Abstract
In India, over 25,000 people have died from cardiovascular annually over the past 4 years , and over 28,000 in the previous 3 years. Most of the deaths nowadays are mainly due to cardiovascular diseases (CVD). Arrhythmia is the leading cause of cardiovascular mortality. Arrhythmia is a condition in which the heartbeat is abnormally fast or slow. The current detection method for diseases is analyzing by the electrocardiogram (ECG), a medical monitoring technique that records heart activity. Since actuations in ECG signals are so slight that they cannot be seen by the human eye, the identification of cardiac arrhythmias is one of the most difficult undertakings. Unfortunately, it takes a lot of medical time and money to find professionals to examine a large amount of ECG data . As a result, machine learning-based methods have become increasingly prevalent for recognizing ECG features. In this work, we classify five different heartbeats using the MIT-BIH arrhythmia database . Wavelet self-adaptive thresholding methods are used to first denoise the ECG signal. Then, an efficient 12-layer deep 1D Convolutional Neural Network (CNN) is introduced for better features extraction, and finally, SoftMax and machine learning classifiers are applied to classify the heartbeats. The proposed method achieved an average accuracy of 99.40%, precision of 98.78%, recall of 98.78%, and F1 score of 98.74%, which clearly show that it outperforms with the exiting model . Architecture of proposed work is simple but effective in remote cardiac diagnosis paradigm that can be implemented on e-health devices.
Similar content being viewed by others
1 Introduction
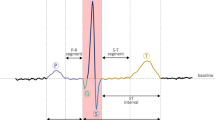
CVD is a widespread health problem that carries a significant risk to humans , especially those in their middle years and later in life. The incidence, disability, and mortality rates are all quite high. Heart disease and stroke are on the rise, and this is a huge public health issue [1]. The most common cause of cardiac death is arrhythmia. A disease in which the rhythm of the heart is disrupted is known as a cardiac arrhythmia [2, 3]. Arrhythmias cause the heart rhythm to beat, too slowly, too fast, or irregularly compared to its normal rhythm. Due to arrhythmia, the heart is not able to supply enough blood to the body parts. As a result, the blood does not show in proper proportion, affecting the functioning of the heart, brain, and other organs of the body. Issues like heart failure and arrhythmias are caused by damaged and weakened heart muscles. Many people with coronary artery disease (CAD) do not have any symptoms until the condition reaches a condition where they experience chest pain and shortness of breath shown in Fig. 1. Thus, early detection of the disease is important before it progresses to an irreversible stage. Therefore, constant monitoring of heartbeat activity is imperative. Determination of arrhythmias is important for adequate medical treatment by recognizing cardiac disorders [4, 5]. An ECG is the non-invasive tool for detecting and monitoring arrhythmias.
ECG signal wave form representation [6]
In addition to the processing of images, voice recognition, and a wide variety of other areas [7, 8], machine learning and deep learning networks have made significant progress in the adjuvant detection of cardiac disease based on ECG data [9]. These advancements can be found in the field of cardiology. When classifying long-term (10 s) ECG data, P-lawiak et al. [10] focus on a deep genetic ensemble of classifiers. To categorize eight distinct rhythms of the heart, Gao et al. [11] employed a powerful long short-term memory recurrent network model. Atal and Singh [12] proposed using an optimization-based deep CNN with the objective of discriminating between five unique heartbeats. While CNN need considerable signal pre-processing before use, deep learning networks can instantly detect the best data patterns and extract relevant characteristics. Deep learning networks also have better non-linear fitting capabilities, making it easier to recognize single-lead, multi-class, and discontinuous ECG signals. CNN is a well-studied and widely used feed-forward neural network in deep learning for arrhythmia classification. CNN has been extensively studied and used in deep learning to classify arrhythmia ECG data. Most of the earliest studies have been classified five types of heart rhythms [13].
CVD micro-classification includes five types of heartbeats: normal, left and right bundle branch block, atrial premature, and premature ventricular contraction. The classification of heartbeats is important, because the dataset contains some more dangerous arrhythmia beats. As a result, it is critical to detect these heartbeats, such as atrial and ventricular beats. The majority of studies in the literature fail to identify these two beats in the dataset. As a result, we propose a deep CNN-based model that more accurately classifies these heartbeats.
