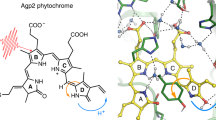
Abstract
Photoisomerization is a fundamental process in several classes of photoreceptors. Phytochromes sense red and far-red light in their Pr and Pfr states, respectively. Upon light absorption, these states react via individual photoreactions to the other state. Cph1 phytochrome shows a photoisomerization of its phycocyanobilin (PCB) chromophore in the Pfr state with a time constant of 0.7 ps. The dynamics of the PCB chromophore has been described, but whether or not the apoprotein exhibits an ultrafast response too, is not known. Here, we compare the photoreaction of 13C/15N labeled apoprotein with unlabeled apoprotein to unravel ultrafast apoprotein dynamics in Cph1. In the spectral range from 1750 to 1620 cm−1 we assigned several signals due to ultrafast apoprotein dynamics. A bleaching signal at 1724 cm−1 is tentatively assigned to deprotonation of a carboxylic acid, probably Asp207, and signals around 1670 cm−1 are assigned to amide I vibrations of the capping helix close to the chromophore. These signals remain after photoisomerization. The apoprotein dynamics appear upon photoexcitation or concomitant with chromophore isomerization. Thus, apoprotein dynamics occur prior to and after photoisomerization on an ultrafast time-scale. We discuss the origin of the ultrafast apoprotein response with the ‘Coulomb hammer’ mechanism, i.e. an impulsive change of electric field and Coulombic force around the chromophore upon excitation.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Phytochromes are photoreceptors of plants, bacteria and fungi that use red and far-red light as a source of information regarding their environment. Light absorption by a bilin chromophore initiates a reaction cascade that finally regulates fundamental processes, such as photosynthesis, flowering, seed germination, and shade avoidance. Phytochromes act as a bistable photoswitch between the Pr (red absorbing) state and the Pfr (far-red absorbing) state. Photoexcitation of the lowest energy Pr state triggers photoisomerization of the bilin chromophore and formation of the first ground-state intermediate Lumi-R on a picosecond time scale, followed by relaxation via Meta-R intermediates to form the Pfr state. Photoexcitation of Pfr, on the other hand, leads to sub-picosecond isomerization and Lumi-F production followed by Meta-F intermediates prior to reformation of Pr [1,2,3,4,5,6,7,8,9,10,11,12,13,14,15]. How these photoisomerizations are brought about is still unclear, however.
The crystal structure of the photosensory module of the cyanobacterial phytochrome Cph1 as Pr [16] like that of plant phytochromes [17], shows a remarkable overall dumbbell structure. The N-terminal PAS-GAF bidomain is tied together by a figure-8 knot, the GAF domain enveloping the chromophore while the PHY domain is held at a distance via a long α-helix. However, a remarkable hairpin loop known as the tongue extends back from the PHY domain to make contact with the GAF domain, thereby sealing the chromophore pocket. It has been suggested that refolding of the tongue pulls on the PHY domain, movement of which activates the physiological signal [18]. Since a crystal structure of Cph1 as Pfr is not yet available, a structural model (Fig. 1a) was built by homology modeling based on the Pfr crystal structure of a distantly-related bacteriophytochrome [19, 20]. This model was used to identify ultrafast chromophore changes during Pfr photoisomerization and heterogeneity in the Pfr ground state [20]. In this study, we compare our published data of 13C/15N labeled apoprotein [20, 21] with the measured data of unlabeled apoprotein, and use the Pfr model (see Fig. 1) as a starting point to identify ultrafast protein changes upon photoexcitation.
a Homology model (βf or down form) of the Pfr state of Cph1 phytochrome [20] with the phycocyanobilin (PCB) chromophore and its electronic transition dipole moment (tdm) represented by a green arrow. b enlarged view into the binding pocket of the chromophore PCB; the blue colored α-helical part was called “capping helix” in DrBphP [37]; here the backbones of arg254, ser206, and backbone of side chains 259–266 are colored in blue, these backbone carbonyl groups exhibit vtdms with a relative angle between 14° and 60° with the electronic transition dipole moment (green vector). Other backbone carbonyl groups are oriented mainly perpendicular to the electronic tdm or are far away from PCB. c enlarged view on the PCB chromophore in the down conformation [20] and arrangement of important amino acids, such as Asp207 (D207)
In general, it is assumed that photoisomerization of the chromophore is the decisive reaction step initiating all subsequent structural changes in the photoreceptor from picoseconds to seconds. This assumption is supported by several investigations using chemically modified chromophores that are unable to photoisomerize (‘locked’ chromophores) and consequently fail to form the photoproduct state, e.g. Pfr [22]. Furthermore, ultrafast investigations on photoreceptors with ‘locked’ chromophores exhibit dynamics on the picosecond to nanosecond time scale, but their impact on the formation of an active state remains elusive [1, 23,24,25,26,27].
