Abstract
Purpose
T cell receptor (TCR) usually determines the specificity and unique function of T cells. Recently, the unconventional T cells with a unique TCR have attracted great attentions because of their clinical importance. TCR Vα7.2+ cells, that consist of the CD161+ mucosal associated invariant T (MAIT) cells and CD161− non-MAIT T cells, have been reported to play crucial roles in immune defenses. However, their characterizations in human blood are still obscure. This study aims to investigate the signatures and functions of circulating TCR Vα7.2+CD161+ MAIT and TCR Vα7.2+CD161− cells under steady state.
Methods
The TCR Vα7.2+CD161+ and TCR Vα7.2+CD161− cells were separately sorted from healthy donor peripheral blood mononuclear cells (PBMCs) and send for single cell RNA sequencing (scRNA-seq). Flow cytometry analysis was used to verify the findings obtained from scRNA-seq analysis.
Results
Our findings demonstrated that there are more TCR Vα7.2+CD161+ cells than TCR Vα7.2+CD161− cells in healthy donor PBMCs and revealed the differences between them. Under steady state, 4 TCR Vα7.2+CD161+ MAIT clusters existed in peripheral blood. Pseudotime analysis further implied the development trajectory of these MAIT cells, which was ordered from CCR7+ resting cluster to LGALS3+ transitional cluster, followed by KLRG1+ cluster and ending with CX3CR1+ terminally differentiated cytotoxic cluster. In addition, our results revealed that TCR Vα7.2+CD161− cells consist of different kind of conventional T cells. These TCR Vα7.2+CD161− non-MAIT cells showed a higher level of Granzyme B expression and upregulated genes associated with cytotoxicity, which implicated their roles in immune defense.
Conclusion
Our findings advanced the understandings of the evolution of circulating MAIT cells. We also preliminarily defined the TCR Vα7.2+CD161− PBMCs as a combination of versatile CD4+ and CD8+ populations with cytotoxicity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The role of T cell receptor (TCR) in lymphocyte-mediated reaction is well known. TCR not only promotes the homeostasis, but also decides the specificity and function of T cells (Courtney et al. 2018; Irla 2022; Shah et al. 2021). To face the complicated challenges, conventional T cells (CD4+ and CD8+) usually acquire a highly diverse TCR repertoire through the rearrangement during thymic development (Irla 2022). However, the unconventional T cells, such as natural killer T (NKT) cells, and γδ T cells, which express a semi-invariant TCR have also been reported to serve critical roles in the immune defenses and autoimmune responses (Constantinides and Belkaid 2021; Li et al. 2022; Pellicci et al. 2020). Among these unconventional populations, mucosal associated invariant T (MAIT) cells which are defined by a unique TCR α chain (TRAV1-2, TCR Vα7.2 in human) and other surrogate markers (CD161, IL-18R), have rapidly got attractions and showed potential clinical importance (Godfrey et al. 2019; Provine and Klenerman 2020). The conserved TCR decides MAIT cells are selectively restricted to MHC class I like molecule MR1 (Legoux et al. 2020). Dominantly, MR1 presents the microbiota-derived vitamin B2 (riboflavin) metabolites to MAIT cells. MAIT cells depend on this presentation to keep development and in turn to constrain the pathogenic invasion (Hinks and Zhang 2020; Legoux et al. 2020).
MAIT cells are enriched in non-lymphoid tissues (liver, lung, intestine), and their percentages were highly different between individuals (Chiba et al. 2021; Provine and Klenerman 2020). This tendentious distribution facilitates MAIT cells eradicate pathogens and even promote the tissue repair (Constantinides et al. 2019; Legoux et al. 2020). Considering peripheral MAIT cells lack the expression of lymphoid homing receptors, the most probable scenario of their migration is transfer from blood to tissues and then to lymph and finally back to the blood (Beura and Masopust 2014; Legoux et al. 2020; Provine and Klenerman 2020). Compared to mice, human blood contains abundant MAIT cells (up to 9%), and their ratio usually decrease in diseases (Godfrey et al. 2019; Provine and Klenerman 2020). Determination the properties of blood MAIT cells may help us grasp the general alterations of this unconventional population and hint the progression of immune diseases (De Biasi et al. 2021). Although lots of works have done (De Biasi et al. 2021; Park et al. 2019; Vorkas et al. 2022), the characterizations of human blood MAIT cells are still obscure.
In different subset of T cells, similar TCR repertoire usually means the identical reactivity and implies the same functions (Schattgen and Thomas 2018). Except the CD161+ cells (MAIT), TCR Vα7.2+ cells also include CD161− populations (Ben Youssef et al. 2018; Merlini et al. 2019; Park et al. 2019). However, in previous works, TCR Vα7.2+CD161− cells were used as MAIT cells control, while their phenotypes and functions were less studied. It is necessary to make a definition for these TCR Vα7.2+CD161− cells, and find out if there is a relation between the TCR Vα7.2+CD161+ and TCR Vα7.2+CD161− populations.
In this research, we characterized the human blood TCR Vα7.2+ cells in steady state by single cell RNA sequencing (scRNA-seq) and analyzed the CD161+ (MAIT) and CD161− cells in them, separately. Different from previous findings, our work delineated a development trajectory of MAIT cells in the mode of activation. Moreover, the different clusters of physiological TCR Vα7.2+CD161− cells were also studied. These findings demonstrated the heterogeneity of TCR Vα7.2+ cells and will help us understand the development of peripheral MAIT cells.
2 Methods and materials
2.1 Human subjects
Healthy donors (aged from 20 to 40 years old) who wasn’t diagnosed with underlying diseases in the recent half year and had a normal result in the examinations of routine blood test and biochemical test, were recruited in the study. The residues of routine blood tests were collected to prepare the peripheral blood mononuclear cells (PBMCs) using Ficoll based density gradient centrifugation. Briefly, after a centrifugation at 800 g for 15 min (room temperature), the PBMCs were enriched at the interface of plasma layer and ficoll layer. All procedures were performed in compliance with the ethical guidelines and approved by the Institutional Committee for Human Research of Beijing Friendship Hospital, China (approval no. 2020-P2-196). A written informed consent was routinely obtained from all individuals.
