Abstract
As we all know, cancer is one of the leading causes of death worldwide and the second leading cause of death overall. This is why regular screenings or health checks are necessary to detect cancer lesions early. Since bone scan images have become the primary means of detecting the emergence of cancer lesions on bone, high segmentation accuracy is essential for establishing the model of some predefined regions in bone scan images where cancer metastasis was predicted to appear. Consequently, robust localization and identification of the specific region in bone scan images are required for automated metastasis detection. To this end, we propose Efficient-BtrflyNet, a new deep learning-based architecture for skeleton segmentation of whole-body bone scan images. The proposed architecture exploits the benefits of EfficientNet’s model scaling and the encoder–decoder design of butterfly-type networks. We added EfficientNetB7 to the encoder section to obtain more specific features. The proposed architecture simultaneously processes anterior and posterior whole-body bone scan images. Using 37 bone scan images, we evaluated the performance of our proposed skeleton segmentation system using the Dice score. Efficient-BtrflyNet achieves superior segmentation performance compared to the existing representative method.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The second leading cause of death in Indonesia, after cardiovascular disease, is cancer. According to the Global Burden of Cancer Study (Globocan) conducted by the World Health Organization (WHO), the total number of cancer cases in Indonesia in 2020 was 396,914, and the total number of cancer-related deaths was 234,502 cases. In the screening process for early detection of cancerous lesions, radionuclide imaging is a crucial method for examining patients who may have metastatic prostate, breast, or lung cancer [1]. This screening method relies heavily on the visual interpretation of bone scan images but suffers from a lack of quantitative diagnosis. Consequently, quantitative indices called bone scan index (BSI) have been proposed as a valuable marker for tracking disease progression and treatment efficacy [2,3,4,5,6].
As BSI is calculated for each predetermined region of the bone skeleton, the determination of its value is highly dependent on the segmentation results. We can determine whether metastatic lesions are present at specific bone sites in cancer patients based on BSI values [2,3,4,5]. It should be noted that the process of segmenting the skeleton from bone scan images is the initial and indispensable step in all stages of the bone scan image metastases detection system [7,8,9,10]. The bone skeleton should be divided into several predetermined areas for this purpose, including the skull, cervical vertebrae, thoracic vertebrae, lumbar vertebrae, sacrum, pelvis, ribs, scapula, humerus, femur, sternum, and clavicle [6, 11, 12].
With the advent of deep learning in computer vision in 2012 [13], however, the adoption of deep learning methods in medical imaging has increased rapidly, particularly in medical image segmentation [14,15,16]. In addition to being one of the most important research areas in computer vision, the segmentation of medical images has significantly aided in developing multiple effective medical treatments. Despite the remarkable advances in medical image segmentation over the past few years, medical image segmentation based on deep learning still faces a myriad of challenges. One of the main challenges is achieving good segmentation accuracy, which is typically low. This is a crucial issue given that segmentation results in medical images are usually used for diagnosing a disease or detecting the presence of potentially harmful tissue. In addition to the low image resolution, it can be difficult to obtain a large number of medical images. These challenges are crucial factors that must be considered when designing an outstanding medical image segmentation system to meet actual medical needs [15].
Several successful semantic segmentation methods for medical image segmentation have been proposed [17,18,19,20,21,22]. Ronneberger et al. introduced U-Net [17] as a network architecture that employs the encoder-decoder principle. Using a contracting path (to capture context) and a symmetric expansion, their proposed architecture achieves precise localization. They added skip connections between the opposite convolution and deconvolution layer to the previously known up-sampling and down-sampling layers concept. Consequently, they combine characteristics from the contracting and expanding paths, yielding more robust characteristics for semantic segmentation [14].
The emergence of U-Net [17] served as the impetus for the development of a variety of detection and segmentation methods for medical images aimed at resolving various problems. Baheti et al. proposed Eff-UNet [23], which is the architecture for semantic segmentation using the same encoder-decoder approach as U-Net. They chose EfficientNetB5 and EfficientNetB7 for feature extraction on the encoder section because their ImageNet performance is superior to that of other CNN architectures. EfficientNet replaces the stack of convolution, activation, and pooling layers present in the original U-Net. In the decoder section, they employ a decoder architecture similar to the original U-Net. Unlike the encoder and decoder architectures in U-Net, the proposed architecture is asymmetrical, with a deeper contracting path than an expansion path.