Photoexcitation can result in largely altered dipole moments in the electronic excited state. A significant dipole moment increase was observed in the electronically excited state of retinal bound proteins, and the created electric fields were proposed to interact with the protein [28, 29]. The altered electric field at the chromophore was proposed to be the driving force for its isomerization and interact with charged and polarizable apoprotein groups in its near environment [28, 29]. The energy absorbed by the retinal could be used for new bond arrangements in the apoprotein structure affecting proton positions and stabilizing retinal conformations [30]. The interaction with the altered dipole was expected to be on the time-scale of nanoseconds or longer to induce relevant conformational apoprotein changes [29]. These hypotheses could not be confirmed by direct experimental data for a long time. Gross et al. demonstrated apoprotein changes of the amide I band of bR in the electronic excited state of a locked retinal persisting up to the nanosecond time scale [23]. Later on energy relaxation to the apoprotein was demonstrated in other proteins after isomerization [31, 32]. However, the impact of these observations on downstream processes of the photocycle remained elusive.
Recently, ultrafast structural changes on apoprotein side chains were reported prior to and after isomerization of the chromophore in bR, CaChR 1, and Agp2 phytochrome [33,34,35] and have been connected to downstream processes leading to the photoactive state [34, 35]. The underlying mechanism was suggested to be a ‘Coulomb hammer’-like process rapidly changing the electric field around the chromophore upon its photoexcitation, thereby triggering changes in hydrogen bond networks and protonation states [36]. Ultrafast X-ray crystallography measurements were reported for a bacteriophytochrome from Deinococcus radiodurans (DrBphP) in its Pr state at 1 ps and 10 ps after excitation [37]. The structural alterations at 1 ps show a twist of the D-ring, the dissociation of the pyrrole water from the chromophore, movement of ring A and a key aspartate (D207), as well as ultrafast water and backbone movements around the chromophore. Significant backbone movements in DrBphP were located at the “capping helix” (blue colored structural motif in Fig. 1b) [37]. Ultrafast water movement around the chromophore as well as apoprotein changes were reported also for bR by ultrafast X-ray crystallography [38, 39]. Compared to other ultrafast methods, X-ray crystallography reports a huge number of structural rearrangements throughout the apoprotein. However, since these experiments were performed with very high pump intensities, it is unclear whether the structural changes occur in a single photon excitation regime or are a consequence of multi-photon processes [39]. Clarifying whether substantial structural changes prior to and after isomerization occur in photoreceptors on the ultrafast time-scale is fundamental to understanding apoprotein dynamics.
Comparison of X-ray results for the Pr state of DrBphP with other methods at single photon excitation intensities with Cph1 reveals contradictory results for the twist of the D-ring, and movement of ring A [40, 41]. Whether these differences are due to the different phytochromes or methods used is unclear. As contributions of the isomerizing chromophore dominate the vibrational signals, very little is known about ultrafast changes in the apoprotein upon photoexcitation. To address this, we compared ultrafast, polarization-resolved VIS pump—IR probe experiments in the Pfr state of Cph1 with and without 13C/15N labelled apoprotein. By comparing the dynamics in each case, we have been able to identify ultrafast apoprotein changes in the spectral region between 1610 cm−1 and 1750 cm−1 not only after but also prior to photoisomerization.
2 Materials and methods
Preparation of the 13C/15N labeled apoprotein Cph1Δ2 (non-labelled PCB chromophore) was described previously [20, 21, 42,43,44]. Unlabelled holophytochrome was generated via in vitro autoassembly of apoCph1Δ2 with PCB extracted by methanolysis of phycobiliproteins from commercially available Spirulina pellets dissolved in 100 mM potassium phosphate buffer (pH 7.0) following procedures described earlier [45]. The SEC-purified sample was rebuffered into D2O-TESß at pD 7.8 by three passes through a home-built pressure cell. The samples had an optical density of 0.15 OD to 0.4 OD at 710 nm.
We use polarization resolved femtosecond visible (VIS) pump infrared (IR) probe spectroscopy to directly investigate the dynamics of the photoreaction. Measurements on 13C/15N labeled apoprotein were described elsewhere [20]. Measurements on unlabeled holophytochrome were performed in a similar way: pump and probe pulses were generated using home built optical parametric amplifiers. By difference frequency mixing in various steps, we obtained tunable mid-IR probe pulses of 300 fs (FWHM) or shorter at a repetition rate of 1.088 kHz. Simultaneously, laser pulses of ~ 150 fs duration around 720 nm with a bandwidth (FWHM) of ~ 10 nm were generated and used to initiate the Pfr photoreaction (system response ~ 350 fs). At this wavelength no Pr absorption occurs. A 640 nm laser diode was used for the background illumination to enrich the Pfr state. Photoselection experiments were performed by overlapping one pump beam with a diameter of 300 μm, and two probe beams with diameters of 150 μm on the sample. The pulse energy of the pump and probe was 0.55 μJ and 10 nJ, respectively. The excitation efficiency was 15% or less. After passing the sample, both probe pulses were dispersed with an imaging spectrograph and recorded with a 2 × 32 element MCT array detector (resolution ∼1.5 cm−1). The polarization direction of the pump pulse was changed after every scan by a half-wave (λ/2) plate, to switch the relative polarizations between pump and two probe beams from parallel to perpendicular or perpendicular to parallel. Thus, we detect absorption signal for parallel Apar and perpendicular Aper polarization. The signals for isotropic polarization Aiso were taken from Aiso = (Apar + 2 × Aper)/3. The global analysis was performed on the complete dataset with three shared time constants and an offset. Error margins represent 1σ deviations (68.27%). An exhaustive search analysis with a broader error margin of 3σ (99.7%) provides time constants with asymmetric error margins of τ1 = 0.3–0.7 ps, τ2 = 2.1–5.5 ps, and τ3 = 9.6–18 ps.
