Abstract
Colorectal cancer ranks third in global malignancy incidence, and serrated adenoma is a precursor to colon cancer. However, current studies primarily focus on polyp detection, neglecting the crucial discrimination of polyp nature, hindering effective cancer prevention. This study established a static image dataset for serrated adenoma (SA) and developed a deep learning SA detection model. The proposed MSSDet (Multi-Scale Sub-pixel Detection) innovatively modifies each layer of the original feature pyramid’s structure to retain high-resolution polyp features. Additionally, feature fusion and optimization modules were incorporated to enhance multi-scale information utilization, leveraging the narrow-band imaging endoscope’s ability to provide clearer colonoscopy capillary and texture images. This paper utilized 639 cases of colonic NBI endoscopic images to construct the model, achieving a mean average precision (mAP) of 86.3% for SA in the test set. The SA detection rate via this approach has significantly surpassed conventional object detection methods.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Colorectal cancer, the third most common cancer globally [1], poses a significant threat to human health. Colorectal polyps/adenomas are precancerous lesions associated with colorectal cancer, encompassing traditional adenomas and serrated polyps. Serrated polyps include hyperplastic polyps (HP), sessile serrated adenoma/polyp (SSA/P), and traditional serrated adenoma (TSA). For this discussion, both SSA/P and TSA are classified as serrated adenomas (SA). The majority of precancerous lesions stem from colon polyps/adenomas, with the progression pathway leading from adenoma to cancer. Approximately 25%–35% of colorectal cancers develop from SA [2], indicating a higher carcinogenic potential compared to traditional adenomas. The removal of tumor polyps through colonoscopy plays a crucial role in preventing intestinal cancer [3]. However, colonoscopy’s effectiveness is contingent upon operator skill and is not infallible [4]. A recent meta-analysis covering more than 15,000 tandem colonoscopies revealed a high missed rate of 26% for adenomas [5]. In the field of colonoscopy, deep learning mainly focuses on the discovery, localization, and diagnosis of polyps during colonoscopy. However, all studies on the diagnostic properties of colorectal polyps using artificial intelligence have not distinguished SA from proliferative polyps, and there have been no studies focused on the diagnosis of SA using artificial intelligence under NBI.
Therefore this paper aims to address these gaps and makes the following contributions:
-
A gold standard dataset of colorectal cancer, including SA and HP, based on NBI.
-
Proposing an improved method based on multi-scale subpixel convolution that utilizes the new feature pyramid to enhance the performance of the detection task.
-
Designing a new feature fusion and optimization block to reinforce the utilization of multi-scale features.
2 Related Work
2.1 Classification Method for Polyps
Scholars have proposed the WASP (Workgroup serrAted polypS and Polyposis) classification method for polyps [6], which forms the basis for polyp classification. Generally, the diagnostic accuracy of colonoscopy for adenomas is below 90% and even lower for SA.
The automatic detection of polyps is a burgeoning research area [7]. Early studies predominantly employed machine learning methods, focusing on the shape, texture, or color characteristics of polyps [8,9,10,11]. As a result, an integrated architecture has been proposed, combining various polyp features such as color, texture, shape, and temporal information.
Polyp features are derived from a combination of hand-crafted and end-to-end learning based on CNN [12]. Additionally, a region-based CNN (Inception-ResNet) has been trained on colonoscopy images and videos to facilitate polyp detection [13]. A deep learning model, based on the new NBI technology, achieved 94% accuracy in polyp detection after learning from NBI video data of colonoscopic polyps [14]. Subsequently, a multi-threaded deep learning algorithm was proposed, leveraging a small receptive field to focus on local features for polyp detection using diverse morphological data [15]. Deep learning methods and handcrafted global features were utilized for pixel-based, frame-based, and block-based segmentation and detection [16]. Extensive experiments on the Kvasir SEG dataset demonstrated that the latest deep learning method for polyp detection, localization, and segmentation significantly improved performance by optimizing the architecture from the encoder to the decoder [17]. The Endoscopy Center of the Second Hospital of Zhejiang University Medical College iteratively adjusted and strategically modified the proportion of different data types based on the unbalanced distribution of data and the complex shapes of colon polyps within 600,000 colon images. Consequently, the detection accuracy, recall, and F1 score for colonic polyps reached 98.3%, 88.1%, and 92.9%, respectively. Utilizing the YOLO algorithm and ResNet deep convolution neural network, the Digestive Endoscopy Center of the People’s Hospital of Wuhan University achieved an average accuracy of 98% across four test datasets [18]. Recently, a bidirectional multiscale feature fusion structure and ObjectBox detection head have been employed, which increased the recall rate from 91.5 to 98.23% [19].