2.2 Flow cytometry analysis
Antibodies against TCR Vα7.2 (3C10), CD3 (OKT3), CD161 (HP-3G10), IL-18R (H44), CD69 (FN50), CD62L (DREG-56), IFN-γ (4S.B3), IL-17A (BL168), Granzyme B (GB11), Perforin (B-D48), CCR7 (G043H7), KLRG1 (SA231A2), CX3CR1 (2A9-1), Galectin-3 (M3/38), CD4 (OKT4), and CD8A (HIT8a) were purchased from Biolegend. The MR1-tetramer was kindly supplied by NIH Tetramer Core Facility. PBMCs obtained by density gradient centrifugation were used for the further flow cytometry analysis. Before cell staining, Zombie Aqua™ and Human TruStain FcX™ were used to exclude the dead cells and block nonspecific binding. For intracellular staining of cytokines, the cells were stimulated with Cell Activation Cocktail (with Brefeldin A) from Biolegend. Six hours later, the cells were stained with antibodies against surface molecules and prepared according to the instructions of the Cyto-Fast™ Fix/Perm Buffer Set (Biolegend).
2.3 Cell culture
CCR7+ and CCR7− CD3+TCR Vα7.2+CD161+ PBMCs were sorted using flow cytometry and cultured for 5 d with IL-2 (2 ng/mLl) in 96-well plate. Antibodies against CD3 (10 μg/mLl) and CD28 (1 μg/mL) were also added. To determine the proliferation, cells were preparedly incubated with CellTrace Violet (Thermal Fisher) according to the instructions. For examining cytokine productions, cells were further stimulated with Cell Activation Cocktail and stained as described above.
2.4 Sample preparation for single cell RNA sequencing
Circulating TCR Vα7.2+CD161+CD3+7-AAD− and TCR Vα7.2+CD161− CD3+7-AAD− live cells were sorted from 4 male donors respectively using a BD FACSAria™ III Flow Cytometer and sent to Annoroad Gene Technology (Beijing) for 10 × Genomics single cell RNA sequencing. The Chromium Single Cell 3’ GEM, Library & Gel Bead Kit v3 were used according to the instructions for library construction. Subsequently, the qualified library was sequenced using Illumina Novaseq6000 platform.
2.5 Data processing
R package Seurat (version 3.0.2) was used to process files derived from Cell Ranger. Cells with detected genes < 200 and genes expressed in cells fewer than 3 were firstly excluded. According to the gene expressions, we further removed low quality cells with a threshold (detected genes<200 or detected genes>4000 or percentage of mitochondrial genes>20%). Finally, we refined our data by applying a gene expression signature (CD3G > 0 & CD3E > 0 & CD3D > 0).
2.6 Batch correcting of scRNA-seq data by integration analysis
We used Seurat integration analysis to correct the batch effects (Stuart et al. 2019). Before data integration, the standardizations of data and selections of highly variable genes were performed. Then, “FindIntegrationAnchors” and “IntegrateData” were carried out to identify anchors of different samples and integrate samples, respectively.
2.7 Cell clustering and visualization
After using linear transformations to scale the data, we made dimensionality reduction using principal component analysis (PCA). The first 10 dimensions of PCA embeddings were selected to construct a Shared Nearest Neighbor (SNN) Graph by “FindNeighbors” function. Subsequently, we applied the “FindClusters” function (resolution = 0.5) to identify cell clusters by SNN modularity optimization based on Louvain algorithm. And the clustering results were further visualized using uniform manifold approximation and projection (UMAP) (Becht et al. 2019).
2.8 Differential expression and enrichment analysis
Using Wilcoxon rank sum test to compare current cluster with each other, we employed the “FindAllMarkers” function (min.pct = 0.25 & logfc.threshold = 0.25) to identify differentially expressed genes [DEGs, Padjusted (Padj.) < 0.05] for each cluster. The upregulated DEGs are defined as highly expressed genes (HEGs). The HEGs were imported to perform enrichment analysis with differential gene sets annotated with GO database (http://geneontology.org/) or KEGG pathway database (https://www.kegg.jp/kegg/) using clusterProfiler package (v.3.18.1). Enrichment pathways were obtained with Padj. < 0.05.
2.9 Pseudotime analysis
To study and simulate the dynamic biological processes of cells, we adopted Monocle (version 2.18.0) (Trapnell et al. 2014) using differentially expressed genes from each cluster ( Padj.< 0.01) to produce trajectory of cell development. DDRTree and “orderCells” function were used to reduce the dimensionality of dataset and re-order the cells, respectively. Finally, we showed the results by the alterations of feature genes, Seurat clusters, and pseudotime on the trajectory.
2.10 Statistical analysis
GraphPad Prism 8 software was applied to perform the statistical analysis. Student’s t test was used to evaluate the significance of the difference between two groups. For comparisons between multiple groups, the RM one-way ANOVA with Tukey's multiple comparisons test was performed. In addition, Akima spline curves were generated to show the tendency of cell alterations along age. Data were presented as the mean ± s, and P < 0.05 was considered significant (* P < 0.05, ** P < 0.002, ***P < 0.0002, **** P < 0.0001).
3 Results
3.1 Peripheral blood of healthy donors possessed higher percentage of CD161+ cells in TCR Vα7.2+ cells than CD161− cells
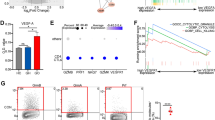
To characterize human blood TCR Vα7.2+ cells, we firstly collected blood sample from healthy donors, and determined the expressions of CD161 and TCR Vα7.2 in CD3+ peripheral blood mononuclear cells (PBMC) (Fig. 1A). Within the limited samples, the percentages of TCR Vα7.2+, TCR Vα7.2+CD161+, and TCR Vα7.2+CD161− cells in CD3+ cells showed no correlations with age. However, the fitted Akima spline curve implied the ratio of TCR Vα7.2+ and TCR Vα7.2+CD161+ cells reached a peak level after age 30, which was consistent with previous reports (Gherardin et al. 2018; Walker et al. 2014) (Fig. 1B). In addition, the statistical analysis also shown that TCR Vα7.2+ PBMCs contained significantly more CD161+ cells (58.33% ± 20.57%) than CD161− cells (39.83 ± 20.04%) (Fig. 1C). And there showed no significant differences between genders in the CD161 expression of TCR Vα7.2+ cells or the percentage of TCR Vα7.2+CD161+ cells in CD3+ T cells. Similarly, as shown in Fig. 1D, the ratio of CD161+ cells in TCR Vα7.2+ populations didn’t correlate with donors ages.
