Abstract
Machine learning helps construct predictive models in clinical data analysis, predicting stock prices, picture recognition, financial modelling, disease prediction, and diagnostics. This paper proposes machine learning ensemble algorithms to forecast diabetes. The ensemble combines k-NN, Naive Bayes (Gaussian), Random Forest (RF), Adaboost, and a recently designed Light Gradient Boosting Machine. The proposed ensembles inherit detection ability of LightGBM to boost accuracy. Under fivefold cross-validation, the proposed ensemble models perform better than other recent models. The k-NN, Adaboost, and LightGBM jointly achieve 90.76% detection accuracy. The receiver operating curve analysis shows that \(k\)-NN, RF, and LightGBM successfully solve class imbalance issue of the underlying dataset.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Diabetes is a metabolic condition primarily caused by aberrant insulin secretion. In the long run, high amounts of glucose (sugar) pose a significant threat to one’s health. Insulin shortage is a major contributing factor, as beta cells in the pancreas fail to produce enough insulin, causing the body to have difficulties managing blood sugar levels (glucose). Type 2 diabetes is common among the majority of individuals who have diabetes. One of the names given to Type 2 Diabetes is Pima Indians’ Diabetes. Heart, renal, and eye problems may result from smoking. About 5.5% of the global population has diabetes, and 90% of those with diabetes have type 2, and that number is anticipated to climb by 48% over the next few years [1, 2]. The presence of diabetes can be identified manually or automatically by medical professionals. Each of these approaches has advantages and disadvantages. Because manual diagnosis does not necessitate a machine-based detection of diabetes, the intervention of a specialized medical professional is essential. It has been seen that early symptoms of diabetes are often so subtle that even a skilled medical practitioner struggles to detect them. On the other hand, data mining is now used in many fields of science, including medicine. Because of advances in artificial intelligence, machine learning and deep learning techniques, automated detection of diabetes is getting popular among medical practitioners [3,4,5,6].
The process of determining a patient’s type of diabetes involves several different tests, making it one of the most challenging tasks faced by medical practitioners. Machine learning techniques have made a huge impact on the healthcare business in recent years. Machine learning is extensively used for diabetes mellitus detection [4]. Diabetes can be diagnosed manually or automatically by a physician. Because manual diagnosis does not rely on a computer-assisted approach, the physician must rely on his or her training, speciality and experience. The symptoms of diabetes at its initial stage can be so subtle that even a skilled physician finds it difficult to identify and diagnose correctly. Although a machine learning-based detection of diabetes cannot take the place of manual intervention and diagnosis by medical practitioners, at least it can be an aid to detect the disease before the actual diagnosis starts. As a result, early diagnosis can be undertaken before the disease worsen. Many artificial intelligence-based diabetes detections and prediction systems have been proposed. They have their advantages and disadvantages. Yuvaraj et al. [7], for example, developed a diabetes prediction application that utilized Random Forest, Decision Tree, and Naive Bayes. It was employed after pre-processing the Pima Indian Diabetes dataset (PID). The authors selected and extracted features using an information gain approach. The random forest technique outperformed the other classifiers with a 94% accuracy rate. Similarly, an automated system for diabetes detection has also been proposed by Negi et al. [8] with the help of Support Vector Machine (SVM). The purpose of this detection approach is to present a universal approach as they have tested their system on multiple diabetes datasets. One of the datasets they tested upon contains 49 clinical information of numerous individuals 102,538, 64,419 of which are positive and 38,115 of which are negative. However, practically it is interesting to see the performance of SVM on such a huge subjects base. Maniruzzaman et al. [9] classified and predicted diabetes using a machine learning paradigm. They classified diabetes using a decision tree, naive Bayes, AdaBoost, and random forest. They examined data from the National Health and Nutrition Examination Survey of diabetes and nondiabetic persons in the United States and obtained encouraging findings using the proposed technique. Using SVM and Naive Bayes, Tafa et al. [10] proposed an integrated and superior model for diabetes prediction. They employed a dataset with eight characteristics and 402 patients, 80 of whom had type 2 diabetes — combining the offered techniques improved prediction accuracy to 97.6%.
The proposed work in this article aims to analyze and build an ensemble model on the Pima Indians diabetes dataset to estimate the risk of developing diabetes for each unique observation based on the independent characteristics. The objective of this paper is to utilize different machine learning classification algorithms and to adopt an ensemble learning approach thereupon. To reduce overfitting in models and to improve robustness over a single model, the ensemble technique is implemented. Ensemble techniques combine multiple individual models and deliver optimized prediction results. For creating an ensemble, the classifiers used here are Naïve Bayes (Gaussian) [11], k-Nearest Neighbor (k-NN) [12], Random Forest (RF) [13], Adaptive Boosting (Adaboost) [14] and a lighter version of Gradient Boosting Machine [15] with hyperparameter optimization to predict diabetes mellitus at an early stage of life. These algorithms are effective in predicting the category of data point when labelled data are available, and it helps in segregating vast quantities of data into distinct values, like 0/1, True/False, or a pre-defined output label class.
This article is divided into several major sections. Section 2 conducts a literature review, highlighting numerous significant existing publications on diabetes prediction systems utilizing various algorithms and models. Section 3 contains the data set description and pre-processing steps, while Sect. 4 has the study’s essential content, which includes the methods and methodology used, as well as the proposed ensemble of classifiers. Section 4 also discusses the work’s findings and inferences. Finally, the study concludes with a discussion of the paper’s primary findings in Sect. 5.