However, existing research on diagnosing colorectal polyps using artificial intelligence often fails to differentiate between colorectal tumor lesions such as polyps and SA under NBI. Furthermore, the literature typically focuses on the performance of a single model evaluated under uniform conditions.
2.2 Self-attention Mechanism
When processing visual images, humans rely on multiple sources of information to achieve better results rather than focusing on a single scene. The self-attention mechanism mimics this process, allowing neural network models to effectively focus on important parts and capture intricate details.
The attention mechanism was initially employed in the encoder–decoder model for machine translation and natural language processing, enabling the network to concentrate on specific encoded semantic vectors or inputs [20]. In 2019, the SENet [21] was introduced as a notable example of incorporating attention mechanisms into computer vision. Its implementation is straightforward, involving the addition of a plug-and-play SE module branch to model the correlation between different channels, resulting in significant improvements in model performance. Subsequently, the authors further enhanced this concept to develop SKNet [22], which allows for the selection of a convolution core of appropriate size. ECA [23] can be regarded as an improved version of SENet, as it removes the fully connected layer used in SENet and replaces it with 1x1 convolutional kernels, reducing the number of model parameters and making it more lightweight. CBAM [24], proposed by Woo et al., improves the feature extraction capabilities of network models without significantly increasing computational complexity or parameters.
2.3 Feature Fusion
Fusing features of different scales is a crucial approach to enhancing performance. Lower-level features offer higher resolution and richer location and detail information, while higher-level features provide stronger semantic information.
FPN [25] constructs a feature pyramid structure with varying sizes by sampling the deep information and element-wise addition of shallow information. PANet [26] shortens the information pathway and enriches the feature pyramid with precise low-level positioning information, thereby enhancing the bottom-up pathway. MLFPN [27] leverages SFAM to fuse multi-level features and obtain a comprehensive feature pyramid for final prediction.
3 Methods
In this paper, inspired by SSD [28], a new object detection model based on multi-scale sub-pixel convolution optimization of artificial intelligence-assisted colonoscopy system is proposed, which is called MSSDet (Multi-Scale Sub-pixel Detection). The feature fusion module and feature optimization module are constructed to form a SA auxiliary detection model under narrow-band imaging endoscopy. After the training set is enhanced by autoaugmentation [29], from 1000 to 5000 images are obtained for training.
Our goal is to identify and categorize SA and HP from NBI images. Figure 1 illustrates the overall architecture of our proposed model. Initially, the NBI images are resized from \(H \cdot w\) to \(224 \times 224\) and fed into the chosen backbone network, which in this study is DetNet [30]. The backbone network generates a feature pyramid consisting of six feature layers through a combination of convolutional layers across six stages. As for polyp detection, modifications are made to the structure of each layer within the original feature pyramid, which has varying resolutions. Specifically, a feature fusion module is introduced to the middle four layers. These four layers, along with the top and bottom layers, undergo feature optimization to generate a new feature pyramid. Subsequently, a \(3 \times 3\) CNN is employed to predict the target box for each layer. Finally, the predicted target boxes are fused using Soft-NMS [31] to obtain the optimal target box.
3.1 Feature Extraction
The endoscopic equipment enters the colon for detection during the actual endoscopic examination. When the endoscopist operates and finds suspicious lesions, he will focus the lens on the specific position. Due to such operational characteristics, the size of polyp lesions on endoscopic images varies greatly. For the detection of SA, multi-scale feature extraction is vital. Therefore, the feature pyramid is used to extract the features of the image.