Determinations of circulating TCR Vα7.2+, TCR Vα7.2+CD161+ and TCR Vα7.2+CD161− cells in different individuals. A The gating strategy of TCR Vα7.2+, TCR Vα7.2+CD161+ and TCR Vα7.2+CD161− cells in human PBMCs. B Analysis of correlations between age and the percentage of TCR Vα7.2+, TCR Vα7.2+CD161+ and TCR Vα7.2+CD161− cells in CD3+ T cells (n = 38). Akima spline curves were generated to show the tendencies of cell percentages alterations in donors with different age. C Flow cytometry analysis of CD161 expression in TCR Vα7.2+CD3+ PBMCs (n = 36). The comparisons between genders of CD161+ cell proportions in TCR Vα7.2+ PBMCs and TCR Vα7.2+CD161+ cell proportions in CD3+ T cells are also shown respectively (n = 11). The Student’s t test was used to compare the differences mentioned above. D Analysis of the correlation between age and CD161+ cell proportions in TCR Vα7.2+ PBMCs (n = 43). An akima spline curve was generated to show the alteration tendency of CD161+ cells. Each experiment was repeated at least three times
3.2 ScRNA-seq revealed the differences between TCR Vα7.2+CD161+ and TCR Vα7.2+CD161− PBMCs
To assess the heterogeneity in TCR Vα7.2+ cells, we separately sorted TCR Vα7.2+CD161+ and TCR Vα7.2+CD161− PBMCs from 4 healthy donors, and sequenced them using 10 × Genomics Chromium system. Through the quality control (200 < feature RNA < 4000&percent mt < 20%), 21,398 feature genes across 30,784 cells were prepared for the further analysis (Figure S1A). After the correction of batch effect, uniform manifold approximation and projection (UMAP) analysis segregated 4 clusters in the integrated TCR Vα7.2+ populations (Fig. 2A). All the 4 donors TCR Vα7.2+ PBMCs included these 4 clusters (Figure S1B). The CD161+ MAIT cells distributed mainly in cluster 0 and cluster 3, while CD161− cells primarily existed in cluster 1 and cluster 2 (Figure S1C). In accordance with the segregated clusters, MAIT cells feature genes, KLRB1(CD161) and ZBTB16 (PLZF) were highly expressed in cluster 0 and cluster 3. And CD4 was shown in cluster 1 and cluster 2, as MAIT cells comprised mainly CD8+ and double negative T cells (Figs. 2B and S2A). As shown in Fig. 2A-C, cluster 0 comprised the most proportion of TCR Vα7.2+ cells (60.74%), showed higher expression of MAIT cell signature genes (Yao et al. 2020), such as CEBPD, SLC4A10, NCR3, and KLRB1. Cluster 1 were cytotoxic cells, which upregulated genes associated with cell killing (GZMH, GZMB, GNLY, NKG7, FCGR3A, and ZEB2). Cluster 2 demonstrating the increased SELL, CCR7, LEF1, TCF7, were defined as resting cells corresponding to naïve or central memory T cells (Fig. 2B and C). In addition, we also observed a cluster of LGALS3+ cells (cluster 3) (Fig. 2B and C). The proportions of these clusters revealed the heterogeneity within TCR Vα7.2+ cells.
ScRNA-seq analysis of TCR Vα7.2+CD161+ and TCR Vα7.2+CD161− cells obtained from peripheral blood of healthy donors. A UMAP visualization of cell clusters obtained from TCR Vα7.2+CD161+ and TCR Vα7.2+CD161− PBMCs. Each plot represented one cell (n = 30,784 cells, cluster 0 = 18,697 cells, cluster 1 = 5473 cells, cluster 2 = 3554 cells, cluster 3 = 3060 cells). B Expressions of selected feature genes in cell clusters identified above. C Heatmap of top 20 highly expressed genes (HEGs) in each cluster. D Comparisons of MR1-tetramer (n = 6), IL-18R (n = 5), CD69 (n = 5), and CD62L (n = 7) expressions in TCR Vα7.2+CD161+ and TCR Vα7.2+CD161− PBMCs were performed. E The percentages of IFN-γ+ (n = 10), IL-17A+ (n = 10), Granzyme B+ (n = 8) cells in TCR Vα7.2+CD161+ and TCR Vα7.2+CD161− populations are shown. The paired t test was performed to compare the differences described above
We next determined the phenotypes of TCR Vα7.2+CD161+ and TCR Vα7.2+CD161− PBMCs. Flow cytometry showed the majority of TCR Vα7.2+CD161+ cells (73.37 ± 12.89%) recognized MR1-tetramers, whereas the CD161− cells barely captured the tetramers. Moreover, the CD161+ populations had a higher expression of IL-18R (Figs. 2D and S2A). These findings indicated the TCR Vα7.2+CD161+ cells may represent MAIT cells. Similar to previous reports (Ben Youssef et al. 2018; Gherardin et al. 2018), CD161+ cells had an increased expression of CD69 and contained fewer CD62Lhi cells, demonstrating the TCR Vα7.2+CD161+ MAIT cells were kept in a pre-activated state (Figs. 2D and S2A). We also detected cytokine productions in TCR Vα7.2+ PBMCs. There was no difference in IFN-γ production between CD161+ and CD161− cells, while CD161+ MAIT cells included more cells with positive expression of IL-17A, although the quantity of IL-17A production was limited. Interestingly, Consistent with our scRNA-seq results, TCR Vα7.2+CD161− cells contained an increased percentage of Granzyme B+ cells (Figs. 2B, E, S2B). This enhanced cytotoxicity implied the CD161− populations may play a potential role in killing pathogens. Together, these data demonstrated the different clusters in TCR Vα7.2+ PBMCs, and compared the differences between TCR Vα7.2+CD161+ MAIT cells and TCR Vα7.2+CD161− cells.
3.3 Distinctive subpopulations existed in human circulating TCR Vα7.2+CD161+ MAIT cells
To characterize MAIT cells in human peripheral blood, we further analyzed the scRNA-seq data of TCR Vα7.2+CD161+ PBMCs. 4 clusters were found in TCR Vα7.2+CD161+ cells derived from the 4 healthy donors (Figs. 3A and S3A). As the largest population (86.77%), cluster 0 expressed higher level of KLRG1, CD69, CCL5, AP-1 subunits (JUN, FOS, FOSB), and MAIT signature genes (SLC4A10, CEBPD), was thought to be the classical activated MAIT cells (Figs. 3B and S3B). Distributed closely with cluster 0, cluster 1 (accounted for 8.62% of TCR Vα7.2+CD161+ PBMCs), showed a distinctive expression of LGALS3 (Galectin-3) and LGALS1, while lack the expression of effector molecules (Figs. 3B and S3B). In addition, highly expressed genes (HEGs) of cluster 1 were mainly enriched in pathways such as T cell activation and lymphocyte differentiation (Fig. 3C). Considering the role of Galection-3 in T cell activation (Amani et al. 2020; Gilson et al. 2019; Srejovic and Lukic 2021), cluster 1 were defined as the “transitional MAIT cells”.