Based on physical criteria, this research proposes a classification system that divides arrhythmia into five different categories. We employed CNN to extract data-driven non-linear features rather than manually constructed features as most prior ECG signal classification research has done. The 1D-CNN model architecture consists of four convolutional, mean pooling, and dense layers that extract different non-linear features from ECG signals and group them into five categories: N, L, R, A, and V, respectively. The proposed method was trained and tested on the MIT-BIH open-source database.
The overall proposed model consists of three steps: first, pre-processing the data using the wavelet transform method, Z-score normalization, and data segmentation, in which the data set is divided into 360 samples and centered on the R-peak. Second, imbalance processing for equilibrium samples from five classes, and finally, the extraction and classification of ECG signals using a 12-layer deep 1D CNN model is used.
1.1 Major Contribution
-
Performed pre-processing on the ECG signal to remove noise and balance the arrhythmic heartbeat classes using wavelet and imbalance processing on the data set.
-
Develop a deep learning-based hybrid model that classifies ECG heartbeats into five micro-classes automatically.
-
Improve the accuracy of proposed work by optimizing the model parameters.
-
It achieves the accuracy of 99.4% which clearly shows that it outperforms with the existing method.
Section 2 describes the review of exiting work performed by different researchers using different method in this field and dataset. Section 3 describes the ECG dataset detail with their beats used in this study, as well as a detailed explanation of pre-processing steps, including denoising, normalization, segmentation, and imbalance processing. Section 4 describes the result and analysis and compares the performance of proposed model with existing work. Finally, in Sect. 5, the work is summarized, led by the conclusion.
2 Related Work
Many experts have been working on automatic detection of cardiac arrhythmia for decades. Most of them used the MIT-BIH arrhythmia database, a typical publicly available arrhythmia database [14]. Table 1 summarizes the different model developed by different researcher on ECG signal classification. The comparison is instructive despite the fact that various researchers made use of different datasets; this is because the categorization is based on the same MIT-BIH and classifies the same heartbeats. Despite this, the comparison is still useful (N, L, R, A, V).
To diagnose arrhythmias in the MIT-BIH database, [13, 15] employed a deep genetic hybrid classifiers on long-term ECG data and found an accuracy of 94.6%. ECG signals are not linear and are not constant in the real world. Higher order statistical (HOS) methods, such as the non-linear dynamic method used in [16], can detect these subtle changes. Principal component analysis is used to reduce the dimensionality of the derived bispectrum features. These axes were used as inputs for a least squares-support vector machine and a four-layer feed-forward neural network that performed automatic pattern recognition. Overall, they had the highest average accuracy (93.48%) of any group in the research.
As the depth of the network increases, the accuracy of deep neural network training continues to decline. However, [17] used a basic CNN model with five layers and found 97.5% accuracy. The technology employs a wavelet transform process based on quadratic waves, to identify individual ECG waveforms and generate a fiduciary marker array. A probabilistic neural network (PNN) is used to classify the data, and it has an accuracy rate of 92.7% [18]. The discrete wavelet transform (DWT) can be used to denoise the signal.
Yu and Chen [19] used DWT to divide the ECG signal into different sub-band components. The ECG signals are then identified using three sets of statistical characteristics extracted from the de-convoluted signals. The feature vectors are classified using PNN. The developed model has a 99.20% accuracy.
3 Materials and Methods
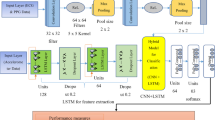
The proposed ECG signals’ classification and arrhythmia detection model is developed in the phase as: pre-processing, feature extraction, and classification using a hybrid classifier. Figure 2 shows the workflow diagram for the proposed work.
