park S, Lee JW. Detection of Coronaviruses Using RNA Toehold Switch Sensors. Int J Mol Sci. 2021;22(4):1772. https://doi.org/10.3390/ijms22041772. PMID.
Wang R, Wang L, Zhao H, Jiang W. A split recognition mode combined with cascade signal amplification strategy for highly specific, sensitive detection of microRNA. Biosens Bioelectron. 2016;86:834–9. https://doi.org/10.1016/j.bios.2016.07.092.
Asadi R, Mollasalehi H. The mechanism and improvements to the isothermal amplification of nucleic acids, at a glance. Anal Biochem. 2021;631:114260. https://doi.org/10.1016/j.ab.2021.114260.
Zhao Y, Chen F, Li Q, Wang L, Fan C. Isothermal amplification of nucleic acids. Chem Rev. 2015;115(22):12491–545. https://doi.org/10.1021/acs.chemrev.5b00428.
Zhang Y, Chen W, Fang Y, Zhang X, Liu Y, Ju H. Activating a DNA nanomachine via computation across cancer cell membranes for precise therapy of solid tumors. J Am Chem Soc. 2021;143(37):15233–42. https://doi.org/10.1021/jacs.1c06361.
Mirzaiebadizi A, Ravan H, Dabiri S, Mohammadi P, Shahba A, Ziasistani M, Khatami M. An intelligent DNA nanorobot for detection of MiRNAs cancer biomarkers using molecular programming to fabricate a logic-responsive hybrid nanostructure. Bioprocess Biosyst Eng. 2022;45(11):1781–97. https://doi.org/10.1007/s00449-022-02785-x.
Chandrasekaran AR, Halvorsen K. Controlled disassembly of a DNA tetrahedron using strand displacement. Nanoscale Adv. 2019;1(3):969–72. https://doi.org/10.1039/C8NA00340H.
Han D, Zhu Z, Wu C, Peng L, Zhou L, Gulbakan B, et al. A logical molecular circuit for programmable and autonomous regulation of protein activity using DNA aptamer-protein interactions. J Am Chem Soc. 2012;134(51):20797–804. https://doi.org/10.1021/ja310428s.
Xiao M, Lai W, Wang F, Li L, Fan C, Pei H. Programming drug delivery kinetics for active burst release with DNA toehold switches. J Am Chem Soc. 2019;141(51):20354–64. https://doi.org/10.1021/jacs.9b10765.
Fox KR, Howarth NR. Investigations into the sequence-selective binding of mithramycin and related ligands to DNA. Nucleic Acids Res. 1985;13(24):8695–714. https://doi.org/10.1093/nar/13.24.8695.
Manou-Stathopoulou S, Lewis MJ. Diversity of NF-κB signalling and inflammatory heterogeneity in Rheumatic Autoimmune Disease. Semin Immunol. 2021;58:10s1649. https://doi.org/10.1016/j.smim.2022.101649.
Zhang Y, Deng Y, Wang C, Li L, Xu L, Yu Y, Su X. Probing and regulating the activity of cellular enzymes by using DNA tetrahedron nanostructures. Chem Sci. 2019;10(23):5959–66. https://doi.org/10.1039/C9SC01912J.
Bi S, Xiu B, Ye J, Dong Y. Target-Catalyzed DNA four-way junctions for CRET imaging of MicroRNA, concatenated logic operations, and self-assembly of DNA nanohydrogels for targeted drug delivery. ACS Appl Mater Interfaces. 2015;7(41):23310–9. https://doi.org/10.1021/acsami.5b07827.
Hoffman AS. Hydrogels for biomedical applications. Adv Drug Del Rev. 2012;64:18–23. https://doi.org/10.1016/j.addr.2012.09.010.
Jonášová EP, Bjørkøy A, Stokke BT. Toehold length of target ssDNA affects its reaction-diffusion behavior in DNA-responsive DNA-co-Acrylamide hydrogels. Biomacromol. 2020;21(5):1687–99. https://doi.org/10.1021/acs.biomac.9b01515.
Cangialosi A, Yoon C, Liu J, Huang Q, Guo J, Nguyen TD, et al. DNA sequence–directed shape change of photopatterned hydrogels via high-degree swelling. Science. 2017;357(6356):1126–30.
Brown CW 3rd, Lakin MR, Horwitz EK, Fanning ML, West HE, Stefanovic D, Graves SW. Signal propagation in multi-layer DNAzyme cascades using structured chimeric substrates. Angew Chem Int Ed Engl. 2014;53(28):7183–7. https://doi.org/10.1002/anie.201402691. PMID: 24890874.
Lopez R, Wang R, Seelig G. A molecular multi-gene classifier for disease diagnostics. Nat Chem. 2018;10(7):746–54. https://doi.org/10.1038/s41557-018-0056-1.
Zhang C, Zhao Y, Xu X, Xu R, Li H, Teng X, et al. Cancer diagnosis with DNA molecular computation. Nat Nanotechnol. 2020;15(8):709–15. https://doi.org/10.1038/s41565-020-0699-0.
Zhu J, Zhang L, Zhou Z, Dong S, Wang E. Aptamer-based sensing platform using three-way DNA junction-driven strand displacement and its application in DNA logic circuit. Anal Chem. 2014;86(1):312–6. https://doi.org/10.1021/ac403235y.
Chang D, Kim KT, Lindberg E, Winssinger N. Smartphone DNA or RNA sensing using semisynthetic luciferase-based logic device. ACS Sens. 2020;5(3):807–13. https://doi.org/10.1021/acssensors.9b02454. PMID: 32124606.