2 Related Works
With the appropriate instance of training and testing, machine learning has provided the most support for disease prediction. Because diabetes cannot be cured, it has a negative impact on our health system. Therefore, machine learning algorithms can be used to detect diabetes in its earliest stages. Alharbi et al. [16] developed an autonomous diabetes prediction model built on top of a Genetic Algorithm (GA)-based feature selection. Diabetes is detected with 97.5% using extreme learning machine. Recently, a modified version of the Weighted Objective Distance-based (WOD) diabetes prediction engine [17] was proposed that takes into account the individuals’ personal health status [18]. The modified WOD makes use of information gained to distinguish positive from negative participants. The WOD-based approach, on the other hand, is confined to binary classification on a tiny dataset. The proposed WOD technique achieves a detectability rate of 93.22%. A Random Forest graph generation-based graph convolutional network (RFG-GCN) [19] has been proposed for diabetes mellitus detection. The Random Forest generates a graph from the structured data using sample correlation, where a 2-layer convolutional network classifies the samples to separate disease from non-disease instances. Although RFG-GC has been used for various clinical diseases, it shows satisfactory results in diabetes detection. Type 2 Diabetes mellitus has been detected in recent past using Chi-Squared test and binary logistic regression [20]. During the preprocessing stage, the Synthetic Minority Over-sampling Technique (SMOTE) helps to balance the underlying data. A cooperative co-evolution framework was also suggested as Clinical Decision Support System that handles Feature Selection (FS) and Instance Selection (IS) as separate subproblems [21]. The wrapper approach is used for both feature and instance selection in this study. The reduced dataset was utilized for training a random forest classifier, which aided clinical decision-making more effectively. Mishra et al. [22] described a novel hybrid attribute optimization technique for removing extraneous data and generating a trustworthy dataset of diabetic features that can be used for more accurate prediction of diabetes. Additionally, the hybrid attribute optimization technique along with neural network successfully estimates the presence of type 2 diabetes in individuals. Additionally, their suggested approach is evaluated on 7 distinct disease datasets to determine the capability of the concerned detector. The proposed model’s performance measures are evaluated against tenfold cross-validation. Their proposed model beat all other comparable research in terms of classification accuracy. The suggested model’s mean precision and recall were 91% and 89.8%, respectively. Thus, the technique may be beneficial for healthcare practitioners in identifying the existence of Diabetes in patients with high accuracy.
There has been a lot of work done to improve diabetes diagnosis and treatment, but the classification of diabetes is still a problem. To increase the performance and accuracy of the model, researchers have taken an ensemble approach by combining individual algorithms/models into one hybrid model [23]. Recently, Ismail et al. [24] explored 35 different classifiers and presented a Bagging-based Logistic Regression (Bagging-LR) approach for the prediction of Type-2 diabetes. The Bagging-LR proved to be the ideal detector, where it took only 0.016 min to detect diabetes mellitus. Bagging-LR employed only 5 prominent features to boost the classification accuracy up to 99%. Random Forest (RF) along with Logistic Regression (LR) and Naïve Bayes (NB) proved to be an effective ensemble, where the classification strategy was decided by soft voting [25]. The LR, RF, and NB ensemble proved to be a better choice compared to AdaBoost and many other bagging approaches. Similar to the soft voting approach, the max voting approach has been used in an ensemble of several classifiers [26]. The max voting-based ensemble also reveals satisfactory detection accuracy of 77.83%. Multiple classifiers ensemble was also presented, where the LR, Linear Discriminant Analysis (LDA), k-NN, Classification and Regression Tree (CART), NB and SVM were combined to form the ensemble [27]. The suggested ensemble draws a detection accuracy of 82.81%. A Quantum-inspired ensemble model has been proposed in the recent past for multi-attribute and multi-agent decision making [28]. The proposed quantum-inspired model yields 90.5% detection accuracy while discriminating between diabetic and non-diabetic instances. An evolutionary framework using the stacking-based ensemble approach has been proposed, k-NN is used as a meta learner to combine base learners [29]. The k-NN based ensemble reveals the highest accuracy of 83.8%, with a sensitivity of 96.1%. Similarly, a parameter-free greedy ensemble approach has been proposed for medical disease classification [30]. For classification, the primary function of the ensemble approach is the combination of various rough set filters’ subsets of attributes, resulting in an optimal subset of attributes. The greedy-based approach reveals 74.9% detection accuracy with NB as the base learner. Bashir et al. proposed an ensemble technique known as Hierarchical Multi-level classifiers Bagging with Multi-objective optimized voting (HM-BagMoov) for disease classification. The proposed HM-BagMoov model employs multi-objective optimization weighted voting ensemble scheme to form the ensemble using seven state-of-the-art classifiers, where the ensemble technique classifies the diabetes dataset with 78.21% detection accuracy. Diabetic Mellitus can be predicted and diagnosed with the help of the Adaboost ensemble learning framework [31]. The Adaboost ensemble model employs a decision stump as its primary classifier. The model was tested against other classifiers to ensure the capability of the system. The Adaboost classification model exhibits an accuracy of 84.19%. A new model for classifying diabetic patients based on their characteristics and medical history has been developed and tested [32]. The study used a random committee classifier as an ensemble method. The presented ensemble was tested on diabetic data using the tenfold cross-validation method, where the ensemble yielded an 81% accuracy rate. A diabetes mellitus prediction mechanism has been proposed for class imbalance datasets with the inherent ability to handle missing values [33]. Naïve Bayes plays a prominent role at preprocessing stage to handle missing values, and the popular adaptive synthetic sampling method (ADASYN) counters the class imbalance issue. Finally, the diabetic patients are detected using the random forest classifier. The combination of ADASYN and Random Forest classifies the diabetic and non-diabetic patients with 87.1% detection accuracy, which is 8.5% higher than the traditional random forest. A summary of literature reviewed pertaining to related works is presented in Table 1.
3 Materials and Methods
The materials and methods section starts with explaining the proposed methodology, followed by the dataset to be used to validate the system. Since the proposed approach is based on ensemble methods; therefore, various ensembles of classifiers are created, and the ensemble reflecting the best result has been proposed as the contribution of this article. To create the ensemble, we have explored Adaboost, LightGBM, k-NN, Random Forest and Naïve Bayes (Gaussian). These classifiers are either decision trees, functions based, or an ensemble itself. A mixture of classifiers having varied decision-making approaches is expected to provide a better result if these classifiers are used alone for the classification task. Moreover, the above-mentioned classifiers are frequently used for medical diagnostics, which is the main reason for considering in the proposed ensemble. Before discussing the proposed ensemble, it is wise to explore the shortlisted classifiers to understand their inherent classification ability. This will provide scope for ascertaining the true capability of the proposed ensemble. All the algorithms discussed in this section assumes \(TR:=\left\{\left({tr}_{1}^{1},{tr}_{1}^{2},\dots \dots , {tr}_{1}^{r}\right),\left({tr}_{2}^{1},{tr}_{2}^{2},\dots \dots , {tr}_{2}^{r}\right),\dots \dots ,\left({tr}_{m}^{1},{tr}_{m}^{2},\dots \dots , {tr}_{m}^{r}\right) \right\},\) a set of training instances with \(m\times r\) dimension and \(TS:=\left\{\left({ts}_{1}^{1},{ts}_{1}^{2},\dots \dots , {ts}_{1}^{r}\right), \left({ts}_{2}^{1},{ts}_{2}^{2},\dots \dots , {ts}_{2}^{r}\right),\dots \dots , \left({ts}_{n}^{1},{ts}_{n}^{2},\dots \dots , {ts}_{n}^{r}\right) \right\},\) a set of testing instances with \(n\times r\) dimension. Both the \({tr}^{r}\) and \({ts}^{r}\) contains the class labels, and all the algorithms return the predicted class labels for the testing instances, which are expected to be overridden at \({ts}^{r}\) attribute.