During an actual endoscopic examination, the endoscopic equipment is inserted into the colon for detection. When suspicious lesions are identified by the endoscopist, the lens will be focused on the specific location. As a result of such operational characteristics, the size of polyp lesions captured in endoscopic images varies significantly. Given the importance of multiscale feature extraction for the detection of SA, a feature pyramid is employed to extract image features.
In high-level feature maps, if the edges of objects are too blurry and small target objects have been lost, the semantic information they carry will also be lost [30]. In polyp detection, the information of small targets and edge information is particularly crucial. In this regard, the high-level feature map keeps the resolution unchanged and forms the new feature pyramid, which is highly beneficial for accurate polyp classification. Therefore, DetNet, which is specifically designed for efficient feature extraction from images, is chosen as the backbone network for this study. DetNet is proposed on the basis of ResNet-50 [32]. It comprises of six stages, with the first four stages identical to those in ResNet-50. In stages 5 and 6, the spatial resolution remains constant and does not decrease. This high-level fixed resolution results in a smaller receptive field than in previous stages. At each stage after adjustment, a feature layer is extracted to form a feature pyramid. Compared with the common feature pyramid, the extracted feature pyramid better preserves the characteristics of capillaries and textures under the high resolution of small receptive fields and better serves the classification of SA.
In addition, the improved residual unit is used to supplement the upper layer feature information, resulting in better utilization of multi-scale feature information. As shown in Fig. 2, the improved unit in this paper adopts the design of dilated convolution [33] as an introductory module to effectively increase the receptive field, when compared to a residual unit of ResNet. Furthermore, the same operation is shared by performing a \(1 \times 1\) convolution to project features into a deeper space. This approach allows for obtaining multi-scale information, which enriches the extraction of polyp feature information.
3.2 Feature Fusion
This paper uses the passthrough layer [34] and the sub-pixel [35] convolution layer instead of downsampling and upsampling. The sub-pixel convolution layer takes the low-resolution feature map as input. High-resolution feature mapping can be obtained by pixel reorganization between convolution kernel multi-channel. It can convert the low-resolution feature map \(n\cdot (C \cdot r \cdot r) \cdot W \cdot H\) to a high-resolution feature map \(n \cdot C \cdot (H \cdot r) \cdot (W \cdot r)\). In the passthrough layer, the high-resolution feature map \(n \cdot C \cdot (r \cdot W) \cdot (r \cdot H)\) is sampled at intervals of rows and columns, resulting in four new feature maps \(n \cdot C \cdot (r \cdot W/2) x (r \cdot H / 2)\). These four feature maps are then connected in series by channels to obtain a new feature map \(n\cdot C \cdot 4 \cdot (r \cdot W/2) \cdot (r \cdot H / 2)\), where n, C, W, H, and r represent the number of images, channels, width and height, and multiples of upsampling or downsampling, respectively (Fig. 3).
Similar to Deconv Transformer (DecT) [36], feature fusion is used to further utilize the feature information provided by convolution layers of different depths. Take the B2 feature map in Fig. 4 as an example. A1, A2, and A3 are the original feature maps directly output by the backbone network, while B2 represents the new feature map obtained after the process of feature fusion. Firstly, A1 adopts the passthrough layer to generate A1 Down. Next, A3 uses a sub-pixel convolution layer to obtain A3 Up. After that, since the sizes of A1 Down, A3 Up, and A2 are the same, they are stacked by channels to produce a new feature map. In particular, the high-level feature layers do not need up- or downsampling and can be directly fused. In the whole fusion process, the sub-pixel convolution layer can automatically learn the mapping relationship through gradient updating. Meanwhile, the passthrough layer can preserve numerous underlying details without the need to learn parameters. Finally, use a 1 * 1 convolution kernel to reduce the dimension of the superimposed new feature map and reduce the number of channels to the same number as A2 to obtain B2. This method can effectively enrich the semantic and texture information in the fused feature map.