You M, Zhu G, Chen T, Donovan MJ, Tan W. Programmable and multiparameter DNA-based logic platform for cancer recognition and targeted therapy. J Am Chem Soc. 2015;137(2):667–74. https://doi.org/10.1021/ja509263k.
Poje JE, Kastratovic T, Macdonald AR, Guillermo AC, Troetti SE, Jabado OJ, et al. Visual displays that directly interface and provide read-outs of molecular states via molecular graphics processing units. Angew Chem Int Ed Engl. 2014;53(35):9222–5. https://doi.org/10.1002/anie.201402698. PMID: 25044570.
Del Grosso E, Irmisch P, Gentile S, Prins LJ, Seidel R, Ricci F. Dissipative control over the toehold-mediated DNA strand displacement reaction. Angew Chem Int Ed. 2022;61(23):e202201929. https://doi.org/10.1002/anie.202201929.
Wu H, Lin X, Zhao M, Zhou X, Liu Y. DNA computing with error correction function in cells for cancer diagnosis and targeted therapy. Sens Actuator B Chem. 2023;378:133167. https://doi.org/10.1016/j.snb.2022.133167.
Morihiro K, Ankenbruck N, Lukasak B, Deiters A. Small Molecule Release and Activation through DNA Computing. J Am Chem Soc. 2017;139(39):13909–15. https://doi.org/10.1021/jacs.7b07831.
Song X, Reif J. Nucleic acid databases and molecular-scale computing. ACS Nano. 2019;13(6):6256–68. https://doi.org/10.1021/acsnano.9b02562.
Xiong X, Zhu T, Zhu Y, Cao M, Xiao J, Li L, et al. Molecular convolutional neural networks with DNA regulatory circuits. Nat Mach Intell. 2022;4(7):625–35. https://doi.org/10.1038/s42256-022-00502-7.
Nikitin MP. Non-complementary strand commutation as a fundamental alternative for information processing by DNA and gene regulation. Nat Chem. 2023;15(1):70–82. https://doi.org/10.1038/s41557-022-01111-y.
Yin J, Wang J, Niu R, Ren S, Wang D, Chao J. DNA nanotechnology-based biocomputing. Chem Res Chin Univ. 2020;36(2):219–26. https://doi.org/10.1007/s40242-020-9086-5.
Lv H, Li Q, Shi J, Fan C, Wang F. Biocomputing based on DNA strand displacement reactions. ChemPhysChem. 2021;22(12):1151–66. https://doi.org/10.1002/cphc.202100140.
Liu H, Hong F, Smith F, Goertz J, Ouldridge T, Stevens MM, et al. Kinetics of RNA and RNA: DNA hybrid strand displacement. ACS Synth Biol. 2021;10(11):3066–73.
Komiya K, Komori M, Noda C, Kobayashi S, Yoshimura T, Yamamura M. Leak-free million-fold DNA amplification with locked nucleic acid and targeted hybridization in one pot. Org Biomol Chem. 2019;17(23):5708–13.
eurofins. What is synthesis scale and synthesis yield? Available from: https://eurofinsgenomics.eu/en/eurofins-genomics/product-faqs/dna-rna-oligonucleotides/general-technical-questions/.
Simmel FC, Yurke B. A DNA-based molecular device switchable between three distinct mechanical states. Appl Phys Lett. 2002;80(5):883–5. https://doi.org/10.1063/1.1447008.
IDT. Oligo synthesis: Why IDT is a leader in the oligo industry. Available from: https://www.idtdna.com/pages/education/decoded/article/oligo-synthesis-why-idt-leads-the-oligo-industry.
Zhong W, Sczepanski JT. Direct comparison of D-DNA and L-DNA strand-displacement reactions in living mammalian cells. ACS Synth Biol. 2020;10(1):209–12. https://doi.org/10.1021/acssynbio.0c00527.
Fern J, Schulman R. Design and characterization of DNA strand-displacement circuits in serum-supplemented cell medium. ACS Synth Biol. 2017;6(9):1774–83. https://doi.org/10.1021/acssynbio.7b00105.
Kabza AM, Kundu N, Zhong W, Sczepanski JT. Integration of chemically modified nucleotides with DNA strand displacement reactions for applications in living systems. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2022;14(2):e1743. https://doi.org/10.1002/wnan.1743.
Wolfe BR, Porubsky NJ, Zadeh JN, Dirks RM, Pierce NA. Constrained multistate sequence design for nucleic acid reaction pathway engineering. J Am Chem Soc. 2017;139(8):3134–44. https://doi.org/10.1021/jacs.6b12693.
Acknowledgements
K.J.S acknowledges the Indian Institute of Technology Delhi, India for the Ph.D. scholarship.
Funding
D.K.A. acknowledges the support from the Science & Engineering Research Board, the Mathematical Research Impact Centric Support (SERB- MTR/2022/000369), and the Indian Institute of Technology Delhi seed grant.
Author information
Authors and Affiliations
Contributions
K.J.S wrote the original manuscript; D.K.A. edited and revised the manuscript; K.J.S and D.K.A. approved the final version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Yes.
Competing interests
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Souza, K.J., Agrawal, D.K. Employing toehold-mediated DNA strand displacement reactions for biomedical applications. Med-X 2, 1 (2024). https://doi.org/10.1007/s44258-024-00015-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s44258-024-00015-5