3.1 Dataset Description
This study uses the MIT-BIH database which was generously donated by MIT. It has been commented upon by various experts and is based on worldwide standards [14]. Researchers rely heavily on the MIT-BIH database to classify arrhythmic heartbeats. The MIT-BIH database contains 48 30-min ECG recordings sampled at 360 Hz. An electrocardiogram always has two leads. Each beat in MIT-BIH is annotated with its class. Table 2 and Fig. 3 display the distribution of MIT-BIH database beats given by cardiologist experts. A variety of abnormal heart rhythms were used, including left and right bundle branch block, atrial premature contraction, and ventricular premature contraction are the types of heart blocks that may occur. In this study, we used each data sample block size of 360, because the sampling rate of ECG signals in MIT-BIH arrhythmia database is 360 Hz. Therefore, in this database, each peak and the entire QRS complex is present in 1 segment. If we change the shape of the segments, the data sample may be missing some peaks (P waves) and QRS complexes that play a major role in identifying arrhythmia classes.
3.2 ECG Signal Pre-processing
Readings obtained from an electrocardiogram (ECG) in a clinical setting may be impacted by a number of factors, the interference of power frequencies, baseline drift, and electromyography (EMG) interference, to name a few. It is necessary to denoise the raw data to improve classification accuracy. Denoising ECGs often involves the use of bandpass filters, low-pass filters, and wavelet transformations [22, 23]. In this inquiry, the electrocardiogram (ECG) data are first prepared with the use of a method called the wavelet transform.
3.2.1 Wavelet Transform
Decomposing non-stationary signals into scale signals that have distinct frequency bands may be accomplished with the use of the wavelet approach. The filter makes use of an adaptive threshold filtering approach [25, 26], and the wavelet function that it employs is the Symlet wavelet function, which is a member of the Symlet wavelet function family [27]. In signal processing, noise signals appear as high-frequency signals, whereas valuable signals appear as low-frequency or smoother signals. The wavelet transform is used to decompose the signals, which results in the high-frequency wavelet coefficients being acquired. After that, the high-frequency wavelet coefficients go through a procedure called threshold processing, which gets rid of the electromyographic noise and the power line interference. After that, the inverse wavelet transform is utilized, so that the signals can be reconstructed. The baseline drift noise may be removed by moving the average filter to a different position. In this work, adds fundamental filtering to the signal, which increases the network’s generalization and reduces signal distortion. Figure 4a and Fig. 4b shows the ECG signal prior to and subsequent to filtering.
3.2.2 ECG Signal Normalization
The Z-score normalization approach was used to standardize all ECG signal values. The purpose of normalization is to equalize the scale of all data points, such that each characteristic is equally important [19]. The following equation is used to calculate the Z-score normalized signal values from the dataset (1). Figure 5 shows the shape of the signal after Z-score normalization
where χ is original value, µ is mean of data, and σ is standard deviation of data
3.2.3 Data Segmentation
In the MIT-BIH dataset, each pulse has a condition that is associated with it. We can determine that there are five distinct types of heart rhythms based on the findings of this analysis. These are normal (N), left bundle branch block (L), right bundle branch block (R), atrial premature beats (A), and premature ventricular beats (PV). To begin, the Pan–Tompkins method [28] is implemented to locate the R-peak. Following the identification of R-peaks in the data, the information is next segmented into 360 random samples shown in Fig. 6.
3.2.4 Imbalance Processing
The distribution of heartbeats in the various classifications of the arrhythmia database is not uniform. The samples in the dataset were not distributed uniformly, which meant that different categories contained varying numbers of samples. Regular beats, for example, contained more data sets than fusion beats. This is known as the problem of class inequality. The unequal training set affects the convolutional neural network’s feature learning [29], thereby decreasing recognition accuracy.
We utilized the dataframe.resample() function of the Pandas library to balance the dataset. Typically, dataframe.resample() is applied to time series data. A time series is a collection of data points that have been chronologically indexed (or listed or graphed). After denoising and segmentation, under-represented classes (those with fewer data) are over-sampled. The groups that are under-represented are over-sampled at random, while the groups that are already well represented are left out (those with more samples). This minimizes the problem of skewed information in the training set. Figures 7a, b shows the original classification chart and after balancing the data class classification.
3.3 Convolutional Neural Network Model
CNNs typically have numerous convolutional and pooling layers to extract data features [30]. Unlike CNNs, convolutional neural networks link neurons locally. Only neurons in close proximity will be interconnected. CNN has the additional advantage of sharing the user’s feature plane. When neurons exchange weights, the number of calculations and connections between layers of the network are reduced. The feature dimension reduction of the pooling layer, which is utilized throughout the CNN, can effectively reduce redundant information, minimize overfitting, and facilitate optimization.