3.1 k-Nearest Neighbor (k-NN)
The k-Nearest Neighbor or, in short, k-NN is a supervised learning classification algorithm [34]. It is a simple way to classify new instances based on similarity measures. In k-NN we can have multiple ways of calculating the distance between two data points to consider which is the nearest neighbour. However, the k-NN consumes more memory as the training data have to be stored entirely on memory. Nevertheless, k-NN is frequently used in medical diagnostics, especially in diabetes detection. The working principle of k-NN has been presented in Table 2 followed by a detailed explanation [35].
The k-NN works on various distance measurement schemes. However, in this case, the Euclidean distance has been taken into consideration. Each training instance of \(TR\) has been sorted in ascending order on distance values realized. Ideally, the class label of the row having a short distance is the predicted label of the corresponding training instance. But according to k-NN, the k number of rows is picked up having short distances to the target instance. The k instances having most class labels become the predicted label of the target instance.
3.2 Naïve Bayes (Gaussian)
Gaussian Naive Bayes is a simple classification algorithm with high functionality and takes less computational time. It is a variant of the Naive Bayes algorithm, which is based on the Bayes theorem that follows Gaussian or normal distribution and is used when the features have continuous values. In the training phase, instances are segregated based on the class labels. The classwise mean and standard deviation are estimated. Further, in the testing phase, each data instance of the training data is processed by estimating the probability density, conditional probability, and posterior probability. The probability density helps to verify the normal distribution of data points; hence, it can be estimated as
Here, \(\sigma \left({tr}^{1 to r}\right)\) and \(\mu \left({tr}^{1 to r}\right)\) represents the standard deviation and mean of all the rows of the training instances estimated during the training phase. Once the probability density is in hand, the conditional probability of each testing instance \({ts}_{i}\) can be estimated as the traditional Naïve Bayes but with probability density.
Finally, the posterior probability is estimated to come across the final decision.
It should be noted that the predicted class label for any test instance \({ts}_{i}\) is the class with maximum posterior probability.
3.3 Light Gradient Boosting Machine (LightGBM)
Gradient Boosting Decision Tree (GBDT) uses an ensemble of several weak decision tree classifiers as a boosting algorithm, thus, having low reliance on the selection of features [36]. Decision trees are the foundation for GBDT, where the predictions made by each tree in the chain are added up to come across the final decision. Throughout the process, a new decision tree is created in each step to fit the difference between the current prediction and the ground truth. The accuracy, efficiency, and interpretability of GBDT made it a popular choice for many researchers [37]. For a training set \(TR=\left\{\left({tr}_{1}^{1},{tr}_{1}^{2},\dots \dots , {tr}_{1}^{r}\right),\left({tr}_{2}^{1},{tr}_{2}^{2},\dots \dots , {tr}_{2}^{r}\right),\dots \dots ,\left({tr}_{m}^{1},{tr}_{m}^{2},\dots \dots , {tr}_{m}^{r}\right)\right\}\) of size \(m\times r\) dimensions, where \(tr\) is the training instances and \({tr}^{r}\) attribute denotes the class labels, then \(f(TR)\) is the optimization and projected goal. The estimated function then \(f(TR)\) for minimizing the loss \(L({tr}^{r}, f(TR))\) would be
and the criteria for iteration of GBDT can be ascertained as
Here, \(k\) represents the iteration number and \({h}_{k}\left(TR\right)\) represents the base decision tree on training set \(TR\). Although the GBDT classification approach is suitable for small data sets, as the number of data sets with many dimensions increases, the GBDT approach struggles to produce suitable results [38]. This is because GBDT is unable to determine the optimal splitting point throughout the decision tree learning process. To overcome the GBDT’s inherent drawbacks, an improvised version of the GBDT has been recently proposed. It combines the concept of Gradient-based One-Side Sampling (GOSS) with the capability of Exclusive Feature Bundling (EFB), popularly referred to as the Light Gradient Boosting Machine (LightGBM) [39]. The LightGBM is another kind of gradient boosting, and light refers to the light version, which allegedly makes this framework for gradient boosting that uses tree-based learning methods faster, distributed, high-performance, and efficient. In a gradient boosting framework, the trees are constructed sequentially, in contrast to a random forest, which constructs a tree for each sample. The framework employs a leaf-wise tree development algorithm that splits the tree leaf-wise if the tree is not balanced. To be precise, the information gain is used to determine the split at each node. The GOSS function in LightGBM determines the splitting point using variance gain. The GOSS function used in LightGBM finds the splitting point with the help of variance gain. The GOSS first sort the training instances \(TR\) in descending order of the absolute gradient values. From the sorted instances, top \(A=\alpha \times 100\%\) instances having larger gradients are selected. Further, the GOSS function randomly samples \(B=\beta \times 100\%\) instances from the rest of the instances. The samples are boosted further using a constant value \(\frac{1-a}{b}\). Finally, the splitting point can be estimated through variance gains \(V\) with the help of a splitting feature \(j\) at any point \(p\) can be estimated as
where \({A}_{u}=\left\{{tr}_{i}\in A:{tr}_{i}^{j}\le p\right\}\), \({A}_{v}=\left\{{tr}_{i}\in A:{tr}_{i}^{j}>p\right\}\), \({B}_{u}=\left\{{tr}_{i}\in B:{tr}_{i}^{j}\le p\right\}\), \({B}_{v}=\left\{{tr}_{i}\in B:{tr}_{i}^{j}>p\right\}\), \({g}_{i}\) is the subset of \(g=\left\{{g}_{1},{g}_{2},\dots \dots \dots , {g}_{n}\right\}\), the negative gradients of the loss function concerning the output of the model. Here, \(\frac{1-a}{b}\) helps to normalize the sum of the gradients.