Samples were placed between two 1 mm thick CaF2 windows of 38 mm diameter in a sample holder with a 50 µm Teflon spacer. The sample was moved continuously in horizontal and vertical direction by a Lissajous-scanner to ensure sample exchange between two successive pump pulses. All experiments were performed at room temperature. The optical density at 640 nm was between 0.3 and 0.6 when the sample was fully in the Pr state. The Pr-Pfr-Pr photocycle was tested before each experiment to ensure that the samples were photoactive.
Polarization resolved measurements use photoselection to induce a transient orientation of excited molecules in the sample. With respect to the pump pulse polarization, the probe pulse is detected for parallel and perpendicular polarizations under identical conditions. The dichroic ratio D is defined as the ratio of parallel versus perpendicular polarized absorption signal. The dichroic ratio is a measure for the relative angle between the excited electronic transition dipole moment (tdm) and the detected vibrational transition dipole moment (vtdm). Since the electronic tdm is fixed within the excited molecule, the relative angle can provide orientational information on the detected vibrational group within the protein [40]. The electronic tdm of the S0 → S1 transition of the Pfr state is shown as a green arrow in Fig. 1.
2.1 Vibrational frequencies and transition dipole moments
We used the published “down conformation” model from Stensitzki et al. [20] as a structural model of Cph1 in the Pfr state. In this structure, the Asp207 is modeled with an anionic COO group. In addition to it, a structural model was generated with a protonated Asp207 by manually protonating the carboxylic side chain. The DFT/CHARMM optimized structures of PCB were used as input for frequency calculations and computation of electronic and vibrational transition dipole moments. In all these calculations the protein environment is described as a cloud of atomic point charges from the CHARMM force field. The procedure is described in detail [46, 47]. Tdm’s of electronic transitions were computed by time-dependent (TD)-DFT theory implemented in Gaussian09 software [48]. The relative angle between the electronic tdm and specific vtdm’s can be estimated directly from these calculations.
3 Results
Femtosecond polarization resolved VIS-pump IR-probe experiments were performed upon excitation of the Pfr state of Cph1 in D2O around 720 nm and probing in the spectral range from 1750 to 1610 cm−1. In this spectral range carbonyl vibrations are dominant and are easily identified by 13C labeling. For example, the high frequency C = O stretching vibration of a carboxyl group at 1730 cm−1 shifts upon 13C labeling by about 42 cm−1 to ~ 1690 cm−1. Amide I vibrations of a backbone C = O group at 1669 cm−1 shift upon 13C labeling to frequencies of 1626 cm−1. The 15N labeling has only minor influence on the spectral position of the amide I group. Other vibrational groups in this spectral range are high frequency CN vibrations of arginine [49]. Upon 13C/15N labeling a red-shift of about 30 cm−1 is observed and a red-shift of about 70 cm−1 is reported in D2O [50,51,52].
3.1 Absorbance difference spectra
In Fig. 2a and 2c absorbance difference spectra of Cph1 phytochrome are presented at different pump—probe delay times upon photoexcitation for unlabeled and 13C/15N labeled apoprotein, respectively. The PCB chromophore is unlabeled and we expect identical chromophore dynamics in both measurements. Indeed, the dynamics are very similar (see Figs. 2a and 2c), although distinct alterations are visible around 1724 cm−1, 1680 cm−1, and 1635 cm−1. Differences in the spectra can be attributed to the 13C/15N labeling of the apoprotein, resulting in a frequency down-shift of apoprotein vibrations. Polarization resolved averaged absorbance difference spectra for parallel, perpendicular and isotropic probe polarization with respect to pump pulse polarization are shown in Fig. 2b and 2d for unlabeled and 13C/15N labeled apoprotein, respectively. Upon photoexcitation, the vibrational dynamics of the 13C/15N labeled apoprotein show time constants of 0.7 ± 0.3 ps and 5 ± 2 ps in the spectral range from 1750 to 1610 cm−1 [20], where apoprotein contributions are expected to be negligible due to 13C/15N labeling. We averaged in the time window from 10 to 50 ps, after the ultrafast chromophore dynamics are mainly completed [21].