Furthermore, Sun et al. introduced a new Circle-UNet, which is an improvement over U-Net [19] due to the addition of a circular pattern representing residual layers. They demonstrated that recurrent patterns could be effectively captured by residual layers, thereby facilitating the network’s comprehension of certain aspects of the objects. The proposed architecture consists of 101 layers and is classified as a very deep architecture based on the premise that residual layers can only contribute effectively if the layers are very deep. In addition, Guo et al. created SA-UNet [18] for retinal blood vessel segmentation by incorporating spatial attention into the U-Net architecture to improve key features. They suggested using structured dropout convolutional blocks rather than U-Net’s native convolution blocks in the encoder and decoder to prevent over-fitting.
Zhou et al. proposed HAUNet-3D [22], which introduces the attention mechanism at various scales and organizes them hierarchically from the bottom up to address the shortcomings of UNet-based lung nodule segmentation methods. Their proposed method captures features of any size and directs the blending of features from adjacent attention outputs, preserving the benefits of 3D U-Net. Their approach demonstrated superior robustness for heterogeneous lung nodule types and outstanding segmentation performance for small lung nodules.
In addition, recent advancements in transformer-based models have sparked interest in the development of medical image segmentation, particularly the U-Net model. Axial Fusion Transformer U-Net (AFTer-UNet) is a medical segmentation framework proposed by Yan et al. [21]. They adhere to the encoder and decoder structure of the original U-Net to extract image features and achieve segmentation at the pixel level, respectively. In the encoder section, they proposed a fusion mechanism based on the transformer to combine contextual data from adjacent slices along the axial axis. They obtained a high-level semantic information model using this mechanism. This feature group is then fed to the decoder’s segmentation component.
The advancement of the various semantic segmentation methods mentioned above cannot be separated from the results of the ConvNet architecture [24,25,26,27,28]. Howard et al. proposed MobileNets [28], which utilized depth-wise separable convolutions, which were introduced in Xception [29] for the first time. A depth-wise separable convolution consists of two operations: a depth-wise and a point-wise convolution. With depth-wise convolution, the number of output channels equals the number of input channels. Convolution with a kernel size of 1 \(\times\) 1 can combine the depth-wise convolution-created features, whereas point-wise convolution cannot. Zhang et al. proposed a new method called ShuffleNet [25] that combines group convolutions and channel shuffles. A group convolution consists of multiple convolutions, each of which takes a portion of the input channels, while channel shuffles randomly mix the group convolution’s output channels. In addition, Sandler et al. created MobileNetV2 [27], which is based on the ideas of MobileNetV1 [28] and uses depth-wise separable convolution as efficient building blocks. Moreover, Freeman et al. proposed EffNet [24], a slim model-optimized version of MobileNet and ShuffleNet, to address issues with existing models. Their proposed method departs from the increasing use of Convolutional Neural Networks in customer products, so it is anticipated that the model will run efficiently on mobile hardware.
Tan and Le [26] realized that balancing network depth, width, and resolution could enhance performance. Based on their observation, they proposed a new compound scaling method that scales all depth, width, and resolution dimensions uniformly using a set of constants and a compound coefficient. Their proposed architecture, which they dubbed EfficientNet, demonstrated that scaling depth, width, and resolution simultaneously produces superior results.
Meanwhile, Shimizu et al. demonstrated the successful implementation [6] of BtrflyNet to segment the skeleton from bone scan images, a critical step in detecting bone cancer metastases. Sekuboyina et al. proposed a butterfly-shaped network called BtrflyNet to efficiently combine information across reformations by combining two views [30]. Their proposed strategy was predicated on EBGANbased adversarial training. EfficientNet’s recent success has also prompted us to attempt to capitalize on EfficientNet’s strengths. Using EfficientNet’s mobile inverted bottleneck convolution (MBConv) has proven to improve the performance of numerous computer vision tasks. In light of this achievement, we attempted to design a segmentation system that would take advantage of the butterfly-type network (BtrflyNet) and EfficientNet.