Characterization of TCR Vα7.2+CD161+ MAIT cells in circulation of healthy donors. A Cell clusters identified by the Louvain algorithm (resolution, 0.5) in scRNA-seq data of TCR Vα7.2+CD161+ MAIT cells (N = 18,873 cells, cluster 0 = 16,377 cells, cluster 1 = 1627 cells, cluster 2 = 498 cells, cluster 3 = 371 cells). B Selected feature genes are shown in identified MAIT clusters. C GO and KEGG enrichment analysis using the HEGs obtained from clusters described above. D Comparison of CCR7+, CCR7−CX3CR1−KLRG1hi, and CX3CR1+ populations in circulating TCR Vα7.2+CD161+ MAIT cells (N = 12). The expression of Galectin-3 in different MAIT clusters are also shown below (N = 9). E Flow cytometry analysis of Perforin (N = 14), Granzyme B (N = 8), IFN-γ (N = 9), and IL-17A (N = 9) expression in CCR7+, CCR7−CX3CR1−KLRG1hi, and CX3CR1+ MAIT cells. At least three independent experiments were performed and a RM one-way ANOVA with Tukey's multiple comparison was performed to compare the differences described above
Notably, the rest two minor clusters of TCR Vα7.2+CD161+ MAIT cells both showed unique features. Cluster 2 (accounted for 2.6% of TCR Vα7.2+CD161+ PBMCs) selectively upregulated the expressions of GZMB, GZMH, FCGR3A, CX3CR1, NKG7, ZEB2, and GNLY (Figs. 3B and S3B). And pathways (cell killing, T cells mediated cytotoxicity, etc.) enriched by gene ontology (GO) analysis further demonstrated cluster 2 were terminally differentiated cytotoxic MAIT cells (Fig. 3C). Similar to MAIT cells reported by Vorkas et al. (Vorkas et al. 2022), we also identified a small percentage of cells (cluster 3, accounted for 2.0%) sharing the features of resting cells (upregulated Sell, CCR7, and LEF1), unexpectedly expressed increased level of CD4 (Figs. 3B and S3B). In addition, GO analysis implied the cluster 3 had an enhanced ability in lymphocyte proliferation and cell chemotaxis (Fig. 3C). Hence, we supposed these CCR7+ cells may be the repository for TCR Vα7.2+CD161+ cells in peripheral blood, waiting for activations.
To validation the identified clusters, we further performed flow cytometry using the signature markers of each cluster. Expectedly, most TCR Vα7.2+CD161+ MAIT cells possessed a phenotype of KLRG1hi (Fig. 3D). The CCR7+ (resting cells) and CX3CR1+ (which identify the effector memory Granzyme B+CD8+ cells (Böttcher et al. 2015; Gerlach et al. 2016)) populations also evidently existed, while the expression of CCR7 and CX3CR1 were dominantly shown in KLRG1hi MAIT cells (Fig. 3D). Similar to CD8+ T cells reported previously (Kaur et al. 2018), peripheral MAIT cells principally expressed Galectin-3 intracellularly rather than on the cell surface (Figure S3C). The percentage of Galectin-3+ cells was much higher in TCR Vα7.2+CD161+ cells than in TCR Vα7.2+CD161− cells (Figure S3C). Within the peripheral MAIT cells, KLRG1hiCCR7−CX3CR1− MAIT cells expressed the highest level of Galectin-3, while CCR7+ resting MAIT cells showed the minimum expression (Fig. 3D). Given the role of Galectin-3 in T cell activation (Amani et al. 2020; Kaur et al. 2018), these findings suggested the importance of Galectin-3 in the transition of MAIT clusters.
We also examined the cytokine productions in TCR Vα7.2+CD161+ PBMCs. In accordance with scRNA-seq results, CX3CR1+ populations had the highest level of Perforin and Granzyme B expression, while contained fewer cells with positive expression of IFN-γ (Figs. 3E and S3D). There was no difference in IFN-γ production between KLRG1hiCCR7−CX3CR1− and CCR7+CX3CR1− cells. However, CCR7+ CX3CR1− populations included the highest percentage of IL-17A+ cells, although all clusters of MAIT cells produced limited IL-17A (Figs. 3E and S3D). Together, we preliminarily proved the existence of clusters with distinctive phenotypes and functions in TCR Vα7.2+CD161+ PBMCs.
3.4 Pseudotime analysis revealed a development trajectory of TCR Vα7.2+CD161+ cells in human peripheral blood
Given the signature genes and enriched pathways (T cell activation/differentiation) of TCR Vα7.2+CD161+ cluster, we speculated there may be a development trajectory in circulating TCR Vα7.2+CD161+ cells. Hence, we performed a pseudotime analysis. Along the generated trajectory, TCR Vα7.2+CD161+ cells were ordered from CCR7+ resting clusters to LGALS3+ transitional clusters, followed by KLRG1+ majorities and ending with CX3CR1+ terminally differentiated cytotoxic clusters (Fig. 4A and B). This order matched the increased expression of effector molecules and the decreased resting cell features (Fig. 4B).
Development trajectory of TCR Vα7.2+CD161+ MAIT cells revealed by pseudotime analysis. A Transcriptional trajectory (Monocle 2) of circulating MAIT cells colored by Seurat clusters defined above. B Feature genes are shown on the trajectory of circulating TCR Vα7.2+CD161+ MAIT cells. C Flow cytometry analysis of CD4 and CD8 in CCR7+ and CCR7− MAIT cells was performed (n = 13). D Comparison of the proliferating ability using CellTrace Violet between CCR7+ derived and CCR7− derived MAIT cells after 5 days stimulation (n = 6). E–F Flow cytometry analysis of IFN-γ (n = 7) and IL-17A (n = 5) production in CCR7+ derived and CCR7− derived MAIT cells after stimulation. The paired t test was performed to compare the differences described above. Each experiment was repeated at least three times
As CCR7+ populations distributed at the onset point of trajectory sharing the features of resting cells (upregulated Sell, LEF1), we supposed these cells were central memory T cells which contributed to the homeostasis of peripheral MAIT cells. Thus, we determined their phenotypes and functions. Consistent with scRNA-seq analysis, The CD4+ cells percentage was found clearly higher in CCR7+ MAIT cells than in CCR7− classical MAIT cells (Figs. 3B and 4C). As shown in Fig. 4D-F, we next separately sorted the CCR7+ and CCR7− TCR Vα7.2+CD161+ cells and cultured them in vitro. After five days stimulation, the proliferating cells (CellTrace Violet−) were higher in cells derived from CCR7+ cells, while CCR7− population derived cells hardly expanded (Fig. 4D). Moreover, CCR7+ derived cells showed more IFN-γ and IL-17A productions than CCR7− derived cells (Fig. 4E and F). These findings indicated CCR7+ populations share the features of central memory cells and implied their potential roles in keep homeostasis of peripheral MAIT cells when facing challenges. Together, we found a development trajectory of TCR Vα7.2+CD161+ PBMCs, which started with the CCR7+ cells.