Absorption difference spectra of Cph1 in D2O upon photoexcitation around 720 nm at different pump-probe delay times. The PCB chromophore is unlabeled. Left: isotropic polarized absorption spectra for unlabeled protein (a) and 13C/15N labeled apoprotein (c). Right: scaled polarization resolved absorption spectra averaged from 10 to 50 ps delay time in the spectral window around 1720 cm−1. Parallel (black spheres), perpendicular (red open circles), and isotropic (purple line) polarizations with respect to pump pulse polarization for unlabeled apoprotein (c), and 13C/15N labeled apoprotein (d). Averaged data in (c) are scaled by a factor of two. Differences between labeled and unlabeled apoprotein are clearly visible. Additional protein contributions shift the apparent band positions at early delay times; after photoreaction at 50 ps the position of the peaks (grey lines) are identical at the given resolution. The dataset on the 13C/15N labeled apoprotein was discussed elsewhere [20]
3.2 Dynamics of 13C/15N labeled protein
Absorbance difference spectra at selected pump-probe delay times are presented in Fig. 2c for isotropic probe polarization. Dynamics of the Pfr state of Cph1 with 13C/15N labeled apoprotein were investigated and discussed in detail elsewhere [20, 21]. In short, the difference spectra show a strong negative (bleaching) signal at 1708 cm−1 with a small shoulder at 1702 cm−1, as well as a negative signal at ~ 1724 cm−1 at 0.5 ps delay time. These signals at 1724 cm−1 and around 1705 cm−1 appear with the system response, decay with 0.7 ± 0.2 ps and 5 ± 2 ps, and were assigned to C = O carbonyl stretching vibrations of PCB rings A and ring D, respectively. The characteristic band of the Pfr photoisomerization is the vibrational band of the ring D carbonyl stretching vibration. Its negative bleaching band is located at 1708 cm−1 and decays to about 20% of its initial value at 50 ps after photoexcitation [21]. The remaining bleaching signal fraction is a measure of the quantum yield of the photoisomerization. The small shoulder at 1702 cm−1 was also assigned to the C = O stretching vibration of ring D representing a different ground-state conformation and demonstrating heterogeneity of the electronic ground state of PCB [9, 20, 53]. At longer delay times, the bleaching band at 1724 cm−1 decays to zero and is superimposed upon the rise of a positive signal assigned to the carbonyl C = O stretching absorption of ring D in the Lumi-F photoproduct [21]. The photoisomerization of the chromophore is completed at 30 ps with a positive signal at 1724 cm−1 reflecting the isomerized PCB in Lumi-F, and negative signal at 1708 cm−1 from the initial Pfr ground state bleaching. These two bands show an absorption strength peak ratio of about 1:1 as expected for the same C = O stretching vibration before and after photoisomerization [21]. After 30 ps no other signals are detected in the spectral range from 1750 to 1620 cm−1 in 13C/15N labeled Cph1 apoprotein (Fig. 2c).
Polarization resolved absorbance difference spectra averaged from 10 to 50 ps are presented in Fig. 2d around 1720 cm−1. The positive signal at 1724 cm−1 reflecting Lumi-F absorption show very similar absorption strengths for parallel (black dots) and perpendicular (open red circles) polarizations. The dichroic ratio D = Apar/Aper reflects the relative angle between the excited electronic tdm (see Fig. 1) and the probed vtdm at a given wavenumber [24, 54]. The relative angle θ can range between 0° and 90° and can be determined from the dichroic ratio D by θ = arccos([(2D–1)/(D + 2)]½). The dichroic ratio can be directly extracted from the parallel and perpendicular polarized absorbance difference spectra at a given delay time and wavenumber. From Fig. 2d the dichroic ratio for the Lumi-F absorption DLF at 1724 cm−1 and the ring D bleaching signal DBL at 1708 cm−1 can be directly estimated to DLF = 1.3 (θ = 46°) and DBL = 1.5 (θ = 41°), respectively. These values agree with dichroic ratios of DLF = 1.3 ± 0.2 (θ between 41° and 51°) and DBL = 2.0 ± 0.5 (θ between 19° and 41°) determined by simulating the polarization resolved absorption spectra [20].
3.3 Unlabeled protein dynamics
The absorbance difference spectra of the unlabeled apoprotein are dominated by the negative bleaching signal at 1708 cm−1 as presented in Fig. 2a. This bleaching band reflects mainly the dynamics of the carbonyl group of ring D in the parent state. Comparison of the bleaching signal strength upon photoexcitation and after photoexcitation reflects the portion of the excited molecules relaxing back to its parent state and the portion undergoing forward reaction. Similar to the photoreaction of the 13C/15N labeled apoprotein, the bleaching signal decreases 30% between delay times of 0.5 ps to 50 ps (see Fig. 2a and 2c). Moreover, the bleaching transients depicted in Fig. 3b indicate nearly identical behavior. Thus, we can conclude that the ultrafast isomerization dynamics of PCB are identical in labeled and unlabeled apoprotein with a forward quantum yield of about 20%.