This study presents a semantic segmentation of whole-body bone scan images. We employed a deep learning-based approach to achieve high accuracy in skeleton segmentation both in anterior and posterior part. We proposed a modified butterfly-type network added with EfficientNet to improve segmentation accuracy. We combine the advantages of butterfly-type network which can process anterior and posterior images simultaneously with the advantages of EfficientNet to help identify the relevant features of each segment. We conducted the experiment using 37 bone scan images and demonstrated the effectiveness of the proposed system by comparing it with the original BtrflyNet approach [6]. Using the high-level features extracted using EfficientNet in conjunction with the features extracted on the BtrflyNet architecture, we demonstrated that our proposed architecture, Efficient-BtrflyNet, is capable of producing excellent segmentation results.
The remaining structures in this article are as follows: The proposed system is described in Sect. 2; the experimental result and analysis are explained in Sect. 3; the conclusion is presented in Sect. 4.
2 Efficient-BtrflyNet for Skeleton Segmentation
This section describes our proposed architecture and performance evaluation.
2.1 Proposed Architecture
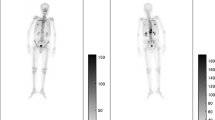
Our work employs a pair of anterior and posterior whole-body bone scan images (Fig. 1) along with their respective segmentation masks. The images were in RGB format with the size of 512 \(\times\) 128 pixels. The segmentation process produced 13 layers corresponding to 12 bones and one background [6]. Figure 2 depicts a guidance for annotating 12 bone classes for both anterior and posterior samples. The 12 bone segments are skull (green), the cervical vertebrae (blue), the thoracic vertebrae (light blue), the ribs (purple), the sternum (pink), the clavicle (light brown), the scapula (brown), the humerus (dark blue), the lumbar vertebrae (yellow), the sacrum (orange), the pelvis (dark green), and the femur (gold).
In this study, we propose Efficient-BtrflyNet to leverage the efficacy of EfficientNet [26] in butterfly-shaped network architecture to produce fine segmentation of bone scan images. According to the human expert, we hypothesize that the anterior and posterior portions of the whole-body bone scan image contain valuable information for producing fine segmentation. The use of a butterfly-shaped network is inspired by BtrflyNet [30], which successfully encoded the use of two views of spine reformation within a single architecture. The encoder–decoder architecture was applied to our proposed architecture, which consisted of convolution and pooling layers with no fully connected layers. Our proposed architecture is illustrated in Fig. 3.
In our proposed architecture, two arms (represented by blue and purple boxes) serve as encoder and decoder, while one body (represented by green boxes) connects these arms. The upper and lower portions of the left arm serve as the encoder for the anterior and posterior bone scan images. Each of the three sub-modules in each section contains a single convolutional layer. To capture more fine-grained patterns, a block of EfficientNet in the form of mobile inverted bottleneck convolution [27] is added to each sub-module.
In contrast, in the decoder section, which consists of three sub-modules, the resolution is incrementally increased through up-sampling and prediction for each pixel. Each sub-module received data from the left arm, which was merged with the up-sampling result from the right arm. Consequently, the network captures patterns at multiple scales. Additionally, the decoder has two parts, the top and the bottom, which serve as decoders for the anterior and posterior bone scan images.
Our work employs a pair of anterior and posterior whole-body bone scan images along with their respective segmentation masks. The images were in RGB format with the size of 512 \(\times\) 128 pixels. The segmentation process produced 13 layers corresponding to 12 bones and one background. The 12 bones are the skull, the cervical vertebrae, the thoracic vertebrae, the lumbar vertebrae, the sacrum, the pelvis, the ribs, the scapula, the humerus, the femur, the sternum, and the clavicle.