3.4 Random Forest
Random Forest (RF) is the best and most adaptable supervised machine learning algorithm in the concept of ensemble technique and hybrid model for the improvement in performance and prediction accuracy. RF algorithm combines many decision trees into an ensemble model using bootstrapping. RF predictions are made on random subsets of features and average voting, rather than giving every classifier a chance to vote in favour of a single class (Table 3).
3.5 Adaptive Boosting (Adaboost)
Adaboost, the abbreviation for adaptive boosting, is used to classify data by pooling the knowledge of many weak learners. Adjusting the weights of each instance means that instances that have been incorrectly classified will be given more weight, while correctly handled instances will be given less weight. Adaboost is the boosting technique that employs an ensemble of decision trees by default (Table 4). However, Adaboost provides flexibility to use many other classifiers as weak learners, where boosting is done by averaging the outputs [40].
The Adaboost assumes a training set \(TR=\left\{\left({tr}_{1}^{1},{tr}_{1}^{2},\dots \dots , {tr}_{1}^{r}\right),\left({tr}_{2}^{1},{tr}_{2}^{2},\dots \dots , {tr}_{2}^{r}\right),\dots \dots ,\left({tr}_{m}^{1},{tr}_{m}^{2},\dots \dots , {tr}_{m}^{r}\right)\right\}\) having \(m\) instances and \(r\) attributes. The \({r}^{th}\) attributes represent the class label such that \({tr}^{r}\in \left\{-\mathrm{1,1}\right\}\). If the number of weak classifiers is denoted as \(\mathcal{F}=\left\{{\mathcal{F}}_{1},{\mathcal{F}}_{2},{\mathcal{F}}_{3},\dots \dots , {\mathcal{F}}_{r-1}\right\}\), such that \({\mathcal{F}}_{k}\left(tr\right)\in \left\{-\mathrm{1,1}\right\}\), the loss function can be defined as
Adaboost as an ensemble principle combines multiple classifiers. For \(j\) number of learners, the target is to minimize the objective function, which can be represented as
Further, for each training instance, the weight has to be updated as
Once the learning process by all the weak learners \(\mathcal{F}\) completes, the final output of Adaboost becomes the linear combination of classification output provided by \(\mathcal{F}\). The predicted output of Adaboost can be presented as
It should be noted that the number of weak learners \(\mathcal{F}\) is based on the number of attributes in the training dataset, excluding the class attribute [41]. Since the number of attributes in \(TR\) is \(r\) including the class attributes, therefore the total number of weak learners are \(r-1\).
3.6 The Proposed Ensemble for Diabetes Detection
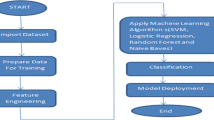
Using the supervised learning algorithms discussed above, various combinations of ensemble methods have been trained and tested. The best ensemble evolved has been proposed as the diabetes detector. The detection process of the ensembles has been presented in Fig. 1, where the blue colour blocks indicate training and the pink color blocks indicate testing blocks.
The identification of suitable ensemble of classifiers has been conducted in four steps, viz., data fold creation, ensemble training, ensemble testing and decision making. In the first stage, the entire dataset was divided into five blocks. For five blocks, five iterations have been made for training and testing of the ensembles, where a single data fold is used for testing and the rest of the four folds are used for training. Secondly, the k-NN, NB(G), RF, Adaboost and LightGBM classifiers are used to prepare nine ensemble schemes, viz., k-NN + NB(G), RF + k-NN, k-NN + Adaboost, LightGBM + k-NN, Adaboost + RF + k-NN, LightGBM + k-NN + RF, LightGBM + k-NN + Adaboost, LightGBM + k-NN + Adaboost + RF and LightGBM + k-NN + Adaboost + RF + NB(G). As we have mentioned earlier, each of the ensembles has been trained with four folds of data and tested with a single fold of data. The average performance of five iterations of training and testing has been realized. The entire process of classification has been conducted using a voting classifier [42, 43] that combines different machine learning classifiers for classification. The final decision about any test instance is decided either through hard voting or soft voting, where the projected probability for the underlying classifiers is used to forecast the class labels. In our case, the soft voting approach is used to obtain the prediction of the ensemble models. For the ensembles mentioned here, soft voting can be achieved as –
Here \({w}_{j}\) is the weight that can be assigned to the jth classifier and \(p\) indicates the predicted probabilities.
It is evident that, combining multiple machine learning algorithms can improve the average prediction performance by either helping tune one another, generalize, or adapt to unknown tasks [44]. However, according to Raschka et al. [45] soft voting method enhances the cumulative predictive results only if the underlying classifiers are well-calibrated. Therefore, in the proposed ensemble utmost care has been taken to tune individual classifiers through various parameter settings. The settings for the classifiers are presented in Table 5.
3.7 Dataset Description
The dataset for this study was collected from the Kaggle website. It is originated from the National Institute of Diabetes and Digestive and Kidney Diseases’ (NIDD) Pima Indian Diabetes Database, which is freely available online. There are 768 instances in this collection, eight attributes, and a class attribute. 500 patients are non-diabetic, and 268 patients are diabetic, i.e., 65.1% of the dataset is healthy and 34.9 percent is diabetes. All patients in this data collection are females aged at least 21 years of Pima Indian ancestry. Eight medical predictor factors and one outcome variable comprise the datasets. The number of pregnancies the patient has had, their BMI, insulin level, age, glucose, blood pressure, skin thickness, and diabetes pedigree function are all predictor variables [46]. Table 6 contains a description of the attributes, whereas Table 7 contains the precise characteristics.
The dataset presented here is mostly cited by numerous researchers due to a smaller number of diagnostic measurements. The only shortcoming of this dataset is that it contains diagnostic measurements of female subjects only. Although we can predict the presence of diabetes for other genders also, since clinical measurements in diabetes are almost similar among genders.
The dataset discussed here can be further explored through feature correlation matrix. We have tried to ascertain correlation between features and the result can be visualized in Fig. 2. According to Fig. 2, the dark color box represents the concern features that are more correlated than the features pertaining to light color cells. As it can be seen very few features are correlated, and most of the features are not correlated, therefore any feature selection procedure is not essential as a preprocessing task. Hence, we have decided to continue the detection process with the current set of features.