Isotropic polarized transients of unlabeled (blue dots), and 13C/15N labelled apoprotein (green circles) are presented on a linear and logarithmic time scale together with simulated transients (blue and green lines). Differences induced by apoprotein labeling are directly visible by comparing the blue and green lines. Isotropic polarized transients at the frequency position of the Lumi-F marker band, and the system response (grey dotted line) (a) and the ground-state marker band (b) are displayed. Polarization resolved transients for parallel (black lines) and perpendicular (red lines) polarization together with isotropic polarized transients (green and blue dots) are presented at 1682 cm−1 (c) and 1630 cm−1 (d)
The missing contribution of the ring D carbonyl bleaching signal at 1708 cm−1 is converted to a new positive signal of ring D carbonyl stretching vibration in the Lumi-F conformation. This positive product signal exhibits a characteristic absorption at 1724 cm−1. At this frequency position, a bleaching signal due to carbonyl stretching absorption of ring A dominates the dynamics at 0.5 ps in Fig. 2a and 2c. This bleaching contribution decays to zero on a few picoseconds. Within a picosecond Lumi-F is formed and its positive contribution dominates for longer delay times. At around 50 ps the positive signature of Lumi-F at 1724 cm−1 is expected to have similar absolute strength compared to the missing contribution of the ring D carbonyl bleaching signal at 1708 cm−1 as reported for labeled apoprotein and shown in Fig. 2c. In contrast, in the unlabeled apoprotein the positive signal at 1724 cm−1 is nearly absent at 50 ps delay time (Fig. 2a). Thus, we expect an additional negative apoprotein contribution around 1724 cm−1 masking the positive contribution of the PCB at longer delay times. Comparison of the transients at 1724 cm−1 for labeled and unlabeled apoprotein are presented in Fig. 3a. The difference between unlabeled (green line) and labeled (blue line) transients gives a rising negative signal within a picosecond that stays constant up to 50 ps. This negative signal reflects the contribution of unlabeled apoprotein. The frequency position at 1724 cm−1 is characteristic of C = O stretching vibrations of carboxylic acid and carbonyl groups. From Fig. 2b the dichroic ratio for the ring D bleaching signal, DBL, at 1708 cm−1 can be directly estimated as ~ 1.3 (θ = 46°), matching the value derived for labeled apoprotein within the error margins. In contrast, the DLF at 1724 cm−1 deviates strongly from the findings in labeled apoprotein. The perpendicular polarized signal is much smaller compared to the parallel one (Fig. 2b). This can be explained by a negative signal with a stronger contribution for perpendicular than parallel polarization. This means the dichroic ratio D < 1 corresponding to a relative angle of the vtdm to the excited electronic tdm between 54° and 90°.
A broad positive signal around 1680 cm−1 is detected upon photoexcitation (Fig. 2a). The positive contribution at 1690 cm−1 with a smaller shoulder from 1680 to 1660 cm−1 was attributed to the ring D carbonyl stretching signal in the electronic excited state in labeled apoprotein [20]. In the unlabeled apoprotein the positive contribution around 1680 cm−1 is broader and more pronounced. We assign this positive feature to the dynamics of chromophore’s ring D and apoprotein dynamics. In addition, a negative signal of similar strength is observed at 1650 cm−1, both signals decaying on a similar time-scale of about 10 ps. This signature could reflect amide I dynamics. Around 1630 cm−1 a broad negative signal decays on a time-scale of about 10 ps. The transient is shown in Fig. 3d.
Differences between the labeled (red lines) and unlabeled (black lines) protein dynamics can also be identified by comparing their absorbance difference spectra in Fig. 4a. The spectra are scaled to the same bleaching signal at 1708 cm−1 at a delay time of 1 ps. At this delay time notable differences are found around 1680 cm−1 and 1640 cm−1 with stronger positive and negative signals in the unlabeled apoprotein, respectively. These differences become smaller at delay times of 15 ps and 50 ps, and only subtle deviations are visible in this spectral range at 50 ps. In contrast, differences at 1724 cm−1 show up at 15 ps and increase up to 50 ps. Furthermore, small differences around 1700 cm−1 are apparent at 15 ps and 50 ps. This indicates apoprotein contributions in addition to chromophore dynamics around 1724 cm−1 and 1708 cm−1.
Absorbance difference spectra of Pfr upon excitation around 720 nm at selected delay times of 1 ps, 15 ps, and 50 ps. a for 13C/15N labeled apoprotein (red lines) and unlabeled apoprotein (black lines); b Decay associated spectra of the dynamics of unlabeled protein for isotropic polarization; for the unlabeled protein an additional time constant of 12 ps was found; the “const” time constant indicates that dynamics do not change on the time-scale of hundreds of picoseconds; it reflects contributions from Lumi-F (positive) and ground state (negative); the spectra for 0.4 ps is not shown due to interferences with the system response
A global analysis of the dynamics of the unlabeled protein provides four components with time constants of 0.4 ± 0.1 ps, 3.5 ± 0.6 ps, and 12 ± 2 ps, and a constant component. The decay associated spectra (DAS) are presented in Fig. 4b. The shortest time component of 0.4 ps (not shown) is affected by our system response of about 0.35 ps and reflects a mixture of the isomerization time constant of 0.7 ps and nonlinear signals due to temporal pump-probe overlap. The DAS for 3.5 ps in unlabeled apoprotein shows similar spectra features compared to the reported DAS for 5.0 ps in labeled apoprotein [20]. Thus, the 3.5 ps component mainly reflects chromophore relaxation dynamics. This is expected, since the chromophore contributes strongest to the IR difference signal due to its direct excitation.
In the investigated spectral range, we found no dynamics of the 13C/15N labeled apoprotein, since the contributions are down-shifted out of the observed spectral window. In the unlabeled apoprotein, we found an additional time constant of 12 ps that we assign to apoprotein dynamics. We expect to find the same time constant at lower frequencies in the labeled apoprotein.