3.5 Characterization of TCR Vα7.2+CD161− PBMCs by scRNA-seq
TCR Vα7.2+CD161− cells were usually used as control for TCR Vα7.2+CD161+ MAIT cells in lots of works (Ben Youssef et al. 2018; Park et al. 2019). However, the definitions for them are still lack. Therefore, we also characterized them by scRNA-seq alone. 7 clusters were demonstrated in TCR Vα7.2+CD161− PBMCs from 4 healthy donors (Figs. 5A and S4A). As shown in Figs. 5B and S4B, the resting cell signatures (CCR7, SELL, LEF1, TCF7) were found in cluster 0 (CD8A+) and cluster 3 (CD4+). GO analysis showed these resting cells upregulated genes associated with translational initiation, confirming they are in a prepared state waiting for motivations (Fig. 5C). Both feature genes and enrichment analysis indicated the rest of cells included activated CD8A+ (cluster 2) and CD4+ (cluster 4) cells which upregulated CD69, FOS, JUN, JUNB. We also observed a group of CD8A+ or CD4+ cytotoxic cells (cluster 1) expressing cell killing related genes (PRF1, GZMB, CX3CR1, ZEB2) and a small portion of cells (cluster 5) sharing the signatures (FOXP3, IL2RA, IKZF2, CTLA4, TIGIT, CTLA4) of Treg cells (Figs. 5B, C, S4B). Different from other activated TCR Vα7.2+CD161− populations, cluster 6 showed an exhausted phenotype (upregulated TIGIT, HAVCR2, CTLA4, LGALS3, CD69, JUNB), which may contribute to their lack of enrichment in lymphocyte proliferation pathways (Figs. 5B, C, S4B).
The heterogeneities of TCR Vα7.2+CD161− PBMCs. A UMAP visualization of cell clusters obtained from TCR Vα7.2+CD161− PBMCs. Each plot represented one cell (n = 22,487 cells, cluster 0 = 8497 cells, cluster 1 = 4653 cells, cluster 2 = 4224 cells, cluster 3 = 2380 cells, cluster 4 = 1773 cells, cluster 5 = 735 cells, cluster 6 = 225 cells). B Selected genes are shown in the TCR Vα7.2+CD161− clusters. C GO analysis using the HEGs of different TCR Vα7.2+CD161− clusters. D Validation of CCR7+, CCR7−CX3CR1−KLRG1hi, and CX3CR1+ populations in TCR Vα7.2+CD161− cells (n = 16). E The comparisons of Granzyme B (n = 7), IFN-γ (n = 13), and IL-17A (n = 9) production in TCR Vα7.2+CD161− clusters described above using flow cytometry. F Flow cytometry analysis of the percentage of Perforin+ cells in CD4+ and CD8+ TCR Vα7.2+CD161− PBMCs (n = 8). Each experiment was repeated at least three times. The RM one-way ANOVA with Tukey's multiple comparison and paired t test were used to made statistical analysis described above, respectively
We further performed flow cytometry analysis to validated the findings of scRNA-seq. As shown in Fig. 5D, KLRG1hiCCR7− activated T cells, CCR7+KLRG1− naïve T cells, CCR7+KLRG1hi central memory T cells, and CX3CR1+KLRG1hi effector T cells were found in TCR Vα7.2+CD161−PBMCs. The ratio of KLRG1hiCCR7−CX3CR1− activated cells were higher in TCR Vα7.2+CD161− T cells than CCR7+ resting cells and CX3CR1+ effector cells (Fig. 5D). Consistent with scRNA-seq results, CX3CR1+ populations expressed the highest level of Granzyme B, while KLRG1hiCCR7−CX3CR1− activated T cells contained more cells with positive expression of IFN-γ compared to the resting or cytotoxic subpopulations (Figs. 5E and S4C). All the TCR Vα7.2+CD161− T cells from healthy donors showed a limited production of IL-17A, which was similar to TCR Vα7.2+CD161+ MAIT cells (Figs. 3E, 5E, and S4C). In addition, a fraction of CD4+ cytotoxic cells expressing perforin was certainly found in TCR Vα7.2+CD161− PBMCs. Although the difference didn’t show a statistical significance, the frequency of CD4+ cytotoxic cells was tendentiously lower compared to CD8+ cytotoxic cells (Fig. 5F). Collectively, these data preliminarily demonstrated that the TCR Vα7.2+CD161− PBMCs were a combination of versatile CD4+ and CD8+ cells. However, more delicate works are needed to give them a further definition.
4 Discussion
MAIT cells play critical roles in pathogen clearance, tissue repairment, autoimmunity, and even cancer (Constantinides et al. 2019; Toubal et al. 2019). Here, we made an explicated characterization of human blood TCR Vα7.2+CD161+ MAIT cells by scRNA-seq. Different from previous works (Vorkas et al. 2022), except the distinctive clusters, we demonstrated a development trajectory of MAIT cells ordered from CCR7+ resting cluster to LGALS3+ transitional cluster, followed by KLRG1+ majorities and ending with CX3CR1+ terminally differentiated cytotoxic cells. In addition, we also preliminarily defined the TCR Vα7.2+CD161− PBMCs and showed their differences from MAIT cells.
Human possessed much higher frequency of MAIT cells in circulation compared to mice (Gherardin et al. 2018; Rahimpour et al. 2015). Through the lifespan of human, MAIT cells arise with age after birth and markedly decrease in elder individuals (Novak et al. 2014; Walker et al. 2014). This tendency may provide the protection from pathogen invasions when youth facing increasing exposures and prevent the autoimmune injury elderly. Within the limited samples, we did not find the correlations of the percentages of TCR Vα7.2+CD161+ and TCR Vα7.2+CD161− cells with age or gender. However, compared to TCR Vα7.2+CD161− cells with a steady percentage, our results implied that TCR Vα7.2+CD161+ MAIT cells in blood reached a peak level after age 30, which was consistent with previous reports (Gherardin et al. 2018; Novak et al. 2014; Walker et al. 2014). Accordingly, we selected samples from 4 male donors (aged 30/32/34/38) for further sequencing analysis.