3.3.1 The Architecture
1D, 12-layer CNN structure is used to describe the five distinct subtypes of cardiac arrhythmia. This structure is utilized to classify the data. The layers that comprise a convolutional neural network are referred to as input, convolution, pooling, fully connected, and output, respectively. The convolutional and pooling layers go beyond the capabilities of typical neural networks by extracting and mapping the characteristics of incoming input. This helps to speed up the learning process and reduce the likelihood of overfitting. 2-D CNN usage in this field of research widely [17, 31] because to its similarities to multilayer perceptrons. To perform uniform interval sampling of one-dimensional time series, [38] our proposal is for a convolutional neural network with 12 layers and a single dimension. This particular network contains one convolutional layer for each and every layer that it possesses. The CNN network makes use of a max-pooling layer; however, the improvement that is being proposed for the CNN network makes use of an average-pooling layer rather than the max-pooling layer that is currently being used in the current CNN network. In addition to those structural improvements, there have been others made. This is just one of several adjustments that have been made recently. The average-pooling layer has the capability of preserving the authenticity of the broad strokes of the input data, which is an essential component for the classification of heartbeats. The industry standard CNN network is one layer thinner than the proposed CNN network, which has one more layer of alternating convolution and pooling than the standard CNN network . This is because the proposed CNN network uses a different algorithm. This is due to the fact that the proposed CNN network would be built on top of the existing CNN network. Table 3 provides a summary of the suggested architecture for the CNN network, which includes a total of eight layers of convolutions and average-pooling that are stacked one on top of the other.
Convolution Layer: When processing one-dimensional ECG data, it is best to use convolution kernels that are just one dimension and do not depend on the feature map of the layer above them to convolve. We are able to retrieve the outflow of the convolution layer using a non-linear activation function in conjunction with an offset convolution kernel [32]. Equation (2) provides the output as
where h is the output vector of the convolutional layer, W is the kernel, and x is the input vector of the ECG signals.
Pooling Layer: In most of the cases, the layer that performs the convolution is the pooling layer. The complexity of the network and the overfitting phenomena can both be mitigated by lowering the number of dimensions used for the convolution layer’s output data. Because of this procedure, the network’s resistance to failure has been significantly improved. The pooling layer either takes an average or a maximum of the features that were produced by the convolutional layer; the techniques that correspond to these two outcomes are called average pooling and maximum pooling, respectively. Equation (3) provides the output as
where o represents the output of the average-pooling vector, x is the input vector obtained from the convolutional layers, and b is the biased vector.
Fully connected layer: After a significant number of convolution layers and pooling layers have been utilized to gather features, after that, the fully connected layer is applied, so that the connections between each and every feature may be strengthened even more. Logistic regression is utilized at the SoftMax layer to continue the classification process [7]. The output of the layer that is fully linked is the weighted sum of the outputs of the layers that are below it. Equation (4) represent the mathematical equation of fully connected layer as
Dropout layer: During the training of CNN layer, there are some chances of over fitting of model, so avoid the overfitting in proposed model we have used the dropout layer in which we remove the same layer to generalize the node value [7]
3.3.2 Training Algorithm
To train the CNN model, a technique backward propagation gradient descent is used. It is used to calculate the network hyper parameters and the loss function, which is the difference between the actual output, that was seen and the predicted output, that should have been observed. There are many different kinds of hyper parameters, some of which are the sample weight coefficient of the pooling layer, the network weight of the fully connected layer, and the offset for each layer. Both forward and backward propagation are required for training a CNN model. During the forward propagation stage of the neural network, the training input is gathered and then used to calculate the output vectors of the hidden and output layers.
For fully connected forward CNN network of \(l-1\) layer with feature \(\mathrm{m}\) is derived as
Then, the computation of feature matrix is
Here, \({X}^{(I-1)}\) is a layer a feature vector of length \(m,{X}^{(I)}\) is a column vector of length \(\mathrm{n},{W}^{(I)}\) is a matrix of \(\mathrm{m}\) rows and \(\mathrm{n}\) columns, \({B}^{(I)}\) is a column vector of length \(\mathrm{n}\), and \({f}^{(l)}\) is a non-linear activation function of layer \(l\).