3.3 Feature Optimization
The computational capacity of neural networks is limited. To enhance the accuracy of neural networks with limited resources, this study draws inspiration from the spatial- and channel-oriented structure discriminative modules (SCSDM) [37]. The key idea is to allocate more resources, represented by weights in neural networks, to the important features of the recognition object. Therefore, this paper adopts attention mechanism to help convolutional neural networks extract effective features and discard useless ones. This paper compares three commonly used attention mechanisms, namely SE, CBAM, and ECA, through experiments. After a thorough comparison, CBAM is ultimately selected as the preferred attention mechanism.
CBAM consists of two independent sub-modules, the channel attention module (CAM), and the spatial attention module (SAM), to fuse attention features in the channel and spatial dimensions, respectively (Figs. 5, 6, 7).
CAM obtains a \(1 \times 1 \cdot C\) correlation vector by using the global average pooling operation to squeeze the input feature map and then excite the feature by putting it into a two-layer full connection layer to obtain an attention score. Afterward, the output is obtained by performing scalar multiplication between the score vector and the original feature map. The lightweight design allows for significant improvement in network performance without any increase in computation.
Firstly SAM adopts average pooling and maximum pooling from the channel dimension and then contacts the feature maps. Furthermore, use the convolution operation to generate the final spatial attention feature map from the spliced feature map.
3.4 Target Detection
Through feature fusion and optimization, a new feature pyramid B is obtained. Each feature map predicts the value used by a \(3 \times 3\) convolution layer, which includes the classification score and the position deviation from the prior box. K prior boxes are predicted at each point of the feature map. For each prior box, two classification scores and four position offsets are predicted. This results in a total of six parameters being outputted for each feature map; thus, the convolution layer behind the feature map has a size of \(3 \times 3 \times 6\). So far, we can obtain six characteristic graphs and output \(m \cdot n \times 6\) predicted values (m and n are the width and height of the feature map).
After obtaining \(m \cdot n \times 6\) predicted values, we need to determine which boundary boxes belong to the correct position classification and which ones are incorrect. The output value of a polyp detection should be a boundary box. Therefore, this paper uses soft-NMS [31] to improve the existing problems of NMS [38].
3.5 Loss Function
Inspired by the SSD algorithm, the overall target loss function is defined as the weighted sum of confidence loss (AS) and location loss (Focal E-IoU). The formula is as follows where x is the input feature, y is the output, l is the predicted box, g is the true box, and n is the number of samples:
Confidence loss adopted AMSoftmax loss [39]. It can better reduce the spacing between the same classes and increase the spacing between different classes. S and m are hyperparameters, where \(f_i\) is the input of the last fully connected layer (representing the i-th sample), and \(W_j\) is the j-th column of the parameter matrix W of the last fully connected layer.
Location loss adopted Focal EIoU loss [40]. It explicitly measures the difference of three geometric factors in the boundary box regression: the overlap area, the center point, and the side length. w and h are the width and height of box, while \(w^c\) and \(h^c\) refer to the smallest enclosing box that covers both boxes. b and \(b^{gt}\) denote the central points of two boxes. \(\rho (\cdot )\) indicates the Euclidean distance.
4 Experiments and Results
4.1 Dataset
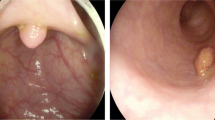
The data used in this paper were collected from the Digestive Endoscopy Center at Shanghai Renji Hospital. The research relied on manual screening, during which images containing misleading or useless information were deleted. For instance, if an image was seriously blurred, out of focus, or had significant changes in illumination due to the camera’s violent motion, it was not selected for the dataset since accurate labeling could not be achieved. Deep neural networks are vulnerable to adversarial attacks, in which a small perturbation to samples can cause misclassification [41]. Therefore some images with fewer defects are retained to improve the robustness of the model under poor and noisy conditions. Due to the scarcity of SA data samples, Trivial Augment [42] was used to expand the number of data samples. The dataset was annotated by senior doctors from the endoscopy center, with polyp positions marked by rectangular boxes and divided into two categories: Figs. 8 and 9. The annotations were saved in VOC format.