Upon photoexcitation, the PCB chromophore is promoted to its electronic excited state. A part of the population isomerizes with 0.7 ps and forms Lumi-F, while the remaining part decays back to Pfr with time constants of 0.7 ps and ~ 5 ps [20]. Excited state signatures of the PCB vibrations are clearly visible at 1695 cm−1 with a declining flank down to 1650 cm−1 (in Fig. 2b). These features are gone at 15 ps delay time. In parallel to the PCB signals apoprotein contributions appear within our time resolution persisting longer than 50 ps. Observed apoprotein dynamics show decaying negative signals at 1724 cm−1, 1708 cm−1, 1660 cm−1, and 1640 to 1600 cm−1, while around 1690 cm−1 we found a positive decaying signal. We tentatively assign the positive and negative signals around 1690 cm−1 and 1660 cm−1, respectively, to an amide I signal of the protein backbone close to the chromophore. The backbone carbonyl groups of the “capping helix” (see Fig. 1b, 1c, and 1d) exhibit a carbonyl stretching vtdm between 14° and 60°. The average impact on several carbonyl groups of the “capping helix” should show a dichroic ratio of about 1.7 (θ = 36°). The polarization resolved transients at 1682 cm−1 (Fig. 3c) show a dichroic ratio of about 1.6 (θ = 39°) at ~ 1 ps delay time, matching the expected value for the carbonyl groups of the “capping helix”. The negative signals in the 12 ps DAS ranging from 1670 to 1600 cm−1 are assigned to apoprotein dynamics of backbone and amino acid side chain contributions. A more precise analysis is not possible here. The negative signals at 1724 and 1708 cm−1 should originate from carboxylic or carbonyl C = O stretching vibrations of amino acid side chains or from high-frequency arginine (CN) stretching vibrations.
The constant DAS component in Fig. 4b reflects changes induced by the photoisomerization. Vibrations formed by the photoproduct show positive features, while absent vibrations from the parent state cause negative features. Positive/negative features at 1724 cm−1( +)/1705 cm−1(−) and at 1680 cm−1( +)/1620 cm−1(−) are apparent. The peak pair at 1724 cm−1( +)/1705 cm−1(−) was assigned to ring D carbonyl stretching vibration in the Lumi-F and ground state, respectively. The intensity ratio of this peak pair was found to be about 1:1 in labeled protein, in contrast to the presented ratio of 1:4 in unlabeled protein. We assign this difference to an additional long-living negative signal around 1724 cm−1 from an apoprotein group. The other peak pair at 1680 cm−1( +)/1620 cm−1(−) was not observed in labeled apoprotein. Thus, we assign this peak pair solely to apoprotein groups, in contrast to the assignment of the positive 1680 cm−1 contribution to a chromophore vibration [50]. Possible candidates for apoprotein vibrations are carbonyl groups from the protein backbone, but other contributions from amino acid side chains are also possible. Further studies have to be undertaken to disentangle individual apoprotein contributions.
In the spectral range between 1750 and 1660 cm−1 mainly absorption bands from C = O stretching vibrations of the apoprotein and PCB, and CN vibrations of arginine are expected. Upon 13C/15N apoprotein labeling, the frequencies of the C = O stretching vibrations downshift by about 40 cm−1, while a downshift of about 30 cm−1 is expected for the high-frequency arginine (CN) stretching vibration [50]. In 13C/15N labeled apoprotein the signal with the highest frequency is observed around 1725 cm−1. If the absorption difference signals between 1730 and 1690 cm−1 derived from PCB alone, the absorbance difference spectra between unlabeled and labeled protein would not be distinguishable in this region. This is clearly not the case when comparing unlabeled and labeled apoprotein in Fig. 2a and 2c, respectively. Hence, we observe apoprotein contributions in the ultrafast dynamics prior to and after photoisomerization.
3.4 Apoprotein contributions
Direct comparison of the transients at 1724 cm−1 in unlabeled and labeled apoprotein in Fig. 3a indicate an instantaneous additional negative signal at this spectral position in unlabeled apoprotein persisting up to 50 ps after excitation. An apoprotein bleaching band in the unlabeled apoprotein around 1724 cm−1 should be visible as a red-shifted bleaching band in the 13C/15N labeled apoprotein around 1680 cm−1. In Fig. 4a the comparison between labeled (red lines) and unlabeled (black lines) absorption difference spectra are shown. At 50 ps significant alterations between labeled and unlabeled apoprotein are only visible around 1724 cm−1. The expected red-shifted negative bleaching band around 1680 cm−1 is missing. It seems that this can only be explained by the existence of another positive band at the position of the expected red-shifted bleaching band cancelling out the negative signal. Such a positive chromophore band at 1680 cm−1 was reported and assigned to a Lumi-F product band [50]. In the spectral range between 1660 and 1400 cm−1 various vibrational bands from the protein backbone and side chains contribute to the overall signal. Studies with different labeling strategies, e.g. with and without chromophore labeling, and mutants will unravel specific signal contributions.