Through scRNA-seq analysis, we revealed peripheral MAIT cells comprised 4 clusters and ordered by CCR7+ resting clusters, LGALS3+ transitional clusters, KLRG1+ activated clusters and ending with CX3CR1+ terminally differentiated cytotoxic clusters in a development trajectory. MAIT cells usually leave thymus with a pre-activated phenotype and lack the expression of lymph-directed CCR7 and CD62L (Dusseaux et al. 2011; Salou et al. 2019). Moreover, MAIT cells in circulation seem further expand to reach the high frequency which is not observed in thymus (Godfrey et al. 2019). Interestingly, similar to MAIT cells reported by Vorkas et al. (Vorkas et al. 2022), we demonstrated a population of CCR7+TCR Vα7.2+CD161+ cells expressing CD4, SELL, and LEF1, distributed at the onset of the development timeline. After stimulation, these CCR7+ derived cells showed augmented proliferation and cytokine productions, while CCR7− MAITs hardly proliferated and produced limited cytokines. It suggested the CCR7+ MAIT clusters may contribute to the homeostasis of peripheral MAIT cells and keep their abundance in blood. Once exposed to the challenges, these central memory cells will be motivated to amplify MAIT-mediated immune reactions. In addition, we also found the CCR7+ MAIT cells contained a higher percentage of CD4+ cells. Indeed, Koay et al. discovered a portion of CD4+LEF1+ MAIT cells in the stage 3 of immature thymic MAIT cells which bridge the stage 2 and the bulk stage 3 thymic MAIT cells. These thymocytes share similar features (CD4, LEF1, TCF7, ITM2A) with our CCR7+ peripheral MAIT cells (Koay et al. 2016, 2019). We inevitably guess the relations between these two populations and suppose a small fraction of CD4+LEF1+ thymic MAIT cells may leave thymus at an early age and constitute or develop into the peripheral CCR7+ MAIT cells. However, the hypothesis still needs to be testified.
Galectin-3 (encoded by LGALS3) is expressed by the activated T cells and essential for the memory CD8+ T cells development (Amani et al. 2020; Kaur et al. 2018). During the activations, the intracellular galectin-3 is recruited to the immune synapse and help constrain the proliferation and cytokine productions (Kaur et al. 2018). Similar to previous findings, the circulating MAIT cells showed clearly upregulated expression of Galectin-3 than TCR Vα7.2+CD161− cells, due to their activated phenotypes. In the KLRG1hi MAIT cells, almost 90% cells expressed Galectin-3, while the CCR7+ populations with higher proliferating ability and CX3CR1+ cytotoxic populations contained slightly fewer cells with positive expression of Galectin-3. These findings suggested the importance of Galectin-3 in the transition of MAIT clusters. However, because of the generally high expression of activation-driven Galectin-3 and lack other surface surrogate markers, we failed to validate cluster 2 MAIT cells precisely by Galectin-3 in the protein level. Works for defining the cluster 2 MAIT cells are still urgent in the future.
The rearrangements proceeded in thymus guarantee the diversity of TCR repertoire and decide the fate of T cells (Irla 2022; Pellicci et al. 2020). T cells expressing similar TCRs may have complementary functions. TCR Vα7.2+ populations contained both CD161+ MAIT cells and CD161− cells (Ben Youssef et al. 2018; Park et al. 2019). Compared to the well-known MAIT cells, TCR Vα7.2+CD161− cells are less studied and their features are obscure. We illustrated the TCR Vα7.2+CD161− PBMCs included naïve clusters, activated clusters, exhausted cluster, and FOXP3+ cluster. Most importantly, we found the TCR Vα7.2+CD161− cells contained a high percentage of cytotoxic cells including both CD4+ and CD8+ populations. It indicated their potential roles in defensing the invasions. Indeed, HIV infected patients displayed significantly higher levels of perforin and a tendentiously upregulated Granzyme B expression in Vα7.2+CD161−CD8+ cells (Merlini et al. 2019), which partially supported our speculations. In addition, after anti-CD3/CD28 stimulation in vitro, TCR Vα7.2+CD161− cells hardly upregulate the level of CD161 expression (data not shown). This finding and works from other group suggested the CD161+ and CD161− cells are different populations (Merlini et al. 2019; Park et al. 2019), although some similar features are shown in them. Further determination of TCR Vα7.2+CD161− cells in the pathological states will help better define them and reveal their clinical applications.
There are limitations in our work. Firstly, the small sample size of scRNA-seq data with male donors and flow cytometry data may hamper the ability to discover the correlation of distinctive TCR Vα7.2+CD161+ and TCR Vα7.2+CD161− clusters with age and gender. Further examinations of the effect of age and gender on these cells are need. Secondly, due to the limited amount of MAIT cells and lack the proper way to simulate MR-1 mediated activation, we did not validate the CCR7+ clusters-derived development trajectory in vitro. Experiments are required to forwardly validate the pseudotime analysis result of circulating MAIT cells.
Taken together, our work characterized the human circulating TCR Vα7.2+CD161+ and TCR Vα7.2+CD161− cells in the physiological state and a development trajectory of MAIT cells order from CCR7+ resting cells to CX3CR1+ terminally differentiated cytotoxic cells was demonstrated to advance the understandings in the evolutions of peripheral MAIT cells. Explorations of different TCR Vα7.2+ clusters in the condition of pathology are required to further underline their importance in immune diseases.
Availability of data and materials
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human: HRA003183) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human.
References
Amani MF, Rolig AS, Redmond WL. Intracellular galectin-3 is essential for OX40-mediated memory CD8(+) T cell development. J Immunol. 2020;205(7):1857–66. https://doi.org/10.4049/jimmunol.1901052.
Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, Newell EW. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2019;37(1):38–44. https://doi.org/10.1038/nbt.4314.
Ben Youssef G, Tourret M, Salou M, Ghazarian L, Houdouin V, Mondot S, Mburu Y, Lambert M, Azarnoush S, Diana JS, Virlouvet AL, Peuchmaur M, Schmitz T, Dalle JH, Lantz O, Biran V, Caillat-Zucman S. Ontogeny of human mucosal-associated invariant T cells and related T cell subsets. J Exp Med. 2018;215(2):459–79. https://doi.org/10.1084/jem.20171739.
Beura LK, Masopust D. SnapShot: resident memory T cells. Cell. 2014;157(6):1488-1488.e1481. https://doi.org/10.1016/j.cell.2014.05.026.
Böttcher JP, Beyer M, Meissner F, Abdullah Z, Sander J, Höchst B, Eickhoff S, Rieckmann JC, Russo C, Bauer T, Flecken T, Giesen D, Engel D, Jung S, Busch DH, Protzer U, Thimme R, Mann M, Kurts C, Schultze JL, Kastenmüller W, Knolle PA. Functional classification of memory CD8(+) T cells by CX3CR1 expression. Nat Commun. 2015;6:8306. https://doi.org/10.1038/ncomms9306.