The data used in this paper are 639 cases of colorectal cancer, containing about 1000 images. The positive and negative samples ratio is about 2:3 (383 positive samples SA and 256 negative samples HP). In this paper, the training, validation, and test sets were created by randomly selecting 75%, 10%, and 15% of cases, respectively. For the test set, specifically, 96 cases were randomly chosen. The proportion of SA and HP was 1:1, with 48 cases, respectively (Table 1).
4.2 Implementation Details
In this experiment, the paper conducted a comparative study between MSSDet and other object detection models, including faster R-CNN [43], YOLOv3 [44], YOLOv4 [45], SSD, RetinaNet [46], RefineDet [47], YOLOv7 [48], YOLOv8 [49]. The super parameter settings and training loss are as follows:
4.3 Ablation Study
This paper improves the performance of target detection under colonoscopy by modifying the SSD backbone network and introducing feature fusion and feature optimization modules. These three components play a critical role, and therefore, the paper conducted various tests to verify the performance gain achieved by these modifications. Using SSD as the baseline, the feature fusion module (FFM) and feature optimization module (FOM) were treated as optional modules. Additionally, three attention mechanisms were tested for the FOM: SE, CBAM, and ECA. The settings for the super parameters were kept identical throughout the experiments, as presented in Table 2.
Overall, the modification of the backbone network results in better feature extraction performance, leading to improved accuracy in polyp classification. Although the improvement is significant compared to the baseline, the mAP performance remains poor, and the number of detected frames and position tracking accuracy is relatively low. However, integrating FFM and FOM modules enhances the performance, with FFM providing a greater boost than FOM. The selection of CBAM for the FOM module further maximizes the performance by effectively utilizing multi-level features and textured vascular information. Optimizing the network from both the channel and spatial aspects contributes to the overall improved performance. The proposed MSSDet model takes full advantage of NBI band information and performs SA detection effectively.
4.4 Results
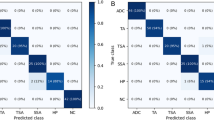
Table 3 shows the performance indicators of all models. Overall, MSSDet achieved the highest average F1 score, and AP value for polyp detection. The detection results are shown in Fig. 10.
SSD-based detectors SSD, RetinaNet, RefineDet, and MSSNet perform well in detecting polyps, because they can obtain better positioning knowledge before generating final predictions by roughly adjusting anchor points. The recall rate of these algorithms is generally higher than the precision rate. Compared with RefineDet, the recall rate of MSSDet has not improved much. The recall rate of RefineDet has been greatly reduced due to its emphasis on the background and targeted improvement of the object. MSSDet may reduce the high-level feature information due to the excessive integration of the detailed features of the low-level feature map, so the missed detection rate is slightly increased.
The evaluation results of the recall rate and the two types of polyps show that the SSD, RetinaNet, RefineDet, and MSSDet based on SSD generally perform well in detecting polyps. By adjusting the anchor points initially, better positioning knowledge can be obtained before generating the final prediction. Faster R-CNN has poor classification of polyps. The possible reason is that there is no targeted adjustment for feature extraction. YOLOv8 is the best performing algorithm in the YOLO family. At the same time, the YOLO series algorithm has excellent performance in the detection of HP, but it is slightly inferior in the detection of SA. The number of predicted frames is lower compared to other models, resulting in missed detections of many polyps. However, the classification accuracy of the detected polyps is higher. This could be attributed to the relatively fixed anchor points (priori boxes) used by these models. Detecting polyps accurately can be challenging due to their unstable size and shape in images.
5 Conclusion
The study presents a multi-scale sub-pixel convolution target detection model to detect and distinguish SA from HP. Firstly, the adjustment of feature extraction can better obtain the shallow and deep texture information. Next, the sub-pixel convolution layer and the passthrough layer are used to optimize the feature map. Finally, the feature fusion and optimization modules are incorporated into the network to learn the feature information of different channels more effectively. The experimental results show that the mAP of MSSDet in detecting SA is up to 86.3%. In addition, MSSDet exhibits strong adaptability to targets with different backgrounds and outperforms common algorithm in accuracy and recall in this case. However, it should be noted that the algorithm complexity leads to a reduction in recognition speed. In future research, particular attention will be given to enhancing the recognition speed while maintaining the same level of accuracy.