There are many convolutional layers present in convolution kernels. By applying the appropriate convolution kernels to the features from the preceding layer, we may generate additional features. After the convolution layer (with learnable convolution kernels \({K}_{\mathrm{1,1}}^{(l)},\dots ,{K}_{i,1}^{(l)},\dots ,{K}_{1,j}^{(l)},\dots ,{K}_{i,j}^{(l)}\) and learnable bias \({B}_{1}^{(l)},\dots ,{B}_{j}^{(l)}\) is executed. Each feature map \(\left({X}_{1}^{(l)},\dots ,{X}_{j}^{(l)}\right)\) is based on the following calculation formula:
Here, \({M}_{j}\) is the collection of feature maps used in the training process, \(\otimes\) is the convolution operation, and \({f}^{(l)}\) is the activation function.
It is common practice for a CNN model to use a pooling layer after the convolutional layer to further compress the features. The output is created through numerous fully connected layers after several convolutional layers and pooling layers have been applied to the input features to turn them into a vector.
Fast convergence is achieved by activating the hidden layer using the linear rectification function (Rectified Linear Unit, ReLU) in this research. The result of the function for the column vector input \({\left({x}_{1},\dots ,{x}_{n}\right]}^{T}\) is
SoftMax activation function is used by the output layer to normalize the output for probability distribution. For an input column vector \({\left({x}_{1},\dots ,{x}_{n}\right]}^{T}\), the function output is
The loss function calculates the absolute value of the discrepancy between the predicted output hat and the actual output math bfy. In this particular piece of work, we make use of the cross-entropy loss function, which, when applied to an n-class classification issue
\(\widehat{{y}^{\mathrm{^{\prime}}}}=\left({\widetilde{y}}_{1},\dots ,{\tilde{y}}_{n}\right]\) represents the predicted output and one-hot encoding of the true class is represented by \(y=\left({y}_{1}^{\mathrm{^{\prime}}},\dots ,{y}_{n}\right]\)
During the forward propagation phase, the actual values of the output layer's output vectors are compared to the expected values. Furthermore, the weights of the network are factored into the loss function calculation. Gradient descent updates the weights of each neuron in each layer, and the loss is transmitted back to the layers from which it originated. This is done to reduce the amount of time required to complete the process. After the gradients of the loss function have been calculated using chain rules, the weight is updated in the opposite direction as the gradient of the loss function. The creation of a cost function for neuron output at each hidden layer allows for continuous tweaking of the network's hyperparameters. Each hidden layer receives this function. When the network achieves the desired error rate, the training is considered complete.
In the proposed model, the following architecture is used, as shown in Fig. 8. The first layer is rather complicated, since it uses a kernel with a size of 13, as well as a total of 16 filters. The output of layer 2 is reduced from 179 to 16 X 179 using an average-pooling layer with a size of 3. The following are the configuration options for Layer 3, which is a convolving of the Feature Map from Layer 2: kernel size 15, filter count 32. We are able to reduce the total number of neurons from 176*16 to 89*32 using a third layer that aggregates averages. At the layer 5 computation level, 64 filters are employed, and the convolution kernel size is 17. By applying a layer of average-pooling with a size of 3 to the output of layer 5, we are able to decrease the output size to 44×64, which is much less than the original. The output of layer 6 is made even more complicated by the 7 layer, in which kernel size is 19 and a with 128 filter number. We will now proceed with the construction of a layer of average pooling that has three tiers in layer 8. Because of this, Layer 9 has a dropout rate of 50%. The 35 neurons that make up the boundary between layer 10 and layer 11 are to blame for this shift in behavior. The last phase consisted of establishing a connection between 5 neurons located in layer 11 and the SoftMax layer. When an average-pooling layer is created, an activation function in the form of a rectifier linear unit (ReLU) is utilized initially. This is done at every opportunity.
3.4 Performance Metrics
The success of the CNN model is measured in terms of % accuracy and loss value, both of which are useful in evaluating performance in multi-class classification problems.