Polarization resolved data show a dichroic ratio D < 1 of the negative signal contribution at 1724 cm−1 after photoreaction. This corresponds to a relative angle in the range of 54°–90° with respect to the electronic tdm. The negative signal is generated instantaneously and persists on a time-scale of 50 ps. Such an apoprotein signal could indicate a deprotonation of a carboxylic acid that stays deprotonated after photoisomerization or a long lasting change of bond character of a carbonyl group [34]. Possible candidates for the signal are the carbonyl stretching vibration of the carboxylic acids of Asp207 and Glu196, as well as the carbonyl stretching vibration of the backbone C = O group of Asp207. All these groups are in the vicinity of the chromophore and can easily be affected by the local electric field change of the chromophore in the excited state. Moreover, their C = O orientations exhibit relative angles in the range of 54°–90° to the excited electronic tdm of the chromophore, consistent with a dichroic ratio D < 1.
Up to now, no high-resolution structure of the Cph1 phytochrome in the Pfr state is available. Thus, conclusive evidence on the protonation state of side groups, such as glutamic and aspartic acids are scarce. Structural modelling on the basis of solid-state magic-angle spinning NMR data provide tentative structural information for the Pfr state in Cph1 [12, 55]. Additional experimental data, such as polarization resolved transient IR data, provide insight into heterogeneity of the ground state. On the basis of these data, two ground-state conformations for the Pfr state were reported, with deviating ring D orientations. The down-conformation exhibits a ring D with an NH group pointing below the PCB plane (see Fig. 1c), while the up-conformation has a ring D orientation with NH group pointing above the PCB plane [20]. In these Pfr ground-state models possible protonation of aspartic acids at pD 7.8 were not included. On the basis of our observation of a bleaching band around 1724 cm−1, we expect Asp207 to be protonated in the Pfr state. We changed the protonation state of Asp207 in our model (see Fig. 1c). One CO group of Asp207 is hydrogen-bonded with the NH group of PCB ring D with a distance of 1.85 Å. This C = O group is also hydrogen-bonded to Ser474 with a distance of 1.57 Å, and to Tyr263 with a distance of 2.89 Å (see Fig. 1c). We assign the other CO group of Asp207 to be protonated and hydrogen-bonded to Ser206 with a distance of 2.02 Å (see Fig. 1c). The vtdm of the C = O group of Asp207 shows a relative angle to the electronic tdm of 63° compatible with the measured angle between 55 and 90°. Its calculated frequency was at 1719 cm−1 with a relative intensity of about 40% of the ring D carbonyl. These calculated properties match with our experimental observations. The vtdm of the Asp207 backbone carbonyl is oriented 79° to the electronic tdm, also in agreement with the measured angle between 55 and 90°. Its calculated frequency was found at 1728 cm−1 with a relative intensity of about 5% of the ring D carbonyl. The small calculated relative intensity does not match with experimental observations. In addition, the backbone carbonyl group would have to change its properties completely, e.g. become protonated, to induce a long lasting signal without an adjacent positive contribution. Another possible candidate for the signal at 1724 cm−1 is Glu196, since its carbonyl is also oriented roughly perpendicular to the electronic tdm. However, its carboxylic group is about 12 Å from the oxygen of ring D, making it rather unlikely to be responsible for the instantaneous bleaching signal at 1724 cm−1.
There are a few groups that could be responsible for the observed apoprotein dynamics at 1724 cm−1. We favor the scenario, where a protonated Asp207 becomes deprotonated upon photoexcitation.
The positive band around 1670 cm−1 in unlabeled apoprotein exhibits a dichroic ratio of about 1.8 pointing to a vtdm with a relative angle of about 34° to the electronic tdm. The spectral position matches frequencies of the α-helical amide I vibration. The capping helix (see Fig. 1) consists of many amide I vibrations with relative angles of their C = O group to the electronic tdm between 14° and 60°. Since the vtdm deviates by about 10° from the C = O group direction of the amide group, amide groups in the capping helix are suitable candidates for the observed signal around 1670 cm−1 in unlabeled apoprotein. Most of the other amide groups in the phytochrome exhibit relative angles around 80° to the electronic tdm, and thus do not match the observed signals. The short distance between the capping helix and the chromophore supports such an assignment, since the coupled dipoles of the amide I groups are affected more strongly by an excited state dipole change the closer they are.
Negative signals around 1630 cm−1 are probably due to apoprotein vibrations due to their substantial change upon apoprotein labeling. Nevertheless, assignment is hampered by increasing possibilities of overlapping signals. Further deconvolution of the signal contributions can be performed by including isotopic labeling of the PCB chromophore in future experiments.
4 Discussion
4.1 Ultrafast protein dynamics
Our investigations support the notion that ultrafast photoisomerization of unlabeled PCB chromophore exhibit the same dynamics and quantum yield in unlabeled and 13C/15N labeled apoprotein. Comparison of 13C/15N labeled apoprotein and unlabeled apoprotein reveal ultrafast apoprotein dynamics prior to and after photoisomerization of the PCB chromophore, as revealed by isotope shifts.