3.8 Effectiveness of Chosen Algorithms for the PIMA Dataset
The selection of supervised classification algorithms discussed in Sect. 3.6 has been undertaken keeping in view the dataset size and number of features in hand. A similar study has been conducted in recent past pertaining to selection of classifiers on varying datasets, which ensures the ideal classifiers specific to dataset size and number of features in hand [47]. The study revealed that k-Nearest Neighbor (k-NN), Naïve Bayes (Gaussian), Adaptive Boosting (Adaboost) and Random Forest (RF) are the leading performer on the similar dataset size which we are using in this research work. Additionally, we are using LightGBM as an additional boosting classifier to validate our objective. However, the behavior of the aforementioned algorithms for the PIMA dataset is still unknown. Although the PIMA dataset contains eight features, the most beneficial features for type-2 diabetes detection is still unknown. A little attempt has been made earlier for identification strength of the features of PIMA dataset for a specific classification algorithm [48]. In this section an extensive analysis has been conducted for all the shortlisted classifiers under study. In order to carry out the feature analysis we have evaluated the permutation importance of each features using ELI5 library [48]. The permutation importance score of each feature for each classifier is presented in Table 8
In Table 8, the features highlighted with deep green color background are most important and the features having background color white is least important for the concern classifier. In the PIMA dataset the feature Glucose evolved as the most significant feature for all the classifiers under study by scoring the highest permutation importance score. On the other hand, the Skin Thickness parameter is least significant for diabetes detection. Both the AdaBoost and LightGBM provide excellent permutation score on 7 features of the PIMA dataset except Skin Thickness. The performance score of AdaBoost and LightGBM signifies both the classifiers can detect presence of diabetes with almost all the features of PIMA dataset.
The number after the ± indicates how outcome of a classifier changes from one-reshuffling to the other. The negative weights reveal that the predictions on the shuffled data appears to be more accurate than the actual data. In a nutshell, all the five classifiers chosen here are state of the art and suitable for diabetes detection using PIMA dataset.
4 Results and Analysis
This section details the suggested diabetes detection system’s findings and analysis. Different performance metrics are used to evaluate the performance of various machine learning algorithms and the suggested ensemble approaches. We compare and analyze the performance of NB(G), k-NN, LightGBM, RF, and Adaboost using accuracy, recall, precision, F1 score, AUC curve, and ROC curve. Accuracy is a helpful evaluation metric since it quantifies the proportion of correctly diagnosed diabetic events to the total number of considered diabetic events. However, having an acceptable detection accuracy alone may not be sufficient to evaluate the model’s performance, as it does not consider incorrectly predicted cases. Therefore, other performance matrices such as recall, precision, and F1 score must be calculated. Precision is defined as the number of correctly classified diabetes instances to the actual number of diabetic instances. Similarly, recall denotes the ratio between the number of correct positive results divided by the number of all relevant samples present in the data set. Additionally, the F1 score or F1 measure is the mean of precision and recall harmonically expressed. The optimal value for Precision, Recall, and F1 score is 1. Finally, the Receiver Operating Curve is the primary performance parameter discussed in this paper. The receiver operating characteristic (ROC) curve is frequently used to solve classification problems at various threshold levels. The Area Under the Curve (AUC) illustrates the trade-off between true positive and false positive rates for each conceivable cut-off value for a test or a set of tests. AUC is a measure of performance that is aggregated across all possible categorization levels.
4.1 Results of the Proposed Hybrid Model
An ensemble voting classifier is implemented to build the proposed ensemble model [42, 43]. The ensemble models are called meta-algorithms as they combine multiple machine learning techniques into one predictive model. In this case, various combinations of k-NN, Naïve Bayes (G), LightGBM, Adaboost, and Random Forest algorithms are implemented as ensembles with certain hyperparameters tuning. The soft voting method is used as the ensembles’ decision. For every implementation of the ensemble model fivefold cross-validated performance metrics are evaluated. Table 9 represents the cross-validation results of various ensembles. In each combination, we ensure that at least one traditional classifier and one ensemble technique are combined. In this way, the correct hybrid of ensembles can be presented as the proposed model.
From the above-presented ensemble models in Table 9, a combination of LightGBM + k-NN + Adaboost provided an accuracy of 90.76% against fivefold cross-validation followed by LightGBM + k-NN at 90.62% fivefold cross-validated accuracy. As seen from Table 10, it is noticed that ensembles models LightGBM + k-NN and LightGBM + k-NN + Adaboost gave well-nigh correspondent results in all the performance matrices. In the earlier analysis of the classifiers, we have seen that k-NN was struggling in detection accuracy, precision, recall and F1-Score. However, the same k-NN reveals a better AUC when the ROC was plotted. This reveals the ability of the k-NN even in the class imbalance environment. Similarly, we have plotted the ROC of all the ensembles mentioned in Table 9. Figure 3 represents the ROC of all the ensembles.
The ROCs are shown here, contradicting what we have observed in Table 10. From Fig. 3 we found that the LightGBM + k-NN + Adaboost + RF and LightGBM + k-NN + RF outperform LightGBM + k-NN + Adaboost and LightGBM + k-NN ensemble. In a dataset with an uneven distribution of classes, the rarest class only accounts for a tiny fraction of the entire data. Due to lack of diversity, few classification algorithms are biased toward predicting the majority class since their loss functions seek to perform well in computing error rates without considering the distribution of the feature [49,50,51]. This is what happened in this case. The datasets used here hold 768 instances out of which 500 patients are non-diabetic and 268 are actual patients. This shows the dataset is imbalanced, and therefore, the ROC for LightGBM + k-NN + Adaboost + RF and LightGBM + k-NN + RF exhibits better results even though they have low detection accuracy as compared to LightGBM + k-NN + Adaboost and LightGBM + k-NN ensembles. On the other hand, the benefits of LightGBM + k-NN + Adaboost and LightGBM + k-NN ensemble are due to the presence of a fewer number of classifiers in the model. The appealing part is that with less ensemble of classifiers, the LightGBM + k-NN + Adaboost and LightGBM + k-NN present a computationally efficient model compared to other ensembles. Therefore, the LightGBM + k-NN + Adaboost and LightGBM + k-NN models are the better choices in diabetes detection, where LightGBM + k-NN + Adaboost shows the highest detection accuracy of 90.76%.