3.4.1 Accuracy
The overall accuracy of the model is displayed, which is the proportion of samples that were correctly labeled by the classifier [24]. Equation (12) for determining accuracy as
Here, TP is True positive, TN is True negative, FP is False positive, and FN is False negative. The number of correct predictions divided by the total number of input data is the ratio.
3.4.2 Loss
A loss is a numerical value that reflects how far the model's prediction was off in a certain case. If the model's forecast is perfect, the loss is zero; otherwise, the loss is greater. The goal of training a model is to find a set of weights and biases that, on average, result in a minimal loss across all cases. Figure 9 shows a high loss model on the left and a low loss model on the right.
-
The arrows represent loss.
-
The blue lines represent predictions.
Left model-high loss; right model-low loss [24]
3.4.3 Precision
It informs you what percentage of optimistic forecasts were truly positive
3.4.4 Recall
It tells you what proportion of all positive samples the classifier predicted correctly. Other synonyms for it include True-Positive Rate (TPR), Sensitivity, and Probability of Detection. Use Eq. (14) to compute Recall
3.4.5 F -Score
It is a measure that combines precision and recall. It is the harmonic mean of precision and recall in mathematics. It is computed by Eq. (15) as
4 Results and Discussion
The MIT-BIH database was used to evaluate the performance of the provided technique. With a batch size of 36, the training of the model comprises a total of 60 epochs. The model accuracy curves and loss curves with respect to the number of epochs are shown in Figs. 10, 11. Where, in Fig. 10, the x-axis represents the number of epochs used by the softmax layer, and the y-axis represents the accuracy of the model obtained during training and testing on data samples. Similarly, in Fig. 11, the y-axis represents the training and test loss of the model using the Adam optimizer in the softmax layer. As the accuracy and loss become saturated, these graphs suggest that the model has been correctly trained after the ideal value of network weights has been obtained. Furthermore, the test loss looks to be virtually identical to the training loss, indicating that the model has been fine-tuned to minimize loss.
Table 4 compares the performance of dataset in which we have used 50, 40, 30, 20, and 10%, data in test dataset. This table shows the total accuracy and loss for each train–test ratio that was calculated. The CNN model’s overall best testing accuracy for 5 class classification was 99.40 percent, which was found by utilizing an 80:20 training test ratio.
The total classification accuracy rate using 12-layer deep 1D-CNN employing an 80:20 train–test ratio is 99.40%. Each class’s individual accuracy is also computed and displayed in Table 5.
For the class of right bundle block (R) beats, the model gets the highest level of accuracy with 99.86% accuracy, and normal (N) beats have the lowest accuracy with 99.19%. In Fig. 12, the confusion matrix associated with the presented CNN model is likewise obtained and illustrated. The confusion matrix provides a succinct summary of the categorization of the various individual classes. The CNN is made up of a feature extractor and a classifier, with the convolution and pooling layers acting as feature extractors and the SoftMax layer acting as a classifier. In this figure, we have obtained the best-performing confusion matrix on the dataset when the data are split in an 80–20 train–test ratio using a softmax layer with a CNN model.
We modified the SoftMax layer to support vector machine classifiers, random forest classifiers, and k-nearest neighbor classifiers to compare the performance of each technique to SoftMax. Table 6 shows comparison between SoftMax, SVM, random forest and k-nearest neighbor classifiers based on overall accuracy, typical recall, typical precision, and typical F1 score.
4.1 Comparison with Existing Approaches
Table 7 shows a comparison of the present work with several other existing approaches. When compared to other proposed approaches and published experimental findings, we can see that the proposed strategy enhances heartbeat classification accuracy. This not only is connected to the notion of applying the wavelet transform to cope with noisy ECG signals, but this also demonstrates that the enhanced CNN model also is adequate for 1D signal classification. In these comparison papers, components are compared in depth.