In our proposed architecture, the left arm is divided into the upper and lower portions, with three sub-modules each. The three sub-modules at the bottom are responsible for convolution and pooling, similar to the three sub-modules at the top. In the first sub-module of the upper left arm, each input image undergoes a 3 \(\times\) 3 convolution with the same padding, batch normalization, and ReLU as the activation function. The 32-bit output channel will be combined with the right arm’s up convolution results. The input image is also provided to Block 1 of EfficientNet in the second sub-module of the upper left arm, resulting in 32 output channels. A 3 \(\times\) 3 convolution is then performed to generate an output channel of 48, which is further combined with the result of the right arm’s up convolution. In the third sub-module, a 32-by-32 feature map is passed to Block 2 of EfficientNet, resulting in a 48-channel output. This sub-module applies a 3 \(\times\) 3 convolution to the feature map to generate 80 channels, which are later combined with the up-convolution results from the right arm. This 48-by-48-pixel feature map is also provided to Block 3 of EfficientNet, which will generate 80 channels. This feature map is combined with the feature map generated from the left arm’s lower portion.
The output of the left arm (top and bottom) is combined to produce a 160-channel feature map for the body part (green boxes). Then, 3 \(\times\) 3 convolution and 2 \(\times\) 2 max pooling are applied. The same convolution is applied a second time, followed by max-pooling, and then a second convolution is applied to yield 1024 channels. The number of output channels for each convolution is doubled from its input channel count. In addition, the up-convolution is applied, followed by a left-side join and the 3 \(\times\) 3 convolution. 512 channels are the number of output channels for this convolution. An up-convolution follows a left-side join, and the right arm is given a 3 \(\times\) 3 convolution.
The final step is to apply a 1 \(\times\) 1 convolution with the same padding to map each output to 13 channels representing 13 segmented regions. This process is visible in the upper and lower portions of the right arm. The codes to our implementation of the proposed Efficient-BtrflyNet are publicly available at our GitHub repository.Footnote 1
2.2 Performance Evaluation
The loss function to be minimized in the training of skeleton segmentation is cross-entropy as in Equation (1) as follows:
where n is the number of segments, \(t_i\) is the truth label, and \(p_i\) is the Softmax probability for the \(i^{th}\) segment. Then, overall loss L is calculated as the sum of the total loss from anterior and posterior parts.
To assess the segmentation system’s performance, we employ a Dice score as in Equation (2), which measures the similarity or overlapping of two segments, as follows:
A is the predicted segment and B is the ground truth segment. To accurately evaluate the system, this calculation excludes the background segment. The final Dice score is determined by averaging the Dice scores of each segment.
3 Experimental Results and Discussion
This section describes the data, experiment setup, and analysis on the experimental results.
3.1 Dataset Description
This experiment utilized data from two groups: Indonesian and non-Indonesian. We chose the two data sets because there may be differences in the bone geometry size between Indonesian and non-Indonesian data. The images were collected from 18 Indonesian and 19 non-Indonesian respondents. Each image consists of an anterior and a posterior part, making the total number of Indonesian and non-Indonesian images 36 and 38, respectively. The Indonesian bone scan images were obtained in DICOM format from the University of Padjadjaran’s Faculty of Nuclear Medicine [12, 31]. The non-Indonesian images were obtained through a Google Images crawl. Figure 4 depicts examples of unprocessed data for both groups.
For training process, first, we annotated each segment with different colors, as depicted in Fig. 2. We also performed normalization on the original RGB colors to speed up the training process. Table 1 shows the RGB normalization result for each segment, while Fig. 5 shows an example of normalization result from an RGB mask image.
3.2 Parameters of skeleton segmentation training
A total of 29 images were used for training, while eight were used for testing. During the image preprocessing phase, we utilized horizontal flipping. The training is conducted over 300, 500, and 800 epochs, with adaptive moment estimation (Adam) [32] as the optimizer, and cross-entropy as the loss function. The parameters of Adam were set to \(\alpha\)={0.001, 0.0001, 0.00001}, \(\beta _1\)=0.9, \(\beta _2\)=0.999, \(\varepsilon\)=10\(^{-8}\), and weight_decay=0. EfficientNetB7 is the EfficientNet architecture employed in this experiment. We also investigated the pre-trained model based on adversarial training (advprop) [33] in applying EfficientNet.