Chiba A, Murayama G, Miyake S. Characteristics of mucosal-associated invariant T cells and their roles in immune diseases. Int Immunol. 2021;33(12):775–80. https://doi.org/10.1093/intimm/dxab070.
Constantinides MG, & Belkaid Y. Early-life imprinting of unconventional T cells and tissue homeostasis. Science. 2021;374(6573):eabf0095. https://doi.org/10.1126/science.abf0095.
Constantinides MG, Link VM, Tamoutounour S, Wong AC, Perez-Chaparro PJ, Han SJ, Chen YE, Li K, Farhat S, Weckel A, Krishnamurthy SR, Vujkovic-Cvijin I, Linehan JL, Bouladoux N, Merrill ED, Roy S, Cua DJ, Adams EJ, Bhandoola A, Scharschmidt TC, Aube J, Fischbach MA, & Belkaid Y. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science. 2019;366(6464). https://doi.org/10.1126/science.aax6624.
Courtney AH, Lo WL, Weiss A. TCR signaling: mechanisms of initiation and propagation. Trends Biochem Sci. 2018;43(2):108–23. https://doi.org/10.1016/j.tibs.2017.11.008.
De Biasi S, Gibellini L, Lo Tartaro D, Puccio S, Rabacchi C, Mazza EMC, Brummelman J, Williams B, Kaihara K, Forcato M, Bicciato S, Pinti M, Depenni R, Sabbatini R, Longo C, Dominici M, Pellacani G, Lugli E, Cossarizza A. Circulating mucosal-associated invariant T cells identify patients responding to anti-PD-1 therapy. Nat Commun. 2021;12(1):1669. https://doi.org/10.1038/s41467-021-21928-4.
Dusseaux M, Martin E, Serriari N, Péguillet I, Premel V, Louis D, Milder M, Le Bourhis L, Soudais C, Treiner E, Lantz O. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. 2011;117(4):1250–9. https://doi.org/10.1182/blood-2010-08-303339.
Gerlach C, Moseman EA, Loughhead SM, Alvarez D, Zwijnenburg AJ, Waanders L, Garg R, de la Torre JC, von Andrian UH. The chemokine receptor CX3CR1 defines three antigen-experienced CD8 T cell subsets with distinct roles in immune surveillance and homeostasis. Immunity. 2016;45(6):1270–84. https://doi.org/10.1016/j.immuni.2016.10.018.
Gherardin NA, Souter MN, Koay HF, Mangas KM, Seemann T, Stinear TP, Eckle SB, Berzins SP, d’Udekem Y, Konstantinov IE, Fairlie DP, Ritchie DS, Neeson PJ, Pellicci DG, Uldrich AP, McCluskey J, Godfrey DI. Human blood MAIT cell subsets defined using MR1 tetramers. Immunol Cell Biol. 2018;96(5):507–25. https://doi.org/10.1111/imcb.12021.
Gilson RC, Gunasinghe SD, Johannes L, Gaus K. Galectin-3 modulation of T-cell activation: mechanisms of membrane remodelling. Prog Lipid Res. 2019;76: 101010. https://doi.org/10.1016/j.plipres.2019.101010.
Godfrey DI, Koay HF, McCluskey J, Gherardin NA. The biology and functional importance of MAIT cells. Nat Immunol. 2019;20(9):1110–28. https://doi.org/10.1038/s41590-019-0444-8.
Hinks TSC, Zhang XW. MAIT cell activation and functions. Front Immunol. 2020;11:1014. https://doi.org/10.3389/fimmu.2020.01014.
Irla M. Instructive cues of thymic T cell selection. Annu Rev Immunol. 2022;40:95–119. https://doi.org/10.1146/annurev-immunol-101320-022432.
Kaur M, Kumar D, Butty V, Singh S, Esteban A, Fink GR, Ploegh HL, & Sehrawat S. Galectin-3 regulates γ-Herpesvirus specific CD8 T cell immunity. iScience. 2018;9:101–119. https://doi.org/10.1016/j.isci.2018.10.013.
Koay HF, Gherardin NA, Enders A, Loh L, Mackay LK, Almeida CF, Russ BE, Nold-Petry CA, Nold MF, Bedoui S, Chen Z, Corbett AJ, Eckle SB, Meehan B, d’Udekem Y, Konstantinov IE, Lappas M, Liu L, Goodnow CC, Fairlie DP, Rossjohn J, Chong MM, Kedzierska K, Berzins SP, Belz GT, McCluskey J, Uldrich AP, Godfrey DI, Pellicci DG. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat Immunol. 2016;17(11):1300–11. https://doi.org/10.1038/ni.3565.
Koay HF, Su S, Amann-Zalcenstein D, Daley SR, Comerford I, Miosge L, Whyte CE, Konstantinov IE, d'Udekem Y, Baldwin T, Hickey PF, Berzins SP, Mak JYW, Sontani Y, Roots CM, Sidwell T, Kallies A, Chen Z, Nüssing S, Kedzierska K, Mackay LK, McColl SR, Deenick EK, Fairlie DP, McCluskey J, Goodnow CC, Ritchie ME, Belz GT, Naik SH, Pellicci DG, & Godfrey DI. A divergent transcriptional landscape underpins the development and functional branching of MAIT cells. Sci Immunol. 2019;4(41). https://doi.org/10.1126/sciimmunol.aay6039.
Legoux F, Salou M, Lantz O. MAIT cell development and functions: the microbial connection. Immunity. 2020;53(4):710–23. https://doi.org/10.1016/j.immuni.2020.09.009.
Li M, Cheng H, Tian D, Yang L, Du X, Pan Y, Zhang D, Mei X. D-mannose suppresses γδ T cells and alleviates murine psoriasis. Front Immunol. 2022;13: 840755. https://doi.org/10.3389/fimmu.2022.840755.
Merlini E, Cerrone M, van Wilgenburg B, Swadling L, Cannizzo ES, d'Arminio Monforte A, Klenerman P, & Marchetti G. Association between impaired Vα7.2+CD161++CD8+ (MAIT) and Vα7.2+CD161-CD8+ T-Cell populations and gut dysbiosis in chronically HIV- and/or HCV-infected patients. Front Microbiol. 2019;10:1972. https://doi.org/10.3389/fmicb.2019.01972.
Novak J, Dobrovolny J, Novakova L, Kozak T. The decrease in number and change in phenotype of mucosal-associated invariant T cells in the elderly and differences in men and women of reproductive age. Scand J Immunol. 2014;80(4):271–5. https://doi.org/10.1111/sji.12193.