5 Conclusion
In today’s environment, CVD is a big health issue. ECG is extremely important in the early detection of cardiac arrhythmia. Unfortunately, specialist medical resources are scarce, making virtual identification of ECG signal difficult and time-consuming. This study we develop a deep learning CNN model that classifies ECG heartbeats into five micro-classes automatically. Because this model is fully automated, no additional systems, such as feature extraction, feature selection, or classification, are required. When the model has been properly trained, it may be used to the process of predicting ECG signals in patients who have arrhythmia. Experiments performed on the arrhythmia database (also known as the MIT-BIH database) show that our approach is more effective and efficient in classifying ECG signals. Due of its reduced computing cost, the CNN model that was proposed may be used for categorizing long-term ECG data and identifying sickness episodes in real time. In addition, one can apply these DL models to the analysis of other existing signal databases such as electroencephalogram signals and electromyogram signals that are also important in the healthcare system to diagnose potential disease in humans being. According to the results, our proposed method achieved an average accuracy of 99.40%, precision of 98.78%, recall of 98.78%, and F1 score of 98.74%. It is a simple but effective paradigm for remote cardiac diagnosis of patients that can be implemented on e-health-based devices. These applications are possible because of the model’s flexibility. It is of great use to wearable technology.
Data Availability
Not applicable.
Abbreviations
- ECG:
-
Electrocardiogram
- CVD:
-
Cardiovascular diseases
- CNN:
-
Convolutional neural network
- PCA:
-
Principal component analysis
- SVM:
-
Support vector machine
- KNN:
-
K-nearest neighbor
- DWT:
-
Discrete wavelet transform
- NN:
-
Neural network
- N:
-
Normal
- L:
-
Left bundle branch block
- R:
-
Right bundle branch block
- V:
-
Premature ventricular contraction
- A:
-
Atrial premature contraction
References
Acharya, U.R., Suri, J.S., Spaan, J., Krishnan, S.M.: Advances in cardiac signal processing. Springer, Berlin, Heidelberg (2007)
Hadhoud, M., Ma, M.I., Eladawy, A., (2006) Farag: Computer aided diagnosis of cardiac arrhythmias. The 2006 International Conference on
Singh, S.: Classification of ECG arrhythmia using recurrent neural networks. Procedia. Comp. Sci. 132, 1290–1297 (2018)
Chazal, D., Philip, M.O., Dwyer, R.B.: Reilly: automatic classification of heartbeats using ECG morphology and heartbeat interval features. IEEE Trans. Biomed. Eng. 51, 1196–1206 (2004)
Alonso-Atienza, F.: Detection of life threatening arrhythmias using feature selection and support vector machines. IEEE Trans. Biomed. Eng 61, 832–840 (2014)
Khadra, L., Al-Fahoum, A.S., Al-Nashash, H.: Detection of life- threatening cardiac arrhythmias using the wavelet transformation. Med. Biol. Eng. Comp. 35, 626–632 (1997)
Wu, M., Lu, Y., Wong, Y.W., Y, S.: A study on arrhythmia via ECG signal classification using the convolutional neural network. Front. Comput. Neurosci. 14, 564015–564015 (2021)
Kandala, R.N., Dhuli, R., P-lawiak, P., Naik, G.R., Moeinzadeh, H., Gargiulo, G.D.: Towards real-time heartbeat classification: evaluation of nonlinear morphological features and voting method. Sensors 19, 5079–5079 (2019)
Zubair, M., Kim, J., Yoon, C.: ,(2016) An automated ECG beat classification system using convolutional neural networks. In: 2016 6th International Conference on IT Convergence and Security (ICITCS), 1–5
P-lawiak, P., Abdar, M., Plawiak, J., Makarenkov, V., Acharya, U.R.: DGHNL: A new deep genetic hierarchical network of learners for predic- tion of credit scoring. Inform. Sci. 516, 401–418 (2020)
Gao, J., Zhang, H., Lu, P., Wang, Z.: An effective LSTM recurrent network to detect arrhythmia on imbalanced ECG dataset. J. Healthcare. Engin. 11, 6320651–6320651 (2019)