Table 2 shows the result of training on the use of learning rate \(\alpha\)={0.001, 0.0001, 0.00001} in the BtrflyNet model. The trend of decreasing train_loss values and increasing train_dice values looks significant when using a learning rate from 0.001 to 0.0001. Meanwhile, a larger number of epochs and a longer training time are required during training with \(\alpha\)=0.00001 to achieve convergence. There is no significant change in the train_loss value, but the train_dice value increases quite well. Therefore, this model is taken as the best model on BtrflyNet.
On the other hand, the trend of decreasing train_loss values and increasing train_dice values in the Efficient-BtrflyNet training model is also significant when using a learning rate from 0.001 to 0.0001, which can be seen in Table 3. In this Efficient-BtrflyNet model, the number of epochs is greater, and longer training time is also needed during training with \(\alpha\)=0.00001 to achieve convergence. There is no significant change in the train_loss value, but the train_dice value increases quite well. Therefore, this model is taken as the best model on Efficient-BtrflyNet.
We also applied pre-trained model based on adversarial training (advprop) [33] in applying Efficient-BtrflyNet. Experiments were carried out at \(\alpha\)={0.001, 0.0001, 0.00001}, with the advprop parameter set to TRUE if the model uses advprop, and FALSE if the model does not use advprop. The experimental results can be seen in Table 4, showing that the use of a pre-trained model based on adversarial training (advprop=TRUE) is able to produce better loss and dice values compared to not using a pre-trained model based on adversarial training (advprop=FALSE). Based on these results, the Efficient-BtrflyNet model used is the one using advprop.
3.3 Results
We compare the testing performance of our proposed architecture to that of the BtrflyNet architecture developed by Shimizu et al. [6] for bone scan image segmentation. The proposed Efficient-BtrflyNet implementation is publicly available at our GitHub page.Footnote 2 We present the experimental results at Tables 5, 6, and 7. Table 5 shows the comparison of average Dice score of anterior and posterior segments, while Tables 6 and 7 show Dice scores for each segment of anterior and posterior results, respectively.
In general, our proposed architecture, Efficient-BtrflyNet, generates segments with higher Dice scores than BtrflyNet, both in anterior and posterior parts (shown in Tables 6 and 7). However, in some segments, it appears that BtrflyNet is able to produce segments with better Dice scores than Efficient-BtrflyNet, which are Clavicle and Scapula segments in the anterior, as well as Lumbar Vertebrae and Sacrum segments in the posterior. Nonetheless, the Dice score gaps between BtrflyNet and proposed Efficient-BtrflyNet are generally less than 0.01.
Furthermore, we visually demonstrate segmentation results on the eight testing images that can be seen in Figs. 6 and 7. Top and middle rows of the figures show segmentation outputs using BtrflyNet and Efficient-BtrflyNet, respectively. While the bottom row shows the corresponding ground truth annotations.
The cervical vertebrae segment had the lowest dice value in the anterior part (see Table 6) for both the BtrflyNet (Dice=0.4316) and Efficient-BtrflyNet (Dice=0.5337) models. As depicted in Fig. 8, the cervical vertebrae segment located between the skull and thoracic vertebrae segments has a very small surface area, making it difficult for the model to capture the segments precisely. Conversely, the results of the skull segments that occasionally merge with the cervical vertebrae segments reduce the segment’s detected area as a cervical vertebrae segment. In contrast, the cervical vertebrae segment performs well in the posterior part due to its clearer appearance and larger segment area in the posterior part.
There are almost no anterior or posterior segmentation problems in the results of skull segmentation. Figure 9 depicts the image with the worst segmentation results, where the anterior skull segment is poorly segmented. Figure 9c demonstrates that the Efficient-BtrflyNet model generates a smaller unsegmented area than the BtrflyNet model (Fig. 9b). If we examine the original image, we can see that the skull has a gray color that is difficult to distinguish from the background, making it challenging to obtain a good segment in this area.
A similar issue occurs in one of the femur segment results, as depicted in Fig. 9, with blue-circled region. The gray color of the femur from the initial bone scan image is indeed difficult to identify, so the model cannot accurately predict the femur segment.