The performance metrics of the LightGBM + k-NN + Adaboost ensemble model are experimentally evaluated in terms of the number of folds in the cross-validation procedure. The number of runs over the training data (epochs) is set to 1500 fits, which were achieved by fitting five folds for each of 300 candidates in 2.1 min. Nonetheless, this alpha hyperparameter tuning is evaluated up to 3000 fits by fitting up to ten folds for 300 candidates, with the greatest scores observed at 1500 fits, or fitting five folds for each of 300 candidates. Except for the fivefold cross-validated scores, all other cross-validated data indicated a minor decline in performance measures. Considering time and over-fitting, fivefold cross-validated findings are chosen for their overall higher scores and efficient model. This procedure was applied to each ensemble model presented, yielding the arithmetic mean of performance metrics. Figure 3 illustrates the output of each fold.
The discrimination threshold graph of LightGBM + k-NN + Adaboost is also presented in Fig. 4. Similarly, the visualization of precision, recall, F1 score, and queue rate concerning the discrimination threshold also justifies the arguments about the selection of LightGBM + k-NN + Adaboost as the best ensemble (Fig. 5).
It should be noted that the detection accuracy, recall and F1-score have been increased with an increase in folds, thus boosting the overall result of LightGBM + k-NN + Adaboost ensemble.
4.2 Comparison of the Proposed Hybrid Model with Other Related Models
In the previous analysis, an ensemble of LightGBM + k-NN + Adaboost has been evolved as the best diabetes detection mechanism. In this section, various state-of-the-art existing ensemble approaches found in the literature are taken for comparison. In this regard, kMeans feature selection and Bagging (Logistic Regression) [24], soft voting of Logistic Regression, Random Forest and Naïve Bayes [25], max voting of Logistic Regression, Decision Tree, Support Vector Machine, k-NN and Naïve Bayes [26], ensemble of multiple classifiers including decision tree, naïve bayes, k-NN, Logistic Regression, etc. [12], classification via Quantum-inspired classifier configurations [28], cooperative co-evolution framework and Random Forest [21], synthetic Minority Over-sampling Technique and decision forest [20], stacking-based evolutionary ensemble learning system [29], parameter-free greedy ensemble approach and Random Forest [30], ensemble of decision tree and logistic regression with majority voting [23], adaptive synthetic sampling for class imbalance and Random Forest for classification [33], Adaboost ensemble learning framework with decision stump as primary classifier [31], and boosting technique in ensemble of random committee classifier [32] ensemble methods are shortlisted. All of these methods use the voting principle for decision-making and are implemented on the same dataset as that of our proposed approach. The comparative results are presented in Table 11. The n.d. represents the value for the said field that has not been documented in the concerned literature.
While comparing our suggested diabetes detection strategy with other relevant approaches, we observed that the proposed approach yields maximum detection accuracy. The proposed approach also took eight features of the underlying data like that of the existing approach. Nevertheless, an interesting thing has been observed about the stacking-based evolutionary ensemble learning system proposed by Singh et al. [29] approach. The stacking approach shows the highest ever 96.10% of recall (sensitivity). This provides a great scope for successfully detecting positive patients among the group of positives on which the test has been performed. However, our proposed ensemble approach shows similar or better recall compared to existing approaches.
We have discussed numerous ensemble approaches in this article, combining k-NN, Naive Bayes (Gaussian), and three other ensemble approaches, namely LightGBM, Adaboost, and Random Forest. The LightGBM + k-NN + Adaboost technique has the highest detection accuracy of all the ensembles, but LightGBM + k-NN has a slightly lower detection rate but is more computationally efficient due to a reduced number of classifiers. Conversely, the ensemble models LightGBM + k-NN + Adaboost + RF and LightGBM + k-NN + RF imply a larger area under the receiver operating curve. Each of the ensemble models presented in this section has a unique set of applications and situations. The LightGBM + k-NN + Adaboost ensemble is recommended for a variety of applications, including diabetes diagnosis. If performance is a priority, LightGBM + k-NN is the optimal solution because of the reduced number of classifiers. Similarly, if the training data contain a significant class imbalance between diabetic and non-diabetic patients, both the LightGBM + k-NN + Adaboost + RF and LightGBM + k-NN + RF ensemble approaches are beneficial.
5 Conclusion
Several Machine Learning methods, including LightGBM, k-NN, Nave Bayes (Gaussian), Random Forest, and Adaboost, were used in this study to predict diabetes. Individual classifiers have also been compared to the ensemble models described in this paper. Performance measures like classification accuracy, precision, recall, F1-score, and receiver operating curve are analyzed. For both individual and ensemble models, a fivefold cross-validation procedure has been introduced. Adaboost and Random Forest had the best detection accuracy in the individual classifier analysis. As an ensemble model, the models LightGBM + k-NN + Adaboost and LightGBM + k-NN achieved a near-fivefold cross-validated accuracy of 90%. The standard deviation of ensemble model performance measures has been somewhat increased. The fivefold cross-validation yields the best results for all ensemble models. The LightGBM + k-NN + Adaboost technique has the best detection accuracy, while the LightGBM + k-NN approach has a slightly lower detection result but is more computationally efficient because it has a smaller number of ensemble classifiers to process. However, the area under the receiver operating curve dictated by the LightGBM + k-NN + Adaboost + RF and LightGBM + k-NN + RF ensemble models is better. We recommend using the LightGBM + k-NN + Adaboost ensemble in diverse fields, including diabetes detection. It is preferable to use k-NN + LightGBM ensemble, as they are computationally efficient due to the lower number of classifiers. Using the LightGBM + k-NN + Adaboost + RF or the LightGBM + k-NN + RF ensemble techniques is equally effective when the training data show a major class imbalance between diabetic and non-diabetic individuals.
Availability of Data and Materials
The datasets analysed during the current study are available in the public repository, URL https://www.kaggle.com/uciml/pima-indians-diabetes-database.