Atal, D.K., Singh, M.: Arrhythmia classification with ECG signals based on the optimization-enabled deep convolutional neural network. Comp. Methods Prog. Biomed 196, 105607–105607 (2020)
Yildirim, O., P-lawiak, P., Tan, R.S., Acharya, U.R.: Arrhythmia detec- tion using deep convolutional neural network with long duration ECG signals. Comp. Biol. Med. 102, 411–420 (2018)
Moody, G.B., Mark, R.G.: The impact of the MIT-BIH arrhythmia database. IEEE Engin. Med. Biol. Mag 20, 45–50 (2001)
Acharya, U.R., Fujita, H., Oh, S.L., Hagiwara, Y., Tan, J.H.: Adam, M: Application of deep convolutional neural network for automated detection of myocardial infarction using ECG signals. Inform. Sci 415, 190–198 (2017)
Martis, R.J., Acharya, R., Mandana, A., Ray, C.K.: Chakraborty: Cardiac decision making using higher order spectra. Biomed. Signal Process. Control 8(2), 193–203 (2013)
Li, D., Zhang, J., Zhang, Q., Wei, X. (2017) Classification of ecg signals based on 1d convolution neural network. 2017 IEEE 19th International Conference on eHealth Networking, Applications and Services (Healthcom), 1–6
Guti´errez-Gnecchi, J.A., Morfin-Magan˜a, R., Lorias-Espinoza, D. (2017) DSP- based arrhythmia classification using wavelet transform and probabilistic neural network. Biomedical Signal Processing and Control 32: 44–56
Yu, S.N., Chen, Y.H.: Electrocardiogram beat classification based on wavelet transformation and probabilistic neural network. Pattern Recogn. Lett 28, 1142–1150 (2007)
Mahdhaoui, Hassen, et al. 2021 1D Convolutional Neural Network Based ECG Classification System for Cardiovascular Disease Detection. No. 6066. EasyChair
Osowski, S., Linh, T.H.: ECG beat recognition using fuzzy hybrid neural network. IEEE Trans. Biomed. Eng. 48(11), 1265–1271 (2018)
Ismaiel, F.O., Mohamed.: Classification of cardiac arrhythmias based on wavelet transform and neural networks. Sudan University of Science and Technology, Diss (2015)
Zadeh, A.E., Khazaee, A.: High efficient system for automatic classifi- cation of the electrocardiogram beats. Ann. Biomed. Eng. 39(3), 996–1011 (2011)
Lin, C.H., Du, Y.C., Chen, T.: Adaptive wavelet network for multiple cardiac arrhythmias recognition. Expert. Syst. Appl 34, 2601–2611 (2008)
Alfaouri, M., Daqrouq, K.: ECG signal denoising by wavelet transform thresholding. Am. J. Appl. Sci. 5, 276–281 (2008)
Awal, M.A., Mostafa, S.S., Ahmad, M., Rashid, M.A.: An adaptive level dependent wavelet thresholding for ECG denoising. Biocyber. Biomed. Engin. 34, 238–249 (2014)
Singh, B.N., Tiwari, A.K.: Optimal selection of wavelet basis function applied to ECG signal denoising. Dig. Sign. Proc 16, 275–287 (2006)
Pan, J., Tompkins, W.J.: A real-time QRS detection algorithm. IEEE Trans. Biomed. Engin 3, 230–236 (1985)
Masko, D., Hensman, P.: (2015). http://urn.kb.se/resolve?urn=urn:nbn: se:kth:diva-166451
Yamashita, R., Nishio, M., Do, R.K.G.: Convolutional neural networks: an overview and application in radiology. Insights Imaging 9, 611–629 (2018)
Wei, Y., Xia, W., Lin, M., Huang, J., Ni, B.: Dong, J: HCP: A flexible CNN framework for multi-label image classification. IEEE Transac. Patt. Anal. Mach. Intell 38, 1901–1907 (2015)
Namara, K.M., Alzubaidi, H., Jackson, K.J.: Cardiovascular disease as a leading cause of death: how are pharmacists getting involved? Integr. Pharm. Res. Pract 8, 1–1 (2019)
Funding
There is no funding for this research.
Author information
Authors and Affiliations
Contributions
SKP: conceptualization, AK: data curation, SB: formal analysis, AK: investigation, SKP and AK: methodology, TRG: project administration, MA and S: visualization, AK: writing and original draft, and writing, and RRJ: review and editing.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
pandey, S.K., Shukla, A., Bhatia, S. et al. Detection of Arrhythmia Heartbeats from ECG Signal Using Wavelet Transform-Based CNN Model. Int J Comput Intell Syst 16, 80 (2023). https://doi.org/10.1007/s44196-023-00256-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s44196-023-00256-z