Compared to the relevant research on polyp detection, this paper may have a specific gap in terms of the final performance index. However, it is important to highlight that this paper focuses on the classification of polyp categories, which is a valuable contribution. Currently, there are still some challenges to address. Due to the scarcity of SA medical records, the data volume of the dataset in this paper is less than that of similar studies, and the part of the test set is relatively small. While this may not be sufficient to validate the model’s generalization performance, it has demonstrated the feasibility of classifying polyp nature. By continuously improving the dataset and increasing its size, it will be possible to design a more specialized convolutional neural network model for polyp classification. Reviewing the rapid development of computer vision in recent years, the availability of datasets plays an important role. For polyp classification, the subtle features of polyps are key factors. Therefore, the next research direction should focus on utilizing these subtle features effectively and discovering the relationship between radiological image features and the pathological gold standard.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Change history
14 May 2024
A Correction to this paper has been published: https://doi.org/10.1007/s44196-024-00516-6
References
Siegel, R.L., Miller, K.D., Fuchs, H.E., Jemal, A.: Cancer statistics 2021. A Cancer J Clin 71(1), 7–33 (2021)
Snover, D.C.: Update on the serrated pathway to colorectal carcinoma. Human Pathol. 42(1), 1–10 (2011)
Winawer, S.J., Zauber, A.G., Ho, M.N., O’brien, M.J., Gottlieb, L.S., Sternberg, S.S., Waye, J.D., Schapiro, M., Bond, J.H., Panish, J.F., Ackroyd, F.: Prevention of colorectal cancer by colonoscopic polypectomy. N. Engl. J. Med. 329(27), 1977–81 (1993)
Robinson, Malcolm: Colonoscopic withdrawal times and adenoma detection during screening colonoscopy. Intern. Med. Alert 29(4), 29–30 (2007)
Zhao, S., Wang, S., Pan, P., Xia, T., Chang, X., Yang, X., Guo, L., Meng, Q., Yang, F., Qian, W.: Magnitude, risk factors, and factors associated with adenoma miss rate of tandem colonoscopy: a systematic review and meta-analysis. Gastroenterology 56(6), 1661–1674 (2019)
Ijspeert, J.E.G., Bastiaansen, B., Leerdam, M.V., Meijer, G.A., Van Eeden, S., Sanduleanu, S., Schoon, E.J., Bisseling, T.M., Spaander, M.C., Van Lelyveld, N.: Sa1559 development and validation of the wasp-classification system for optical diagnosis of adenomas, hyperplastic polyps and sessile serrated adenomas/polyps. Gut 65(6), 963–970 (2015)
Lee, C.K., Park, D.I., Lee, S.H., Hwangbo, Y., Eun, C.S., Han, D.S., Cha, J.M., Lee, B.I., Shin, J.E.: Participation by experienced endoscopy nurses increases the detection rate of colon polyps during a screening colonoscopy: a multicenter, prospective, randomized study. Gastrointest. Endoscopy 74(5), 1094–1102 (2011)
Cheng, S.C., Huang, Y.M., Wong, S., Tjandra, D.: Computer-aided tumor detection in endoscopic video using color wavelet features,sa karkanis, dk iakovidis, de maroulis, da (2003)
Mamonov, A.V., Figueiredo, I.N., Figueiredo, P.N., Tsai, Y.: Automated polyp detection in colon capsule endoscopy. IEEE Trans. Med. Imag. 33(7), 1488–1502 (2013)
Ameling, S., Wirth, S., Paulus, D., Lacey, G., Vilario, F.: Texture-based polyp detection in colonoscopy. DBLP (2009)
Bernal, J., Tajbakhsh, N., Sanchez, F.J., Matuszewski, B.J., Chen, H., Yu, L., Angermann, Q., Romain, O., Rustad, B., Balasingham, I.: Comparative validation of polyp detection methods in video colonoscopy: Results from the miccai 2015 endoscopic vision challenge. IEEE Trans. Med. Imag. 36(6), 1231–1249 (2017)