Abbreviations
- k-NN:
-
K-Nearest Neighbors
- RF:
-
Random Forest
- NB:
-
Naïve Bayes
- GBM:
-
Gradient Boosting Machine
- SVM:
-
Support Vector Machine
- GA:
-
Genetic Algorithm
- WOD:
-
Weighted Objective Distance-based
- RFG-GCN:
-
Random Forest graph generation-based graph convolutional network
- SMOTE:
-
Synthetic Minority Over-sampling Technique
- IS:
-
Instance Selection
- FS:
-
Feature Selection
- LR:
-
Logistic Regression
- LDA:
-
Linear Discriminant Analysis
- CART:
-
Classification and Regression Tree
- HM-BagMoov:
-
Hierarchical Multi-level classifier Bagging with Multi-objective optimized voting
- ADASYN:
-
Adaptive synthetic
- LightGBM:
-
Light Gradient Boosting Machine
- GBDT:
-
Gradient Boosting Decision Tree
- GOSS:
-
Gradient-based One-Side Sampling
- EFB:
-
Exclusive Feature Bundling
- Adaboost:
-
Adaptive Boosting
- NIDD:
-
National Institute of Diabetes and Digestive
- BMI:
-
Body Mass Index
- BP:
-
Blood Pressure
- ST:
-
Skin Thickness
- DPF:
-
Diabetes Pedigree Function
- ROC:
-
Receiver operating characteristic
- AUC:
-
Area Under the Curve
References
Kopitar, L., Kocbek, P., Cilar, L., Sheikh, A., Stiglic, G.: Early detection of type 2 diabetes mellitus using machine learning-based prediction models. Sci. Rep. 10(1), 1–12 (2020)
Cho, N.H., et al.: IDF Diabetes Atlas: Global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res. Clin. Pract. 138, 271–281 (2018). https://doi.org/10.1016/j.diabres.2018.02.023
Khandakar, A., et al.: A machine learning model for early detection of diabetic foot using thermogram images. Comput. Biol. Med. 137, 104838 (2021). https://doi.org/10.1016/j.compbiomed.2021.104838
Chaki, J., Thillai Ganesh, S., Cidham, S.K., Ananda Theertan, S.: Machine learning and artificial intelligence based diabetes mellitus detection and self-management: a systematic review. J. King Saud Univ. - Comput. Inf. Sci. (2020). https://doi.org/10.1016/j.jksuci.2020.06.013
Islam, M.M.F., Ferdousi, R., Rahman, S., Bushra, H.Y.: Likelihood Prediction of Diabetes at Early Stage Using Data Mining Techniques, pp. 113–125. Springer, Singapore (2020)
Mercaldo, F., Nardone, V., Santone, A.: Diabetes mellitus affected patients classification and diagnosis through machine learning techniques. Procedia Comput. Sci. 112, 2519–2528 (2017). https://doi.org/10.1016/j.procs.2017.08.193
Yuvaraj, N., SriPreethaa, K.R.: Diabetes prediction in healthcare systems using machine learning algorithms on Hadoop cluster. Cluster Comput. 22(1), 1–9 (2019)
Negi, A., Jaiswal, V.: A first attempt to develop a diabetes prediction method based on different global datasets, In: 2016 Fourth International Conference on Parallel, Distributed and Grid Computing (PDGC), 2016, pp. 237–241 (2016). https://doi.org/10.1109/PDGC.2016.7913152
Maniruzzaman, M., Rahman, M.J., Ahammed, B., Abedin, M.M.: Classification and prediction of diabetes disease using machine learning paradigm. Heal. Inf. Sci. Syst. 8(1), 7 (2020). https://doi.org/10.1007/s13755-019-0095-z
Tafa, Z., Pervetica, N., Karahoda, B.: An intelligent system for diabetes prediction. In: 2015 4th Mediterranean Conference on Embedded Computing (MECO), pp. 378–382 (2015)
Labhade, J.D., Chouthmol, L.K., Deshmukh, S.: Diabetic retinopathy detection using soft computing techniques. In: 2016 International Conference on Automatic Control and Dynamic Optimization Techniques (ICACDOT), pp. 175–178 (2016). https://doi.org/10.1109/ICACDOT.2016.7877573.
Saxena, R.: Role of k-nearest neighbour in detection of diabetes mellitus. Turk. J. Comput. Math. Educ. 12(10), 373–376 (2021)
Benbelkacem, S., Atmani, B.: Random forests for diabetes diagnosis. In: 2019 International Conference on Computer and Information Sciences (ICCIS), pp. 1–4 (2019)
Washburn, P.S.: Investigation of severity level of diabetic retinopathy using adaboost classifier algorithm. Mater. Today Proc. 33, 3037–3042 (2020)
Rufo, D.D., Debelee, T.G., Ibenthal, A., Negera, W.G.: Diagnosis of diabetes mellitus using gradient boosting machine (LightGBM). Diagnostics 11(9), 1714 (2021)
Alharbi, A., Alghahtani, M.: Using genetic algorithm and ELM neural networks for feature extraction and classification of type 2-diabetes mellitus. Appl. Artif. Intell. 33(4), 311–328 (2019). https://doi.org/10.1080/08839514.2018.1560545
Chaising, S., Temdee, P., Prasad, R.: Weighted objective distance for the classification of elderly people with hypertension. Knowledge-Based Syst. 210, 106441 (2020)
Nuankaew, P., Chaising, S., Temdee, P.: Average weighted objective distance-based method for type 2 diabetes prediction. IEEE Access 9, 137015–137028 (2021). https://doi.org/10.1109/ACCESS.2021.3117269
Cao, K., Xiao, Y., Hou, M.: Correlation-driven framework based on graph convolutional network for clinical disease classification. J. Stat. Comput. Simul. 91(15), 3108–3124 (2021). https://doi.org/10.1080/00949655.2021.1921777
Syed, A.H., Khan, T.: Machine learning-based application for predicting risk of Type 2 Diabetes Mellitus (T2DM) in Saudi Arabia: a retrospective cross-sectional study. IEEE Access 8, 199539–199561 (2020)
Christo, V.R.E., Nehemiah, H.K., Brighty, J., Kannan, A.: Feature selection and instance selection from clinical datasets using co-operative co-evolution and classification using random forest. IETE J. Res. 68(4), 1–14 (2020)
Mishra, S., Tripathy, H.K., Mallick, P.K., Bhoi, A.K., Barsocchi, P.: EAGA-MLP—an enhanced and adaptive hybrid classification model for diabetes diagnosis. Sensors 20(14), 4036 (2020)
Sathurthi, S., Saruladha, K.: An analysis of parallel ensemble diabetes decision support system based on voting classifier for classification problem. Electron. Gov. an Int. J. 16(1–2), 25–38 (2020)