On the other hand, both BtrflyNet and Efficient-BtrflyNet produced excellent segmentation results for the ribs segment of anterior part. As shown in Fig. 10, Efficient-BtrflyNet is able to solve segmentation problems better in some instances. Despite the application of BtrflyNet, a number of pixels cannot be identified as ribs segment. In the meantime, this issue is absent from the Efficient-BtrflyNet model.
In the meantime, the posterior rib segment has a segmentation flaw. There are multiple pixels that are not part of the ribs but are predicted to be ribs. Figure 11 demonstrates that there is a distinct region of the ribs that receives the same purple label as the ribs themselves.
The anterior and posterior segmentation of the pelvis is another noteworthy segmentation result. Figure 12 contains several examples. Using the BtrflyNet model (Fig. 12’s upper portion), there are still unsegmented segment areas; However, this condition can be greatly improved by employing Efficient-BtrflyNet (middle part in Fig. 12). On this pelvis segmentation, the presence of metastatic hotspots on the bone scan image has caused a segmentation error. As depicted in Fig. 13, the results of pelvis segmentation are widened by assuming the metastatic hotspot (the red-circled portion) is a part of the pelvis.
In contrast to the research by Shimizu et al. [6], which combines the scapula segment with the rib segment on the posterior part, we hypothesized that the scapula segment would be distinct from the rib segment in this study. A portion of the posterior scapula overlaps the segment of the ribs, making it potentially difficult to identify as part of the scapula. Therefore, we labeled the scapula on this posterior part with two different colors: pink for the scapula adjacent to the ribs and dark brown for the scapula adjacent to the clavicle (see Fig. 2 for clarity). Both of these scapula segments received favorable dice scores (see Table 7). Figure 14 depicts an example of the results of scapula segmentation in the posterior section.
4 Conclusion
Developing semantic segmentation architecture for the segmentation of whole-body bone scan images is a challenging task. Efficient-BtrflyNet, a novel method for segmenting whole-body bone scan images, has been proposed, and its superiority has been successfully demonstrated. Our approach successfully segmented several segments that BtrflyNet was unable to, and improved segmentation results in several segments, as indicated by the increase in dice score.
Based on the experimental results, even though medical images typically have low image resolution, significant future work will involve some advanced image pre-processing to enhance the quality of medical images to reduce segmentation errors. In addition, post-processing is necessary to eliminate inaccurate segments without diminishing the quality of the segmentation results in other parts that are already good. To improve segmentation performance in deep learning-based models, it is also crucial to increase the size of the training dataset.
Data Availability
The implementation codes for this research is publicly available, as also mentioned in the paper, at https://github.com/Ema-Rachmawati/bonescansegment. The dataset used in this research is currently not publicly available.
Abbreviations
- AFTer-UNet:
-
Axial fusion transformer U-Net
- Advprop:
-
Adversarial training
- BSI:
-
Bone Scan Index
- CNN:
-
Convolutional neural network
- DICOM:
-
Digital imaging and communications in medicine
- Globocan:
-
Global Burden of Cancer Study
- HAUNet-3D:
-
Hierarchical attention 3D U-Net
- LR:
-
Learning rate
- ReLU:
-
Rectified linear unit
- RGB:
-
Red–green–blue
- SA-UNet:
-
Spatial attention U-Net
References
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R.L., Torre, L.A., Jemal, A.: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68(6), 394–424 (2018)
Inaki, A., Nakajima, K., Wakabayashi, H., Mochizuki, T., Kinuya, S.: Fully automated analysis for bone scintigraphy with artificial neural network: usefulness of bone scan index (BSI) in breast cancer. Ann. Nucl. Med. 33(10), 755–765 (2019). https://doi.org/10.1007/S12149-019-01386-1