Park D, Kim HG, Kim M, Park T, Ha HH, Lee DH, Park KS, Park SJ, Lim HJ, Lee CH. Differences in the molecular signatures of mucosal-associated invariant T cells and conventional T cells. Sci Rep. 2019;9(1):7094. https://doi.org/10.1038/s41598-019-43578-9.
Pellicci DG, Koay HF, Berzins SP. Thymic development of unconventional T cells: how NKT cells, MAIT cells and γδ T cells emerge. Nat Rev Immunol. 2020;20(12):756–70. https://doi.org/10.1038/s41577-020-0345-y.
Provine NM, Klenerman P. MAIT cells in health and disease. Annu Rev Immunol. 2020;38(1):203–28. https://doi.org/10.1146/annurev-immunol-080719-015428.
Rahimpour A, Koay HF, Enders A, Clanchy R, Eckle SB, Meehan B, Chen Z, Whittle B, Liu L, Fairlie DP, Goodnow CC, McCluskey J, Rossjohn J, Uldrich AP, Pellicci DG, Godfrey DI. Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. J Exp Med. 2015;212(7):1095–108. https://doi.org/10.1084/jem.20142110.
Salou M, Legoux F, Gilet J, Darbois A, du Halgouet A, Alonso R, Richer W, Goubet AG, Daviaud C, Menger L, Procopio E, Premel V, Lantz O. A common transcriptomic program acquired in the thymus defines tissue residency of MAIT and NKT subsets. J Exp Med. 2019;216(1):133–51. https://doi.org/10.1084/jem.20181483.
Schattgen SA, Thomas PG. Bohemian T cell receptors: sketching the repertoires of unconventional lymphocytes. Immunol Rev. 2018;284(1):79–90. https://doi.org/10.1111/imr.12668.
Shah K, Al-Haidari A, Sun J, Kazi JU. T cell receptor (TCR) signaling in health and disease. Signal Transduct Target Ther. 2021;6(1):412. https://doi.org/10.1038/s41392-021-00823-w.
Srejovic IM, Lukic ML. Galectin-3 in T cell-mediated immunopathology and autoimmunity. Immunol Lett. 2021;233:57–67. https://doi.org/10.1016/j.imlet.2021.03.009.
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888-1902.e1821. https://doi.org/10.1016/j.cell.2019.05.031.
Toubal A, Nel I, Lotersztajn S, Lehuen A. Mucosal-associated invariant T cells and disease. Nat Rev Immunol. 2019;19(10):643–57. https://doi.org/10.1038/s41577-019-0191-y.
Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–6. https://doi.org/10.1038/nbt.2859.
Vorkas CK, Krishna C, Li K, Aubé J, Fitzgerald DW, Mazutis L, Leslie CS, Glickman MS. Single-Cell transcriptional profiling reveals signatures of helper, effector, and regulatory MAIT cells during homeostasis and activation. J Immunol. 2022;208(5):1042–56. https://doi.org/10.4049/jimmunol.2100522.
Walker LJ, Tharmalingam H, Klenerman P. The rise and fall of MAIT cells with age. Scand J Immunol. 2014;80(6):462–3. https://doi.org/10.1111/sji.12237.
Yao T, Shooshtari P, Haeryfar SMM. Leveraging public single-cell and bulk transcriptomic datasets to delineate MAIT cell roles and phenotypic characteristics in human malignancies. Front Immunol. 2020;11:1691. https://doi.org/10.3389/fimmu.2020.01691.
Funding
Grants from the National Natural Science Foundation of China (No. 82100670, 82171823, and 81870399), Youth Beijing Scholar (No. 035), and Beijing Nova Program (Z211100002121036) supported this work.
Author information
Authors and Affiliations
Contributions
All listed authors participated meaningfully in the study. They have seen and approved the submission of this manuscript. M.L., H.J. and L.W. participated in performing the research, analyzing the data, and initiating the original draft of the article. T.Z., S.H., G.S., and J.Z. participated in performing the research. D.Z., D.T., C.L., and J.J. established the hypotheses, supervised the studies, analyzed the data, and co-wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The author Dong Zhang is a member of the Editorial Board for Current Medicine. The paper was handled by the other journal Editor and has undergone rigorous peer review process. Author Dong Zhang was not involved in the journal’s review of, or decisions related to, this manuscript. The other authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1.
scRNA-seq of TCR Vα7.2+CD161+ and TCR Vα7.2+CD161− PBMCs. (A) The quality control strategy for scRNA-seq data from TCR Vα7.2+CD161+ and TCR Vα7.2+CD161− cells. (B) The identified clusters are shown in the 4 healthy donors. (C) The distribution of TCR Vα7.2+CD161+ and TCR Vα7.2+CD161− cells are displayed in the clusters described above, respectively. Figure S2. Comparison of the circulating TCR Vα7.2+CD161+ and TCR Vα7.2+CD161− cells. (A) Flow cytometry analysis of MR1-tetramer, CD4, CD8A, CD62L, and CD69 expression in the CD161+ and CD161− populations (n = 5–7). (B) Comparisons of IFN-γ (n = 10), IL-17A (n = 10), and Granzyme B (n = 8) expression in CD161+ and CD161− populations. The experiments were repeated at least three times. Figure S3. Characterization of TCR Vα7.2+CD161+ MAIT cells in circulation. (A) The identified MAIT clusters are shown in the 4 donors. (B) Heatmap of the top 20 HEGs in TCR Vα7.2+CD161+ MAIT clusters. (C) Flow cytometry analysis of the membrane and intracellular Galectin-3 expression in TCR Vα7.2+CD161+ and TCR Vα7.2+CD161− cells. (D) Determination of Granzyme B, Perforin, IFN-γ and IL-17A production in CCR7+, CCR7−CX3CR1−KLRG1hi, and CX3CR1+ MAIT cells (n = 8–14). The experiments described above were repeated at least three times. Figure S4. Characterization of TCR Vα7.2+CD161− PBMCs. (A) The identified TCR Vα7.2+CD161− clusters are shown in the 4 donors. (B) Heatmap of the top 20 HEGs in each TCR Vα7.2+CD161− cluster. (C) Flow cytometry analysis of Granzyme B, IFN-γ and IL-17A production in CCR7+, CCR7−CX3CR1−KLRG1hi, and CX3CR1+ TCR Vα7.2+CD161− PBMCs (n = 7–13). At least three independent experiments were performed.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, M., Jin, H., Wei, L. et al. Single cell transcriptomics reveal the heterogeneities of TCR Vα7.2+CD161+ and TCR Vα7.2+CD161− T cells in human peripheral blood. Curr Med 2, 7 (2023). https://doi.org/10.1007/s44194-023-00026-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s44194-023-00026-1