Ismail, L., Materwala, H., Tayefi, M., Ngo, P., Karduck, A.P.: Type 2 diabetes with artificial intelligence machine learning: methods and evaluation. Arch. Comput. Methods Eng. 29(1), 313–333 (2022). https://doi.org/10.1007/s11831-021-09582-x
Kumari, S., Kumar, D., Mittal, M.: An ensemble approach for classification and prediction of diabetes mellitus using soft voting classifier. Int. J. Cogn. Comput. Eng. 2, 40–46 (2021). https://doi.org/10.1016/j.ijcce.2021.01.001
Rajendra, P., Latifi, S.: Prediction of diabetes using logistic regression and ensemble techniques. Comput. Methods Programs Biomed. Updat. 1, 100032 (2021)
Saxena, S., Mohapatra, D., Padhee, S., Sahoo, G.K.: Machine learning algorithms for diabetes detection: a comparative evaluation of performance of algorithms. Evol. Intell. (2021). https://doi.org/10.1007/s12065-021-00685-9
Ishwarya, M.S., Cherukuri, A.K.: Quantum-inspired ensemble approach to multi-attributed and multi-agent decision-making. Appl. Soft Comput. 106, 107283 (2021)
Singh, N., Singh, P.: Stacking-based multi-objective evolutionary ensemble framework for prediction of diabetes mellitus. Biocybern. Biomed. Eng. 40(1), 1–22 (2020)
Bania, R.K., Halder, A.: R-Ensembler: A greedy rough set based ensemble attribute selection algorithm with k-NN imputation for classification of medical data. Comput. Methods Programs Biomed. 184, 105122 (2020). https://doi.org/10.1016/j.cmpb.2019.105122
Vijayan, V.V., Anjali, C.: Prediction and diagnosis of diabetes mellitus—a machine learning approach. In: 2015 IEEE Recent Advances in Intelligent Computational Systems (RAICS), pp. 122–127 (2015)
Ali, R., Siddiqi, M.H., Idris, M., Kang, B.H., Lee, S.: Prediction of diabetes mellitus based on boosting ensemble modeling. In: International conference on ubiquitous computing and ambient intelligence, pp. 25–28 (2014)
Wang, Q., Cao, W., Guo, J., Ren, J., Cheng, Y., Davis, D.N.: DMP_MI: an effective diabetes mellitus classification algorithm on imbalanced data with missing values. IEEE Access 7, 102232–102238 (2019)
Srivastava, T., Srivastava, T.: Introduction to k-NN, k-nearest neighbors: Simplified. Anal. Vidhya (2014)
Zhang, Z.: Introduction to machine learning: k-nearest neighbors. Ann. Transl. Med. 4(11) (2016)
Song, W., et al.: Design of a flexible wearable smart sEMG recorder integrated gradient boosting decision tree based hand gesture recognition. IEEE Trans. Biomed. Circuits Syst. 13(6), 1563–1574 (2019)
Zhang, Z., Jung, C.: GBDT-MO: Gradient-Boosted Decision Trees for Multiple Outputs. IEEE Trans. Neural Netw. Learn. Syst. 32(7), 3156–67 (2020)
Chen, C., Zhang, Q., Ma, Q., Yu, B.: LightGBM-PPI: predicting protein-protein interactions through LightGBM with multi-information fusion. Chemom. Intell. Lab. Syst. 191, 54–64 (2019)
Ke, G., et al.: Lightgbm: a highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 30, 3146–3154 (2017)
Hertzmann, A., Fleet, D.J., Brubaker, M.: AdaBoost. Univ, Toronto (2015)
Rahim, N.A., Paulraj, M., Adom, A.H.: Adaptive boosting with SVM classifier for moving vehicle classification. Procedia Eng. 53, 411–419 (2013)
Kittler, J., Hatef, M., Duin, R.P.W., Matas, J.: On combining classifiers. IEEE Trans. Pattern Anal. Mach. Intell. 20(3), 226–239 (1998)
Kuncheva, L.I.: Combining Pattern Classifiers: Methods and Algorithms, 2nd edn. Wiley, Hoboken, NJ, USA (2014)
Raschka, S.: MLxtend: providing machine learning and data science utilities and extensions to Python’s scientific computing stack. J. Open Source Softw. 3(24), 638 (2018). https://doi.org/10.21105/joss.00638
Raschka, S: Python machine learning. Packt publishing ltd (2015)
Kaggle: https://www.kaggle.com/uciml/pima-indians-diabetes-database, 2016. https://www.kaggle.com/uciml/pima-indians-diabetes-database (2021). Accessed 9 Sep 2021
Althnian, A., et al.: Impact of dataset size on classification performance: an empirical evaluation in the medical domain. Appl. Sci. 11(2), 796 (2021). https://doi.org/10.3390/app11020796
Kumar, K.: Indian Diabetes Analysis -LIME-Shapley, kaggle.com, 2022. https://www.kaggle.com/code/jagannathrk/indian-diabetes-analysis-lime-shapley
Thabtah, F., Hammoud, S., Kamalov, F., Gonsalves, A.: Data imbalance in classification: experimental evaluation. Inf. Sci. (NY) 513, 429–441 (2020). https://doi.org/10.1016/j.ins.2019.11.004
Leevy, J.L., Khoshgoftaar, T.M., Bauder, R.A., Seliya, N.: A survey on addressing high-class imbalance in big data. J. Big Data 5(1), 42 (2018). https://doi.org/10.1186/s40537-018-0151-6
Bader-El-Den, M., Teitei, E., Perry, T.: Biased random forest for dealing with the class imbalance problem. IEEE Trans. Neural Netw. Learn. Syst. 30(7), 2163–2172 (2019). https://doi.org/10.1109/TNNLS.2018.2878400
Funding
This research received no external funding.
Author information
Authors and Affiliations
Contributions
All authors have designed the study, developed the methodology, performed the analysis, and written the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
'Not applicable' as the study did not require ethical approval.
Consent to Participate
'Not applicable' as the studies not involving humans.
Consent for Publication
As per the journal guidelines and norms.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sai, M.J., Chettri, P., Panigrahi, R. et al. An Ensemble of Light Gradient Boosting Machine and Adaptive Boosting for Prediction of Type-2 Diabetes. Int J Comput Intell Syst 16, 14 (2023). https://doi.org/10.1007/s44196-023-00184-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s44196-023-00184-y