Larson, S.M.: EXINI quantitative bone scan index: Expanded utility for the planar radionuclide bone scan. J. Nucl. Med. 57(1), 5–6 (2016)
Nakajima, K., Edenbrandt, L., Mizokami, A.: Bone scan index: a new biomarker of bone metastasis in patients with prostate cancer. Int. J. Urol. 24(9), 668–673 (2017)
Wakabayashi, H., Nakajima, K., Mizokami, A., Namiki, M., Inaki, A., Taki, J., Kinuya, S.: Bone scintigraphy as a new imaging biomarker: the relationship between bone scan index and bone metabolic markers in prostate cancer patients with bone metastases. Ann. Nucl. Med. 27(9), 802–807 (2013)
Shimizu, A., Wakabayashi, H., Kanamori, T., Saito, A., Nishikawa, K., Daisaki, H., Higashiyama, S., Kawabe, J.: Automated measurement of bone scan index from a whole-body bone scintigram. Int. J. Comput. Assist. Radiol. Surg. 15(3), 389–400 (2020)
Sadik, M., Suurkula, M., Höglund, P., Järund, A., Edenbrandt, L.: Improved classifications of planar whole-body bone scans using a computer-assisted diagnosis system: a multicenter, multiple-reader, multiple-case study. J. Nucl. Med. 50(3), 368–375 (2009)
Sadik, M., Hamadeh, I., Nordblom, P., Suurkula, M., Höglund, P., Ohlsson, M., Edenbrandt, L.: Computer-assisted interpretation of planar whole-body bone scans. J. Nucl. Med. 49(12), 1958–1965 (2008)
Ulmert, D., Kaboteh, R., Fox, J.J., Savage, C., Evans, M.J., Lilja, H., Abrahamsson, P.A., Björk, T., Gerdtsson, A., Bjartell, A., Gjertsson, P.: A novel automated platform for quantifying the extent of skeletal tumour involvement in prostate cancer patients using the bone scan index. Eur. Urol. 62(1), 78–84 (2012)
Kikuchi, A., Kawakami, T.: Future of artificial intelligence and nuclear cardiology. Ann. Nucl. Cardiol. 4(1), 79–82 (2018)
Rachmawati, E., Sumarna, F.R., Jondri, Kartamihardja, A.H.S., Achmad, A., Shintawati, R.: Bone scan image segmentation based on active shape model for cancer metastasis detection. In Proceedings of the IEEE international conference on information and communication Technology, pp. 1–6, (2020)
Rachmawati, E., Jondri, Ramadhani, K.N., Kartamihardja, A.H.S., Achmad, A., Shintawati, R.: Automatic whole-body bone scan image segmentation based on constrained local model’. Bull. Electr. Eng. Inf. 9(6), 2526–2537 (2020)
Krizhevsky, A., Sutskever, I., Hinton, G.E.: ImageNet classification with deep convolutional neural networks. Commun. ACM 60(6), 84–90 (2017)
Litjens, G., Kooi, T., Bejnordi, B.E., Setio, A.A.A., Ciompi, F., Ghafoorian, M., Laak, J.A.V.D., Ginneken, B.V., Sánchez, C.I.: A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017)
Liu, X., Song, L., Liu, S., Zhang, Y.: A review of deep-learning-based medical image segmentation methods. Sustainability 13(3), 1224 (2021)
Fu, Y., Lei, Y., Wang, T., Curran, W.J., Liu, T., Yang, X.: A review of deep learning based methods for medical image multi-organ segmentation. Physica Med. 85, 107–122 (2021)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Proceedings of the international conference on medical Image computing and computer-assisted intervention, pp. 234-241, (2015)
Guo, C., Szemenyei, M., Yi, Y., Wang, W., Chen, B., Fan, C.: SA-UNet: spatial attention U-Net for retinal vessel segmentation. In: Proceedings of the 25th IEEE international conference on pattern recognition, pp. 1236-1242, (2021)
Sun, F., Kumar, V.A., Yang, G., Zhang, A., Zhang, Y.: Circle-U-Net: an efficient architecture for semantic segmentation. Algorithms 14(6), 159 (2021)
Aledhari, M., Razzak, R.: An adaptive segmentation technique to detect brain tumors using 2D UNet. In: Proceedings of the IEEE international conference on bioinformatics and biomedicine, pp. 2328-2334, (2020)
Yan, X., Tang, H., Sun, S., Ma, H., Kong, D., Xie, X.: AFTerUNet: axial fusion transformer UNet for medical image segmentation. In: Proceedings of the IEEE winter conference on applications of computer vision, pp. 3971-3981, (2020)