Shin, Y., Qadir, H.A., Aabakken, L., Bergsland, J., Balasingham, I.: Automatic colon polyp detection using region based deep CNN and post learning approaches. IEEE Access 6, 40950–40962 (2019)
Wang, P., Xiao, X., Glissen Brown, J.R., Berzin, T.M., Tu, M., Xiong, F., Hu, X., Liu, P., Song, Y., Zhang, D.A.: Development and validation of a deep-learning algorithm for the detection of polyps during colonoscopy. Nat. Biomed. Eng. 2(10), 741–748 (2018)
Byrne, M.F., Chapados, N., Soudan, F., Oertel, C., Linares Pérez, M., Kelly, R., Iqbal, N., Chandelier, F., Rex, D.K.: Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut 68(1), 94–100 (2019)
Pogorelov, K., Ostroukhova, O., Jeppsson, M., Espeland, H., Griwodz, C., Lange, T.D., Johansen, D., Riegler, M., Halvorsen, P.: Deep learning and hand-crafted feature based approaches for polyp detection in medical videos, In: 2018 IEEE 31st International Symposium on Computer-Based Medical Systems (CBMS) pp. 381–386 (2018)
Jha, D., Ali, S., Johansen, H.D., Johansen, D.D., Halvorsen, P.: Real-time polyp detection, localisation and segmentation in colonoscopy using deep learning. IEEE Access 9, 40496–40510 (2020)
Yuichi, M., Shin-Ei, K., Masashi, M., Yutaka, S., Hiroaki, I., Kinichi, H., Kazuo, O., Fumihiko, U., Shinichi, K., Yushi, O.: Real-time use of artificial intelligence in identification of diminutive polyps during colonoscopy: a prospective study. Ann. Intern. Med. 169, 357–366 (2018)
Lianlian., W., Dexin, G., Shan, H., Xia, L., Honggang, Y.: A detection model of colorectal polyps based on yolo and resnet deep convolutional neural networks(with video). Zhonghua Xiaohua Nei Jing Zazhi (2022)
Yang, X., Song, E., Ma, G., Zhu, Y., Yu, D., Ding, B., Wang, X.: YOLO-OB: An improved anchor-free real-time multiscale colon polyp detector in colonoscopy (2023)
Cho, K., Merrienboer, B., Gülçehre, Bahdanau, D., Bougares, F., Schwenk, H., Bengio, Y.: Learning phrase representations using rnn encoder-decoder for statistical machine translation. In: EMNLP (2014)
Jie, Shen: Samuel, Albanie, Gang, Sun. Squeeze-and-excitation networks. IEEE transactions on pattern analysis and machine intelligence, Enhua (2019)
Li, X., Wang, W., Hu, X., Yang, J.: Selective kernel networks. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (2020)
Wang, Q., Wu, B., Zhu, P., Li, P., Zuo, W., Hu, Q.: Eca-net: Efficient channel attention for deep convolutional neural networks. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 11531–11539 (2020). https://doi.org/10.1109/CVPR42600.2020.01155
Woo, S., Park, J., Lee, J.-Y., Kweon, I.S.: Cbam: Convolutional block attention module. In: Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y. (eds.) Computer Vision - ECCV 2018, pp. 3–19. Springer, Cham (2018)
Lin, T.-Y., Dollár, P., Girshick, R.B., He, K., Hariharan, B., Belongie, S.J.: Feature pyramid networks for object detection. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 936–944 (2017)
Liu, S., Qi, L., Qin, H., Shi, J., Jia, J.: Path aggregation network for instance segmentation. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, 8759–8768 (2018)
Zhao, Q., Sheng, T., Wang, Y., Tang, Z., Chen, Y., Cai, L., Ling, H.: M2det: A single-shot object detector based on multi-level feature pyramid network. ArXiv: abs/1811.04533 (2019)
Wei, L., Dragomir, A., Dumitru, E., Christian, S., Scott, R., Cheng-Yang, F., Berg, A.C.: Ssd: Single shot multibox detector. Springer, Cham (2016)
Cubuk, E.D., Zoph, B., Mane, D., Vasudevan, V., Le, Q.V.: Autoaugment: Learning augmentation policies from data (2018)
Li, Z., Chao, P., Gang, Y., Zhang, X., Jian, S.: Detnet: A backbone network for object detection (2018)
Bodla, N., Singh, B., Chellappa, R., Davis, L.S.: Soft-nms - improving object detection with one line of code. In: 2017 IEEE International Conference on Computer Vision (ICCV) (2017)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. IEEE (2016)
Liu, J., Li, C., Liang, F., Lin, C., Sun, M., Yan, J., Ouyang, W., Xu, D.: Inception convolution with efficient dilation search. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition (2020)
Redmon, J., Farhadi, A.: Yolo9000: Better, faster, stronger (2016)
Jampani, V., Sun, D., Liu, M.Y., Kautz, J.: Superpixel sampling networks. Springer (2020)
He, Z., Lin, M., Xu, Z., Yao, Z., Chen, H., Alhudhaif, A., Alenezi, F.: Deconv-transformer (dect): a histopathological image classification model for breast cancer based on color deconvolution and transformer architecture. Inf. Sci. 608, 1093–1112 (2022). https://doi.org/10.1016/j.ins.2022.06.091
Chen, Y., Lin, M., He, Z., Polat, K., Alhudhaif, A., Alenezi, F.: Consistency- and dependence-guided knowledge distillation for object detection in remote sensing images. Expert Syst. Appl. 229, 120519 (2023). https://doi.org/10.1016/j.eswa.2023.120519
Neubeck, A., Gool, L.: Efficient non-maximum suppression. In: International Conference on Pattern Recognition (2006)
Wang, F., Cheng, J., Liu, W., Liu, H.: Additive margin softmax for face verification. IEEE Signal Process. Lett. 25(7), 926–930 (2018)
Zhang, Y.F., Ren, W., Zhang, Z., Jia, Z., Wang, L., Tan, T.: Focal and efficient IOU loss for accurate bounding box regression (2021)
Liu, J., Jin, H., Xu, G., Lin, M., Wu, T., Nour, M., Alenezi, F., Alhudhaif, A., Polat, K.: Aliasing black box adversarial attack with joint self-attention distribution and confidence probability. Expert Syst. Appl. (2023). https://doi.org/10.1016/j.eswa.2022.119110
Müller, S., Hutter, F.: Trivialaugment: Tuning-free yet state-of-the-art data augmentation. (2021)
Ren, S., He, K., Girshick, R., Sun, J.: Faster r-CNN: Towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 39(6), 1137–1149 (2017). https://doi.org/10.1109/TPAMI.2016.2577031
Redmon, J., Farhadi, A.: Yolov3: an incremental improvement. arXiv e-prints (2018)
Bochkovskiy, A., Wang, C.Y., Liao, H.: Yolov4: optimal speed and accuracy of object detection (2020)
Lin, T.Y., Goyal, P., Girshick, R., He, K., Dollar, P.: Focal loss for dense object detection. IEEE (2) (2020)
Zhang, S., Wen, L., Bian, X., Lei, Z., Li, S.Z.: Single-shot refinement neural network for object detection. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (2018)
Wang, C.-Y., Bochkovskiy, A., Liao, H.-Y.M.: Yolov7: Trainable bag-of-freebies sets new state-of-the-art for real-time object detectors. In: 2023 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 7464–7475 (2023). https://doi.org/10.1109/CVPR52729.2023.00721
Reis, D., Kupec, J., Hong, J., Daoudi, A.: Real-Time Flying Object Detection with YOLOv8 (2023)
Acknowledgements
Not applicable
Funding
Not applicable
Author information
Authors and Affiliations
Contributions
Jiading Xu (Author 1) was responsible for algorithm model development. Shuheng Tao (Author 2) and Chiye Ma (Author 3) were responsible for data annotation and processing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xu, J., Tao, S. & Ma, C. Detection of Serrated Adenoma in NBI Based on Multi-Scale Sub-Pixel Convolution. Int J Comput Intell Syst 17, 95 (2024). https://doi.org/10.1007/s44196-024-00441-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s44196-024-00441-8