Zhou, F., Luo, F., Efio-Akolly, K., Bbosa, R., Huang, W.C., Zou, J.N., Chen, Y.P., Liu, F.: “HAUNet-3D: a novel hierarchical attention 3D UNet for lung nodule segmentation. In: Proceedings of the IEEE international conference on bioinformatics and biomedicine, pp. 1213-1220, (2021)
Baheti, B., Innani, S., Gajre, S., Talbar, S.: Eff-UNet: a novel architecture for semantic segmentation in unstructured environment. In: Proceedings of the IEEE conference on computer vision and pattern recognition workshops, pp. 1473-1481, (2020)
Freeman, I., Roese-Koerner, L., Kummert, A.: EffNet: an efficient structure for convolutional neural networks. In: Proceedings of the 25th IEEE international conference on image processing, pp. 6-10, (2018)
Zhang, X., Zhou, X., Lin, M., Sun, J.: ShuffleNet: an extremely efficient convolutional neural network for mobile devices. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 6848-6856, (2017)
Tan, M., Le, Q.v.: “EfficientNet: Rethinking model scaling for convolutional neural networks. In: Proceedings of the 36th international conference on machine learning, pp. 6105-6114, (2019)
Sandler, M., Howard, A., Zhu, M., Zhmoginov, A., Chen, L.C.: “MobileNetV2: Inverted residuals and linear bottlenecks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 4510-4520, (2018)
Howard, A.G., Zhu, M., Chen, B., Kalenichenko, D., Wang, W., Weyand, T., Andreetto, M., Adam, H.: MobileNets: efficient convolutional neural networks for mobile vision applications,” Computing Research Repository arXiv preprint, arXiv:1704.04861, (2017)
Chollet, F.: Xception: deep learning with depthwise separable convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 1800-1807, (2016)
Sekuboyina, A., Rempfler, M., Kukačka, J., Tetteh, G., Valentinitsch, A., Kirschke, J.S., Menze, B.H.: Btrfly Net: vertebrae labelling with energy-based adversarial learning of local spine prior. In: Proceedings of the international conference on medical image computing and computer-assisted intervention, pp. 649-657, (2018)
Nugraha, D.B., Rachmawati, E., Sulistiyo, M.D.: Semantic segmentation of whole-body bone scan image using Btrfly-Net. In: Proceedings of the 14th international conference on information technology and electrical engineering, pp. 264-269, (2022)
Kingma, D.P., Ba, J.L.: Adam: A method for stochastic optimization. Comput. Res. Repository arXiv preprint, arXiv:1412.6980, (2014)
Xie, C., Tan, M., Gong, B., Wang, J., Yuille, A.L., Le, Q.v.: Adversarial examples improve image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 819-828, (2020)
Acknowledgements
The authors would like to thank the Department of Nuclear Medicine and Molecular Theranostic, Dr. Hasan Sadikin General Hospital, Faculty of Medicine, Universitas Padjadjaran, Indonesia, for assistance with the data collection and analysis.
Funding
This work was funded by the Ministry of Education, Culture, Research, and Technology, Republic of Indonesia, in the scheme of National Competitive Basic Research with the research grant number of 126/SP2H/RT-MONO/LL4/2022 and supported by Telkom University with research grant number of 357/PNLT3/PPM/2022.
Author information
Authors and Affiliations
Contributions
Conceptualization by Rachmawati; funding acquisition by Rachmawati and Sulistiyo; research’s project manager by Rachmawati; experiment design by Rachmawati; data preparation by Rachmawati and Nugraha; experiment execution by Rachmawati and Nugraha; result visualization by Rachmawati, Nugraha, and Sulistiyo; manuscript drafting and writing by Rachmawati, Nugraha, and Sulistiyo; manuscript finishing and compilation by Sulistiyo. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that there is no competing interest.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rachmawati, E., Sulistiyo, M.D. & Nugraha, D.B. Leveraging Model Scaling and Butterfly Network in the Bone Scan Image Segmentation. Int J Comput Intell Syst 17, 92 (2024). https://doi.org/10.1007/s44196-024-00453-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s44196-024-00453-4