At 1724 cm−1 a protein bleaching signal rises upon photoexcitation. The bleaching signal remains almost constant on a time-scale up to 50 ps. We tentatively assign this band to the C = O stretching vibration of Asp207 carboxylic acid that is deprotonated upon photoisomerization. The deprotonated carboxylic acid is expected to be relevant in down-stream processes such as Arg-Asp salt bridge formation [56]. This assignment is supported by polarization resolved data that indicate a relative angle of the vtdm of the carbonyl group to the electronic tdm of PCB between 55° and 90°, matching the calculated angle of 63°. The impact of this aspartic acid on the photoreaction of the Pr state was discussed for bacteriophytochrome RpBphP3 from Rhodopseudomonas palustris as a proton acceptor in the electronic excited state [57]. In this report the backbone C = O group of Asp207 was involved. This process is unlikely for the observed protein signal at 1724 cm−1, because the signal remains after the decay of the electronic excited state up to 50 ps, but cannot be excluded.
Excited state proton transfer from the chromophore to the apoprotein was observed in the Pfr state of Agp2 phytochrome in the electronic excited state. Its decay on a sub-picosecond time-scale is accompanied by the re-protonation of the chromophore [35]. A similar mechanism seems very unlikely in Cph1 Pfr, however since no continuum band is observed.
Alternative assignments of the 1724 cm−1 protein band cannot be excluded. A possibility would be the arginine salt bridge with the deprotonated propionic side chain at ring B. The high frequency CN vibration of arginine in an Arg:Asp salt bridge was simulated to absorb around 20 cm−1 red-shifted to the C = O stretching vibration of ring D [50]. This is in line with other studies assigning arginine salt bridges at lower frequencies around 1620 cm−1 [58]. Hence, the observed apoprotein band exhibits a frequency probably too high for arginine salt bridges. Other glutamic and aspartic acids, perhaps Glu196 could match the frequency and the measured orientation. Since the response of the signal is concomitant with PCB photoisomerization, we expect a direct coupling to the chromophore or a coupling via hydrogen bonds. This is obvious for Asp207, but not clear for other carboxylic acids in the protein.
The positive signal around 1670 cm−1 in unlabeled protein appears with photoexcitation of the Pfr state and decays with a time constant of about 12 ps to a small positive value at 50 ps. Its vtdm exhibits a relative angle to the electronic tdm of about 35°. The underlying chromophore contribution is negligible. This relative angle agrees with many backbone carbonyl groups of the capping helix that is oriented parallel to electronic tdm of PCB (see Fig. 1b). This corresponds to ultrafast structural changes observed in the capping helix by serial femtosecond X-ray crystallography in the Pr state of DrBphP from Deinococcus radiodurans [37].
4.2 Proposed mechanism
Upon excitation of Pfr, the electron density is increased around ring B and decreased around rings C and D of the chromophore [35]. This almost instantaneous event induces an impulsive electric field change around the chromophore. This process has been called a ‘Coulomb Hammer’, altering the coupling strengths of hydrogen bonds between the chromophore and the surrounding protein [36]. Polarizable and charged protein groups will be directly affected by the electric field change, before reorientation of movable groups, e.g. water, accompanied with screening of the electric field takes place. The effects on individual groups can vary considerably depending on polarizability and are currently difficult to quantify. Further studies are needed to unravel transient effects on individual groups.
We assign the apoprotein bleaching band at 1724 cm−1 tentatively to the deprotonation of Asp207. With excitation and the instantaneous decrease of electron density at ring D and increase around ring B the hydrogen-bonding network between the NH group of ring D, Asp207, Ser474, and Tyr263 is altered. We propose a weakening of the hydrogen bond to ring D, facilitating its rotation. This is necessary, because strong hydrogen bonds can impede photoisomerization—as demonstrated for bacteriophytochrome RpBphP2 and RpBphP3 from Rhodopseudomonas palustris [59]. With the weakening of the hydrogen bond to ring D and beginning rotation of ring D, the hydrogen-bonding network rearranges and Asp207 is deprotonated. Up to now, we were not able to identify the proton acceptor. These investigations can be carried out in a more targeted manner once a high-resolution structure of the Cph1 Pfr state becomes available.
Ultrafast apoprotein responses around 1680 cm−1 were assigned to amide I vibrations. We assigned the interacting amide I groups to the capping helix above and parallel to the chromophore. The electric field change of the chromophore polarizes the α-helix and strengthens the couplings of the amide I groups, thereby increasing their absorption resulting in the observed positive signal around 1670 cm−1 [23].
By comparing unlabeled with labeled apoprotein dynamics we were able to identify ultrafast apoprotein responses prior to and after the photoisomerization of the PCB chromophore in Cph1. This highlights the impact of the excitation induced electric field change on the apoprotein response and supports the ‘Coulomb hammer’ mechanism setting the protein up for photoisomerization of the chromophore and down-stream protein changes.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Heyne, K., et al. (2002). Ultrafast dynamics of phytochrome from the cyanobacterium Synechocystis, reconstituted with phycocyanobilin and phycoerythrobilin. Biophysical Journal, 82, 1004–1016.
Rohmer, T., et al. (2010). Phytochrome as molecular machine: revealing chromophore action during the Pfr -> Pr photoconversion by magic-angle spinning NMR spectroscopy. Journal of the American Chemical Society, 132, 9219–9219. https://doi.org/10.1021/Ja104192u
Foerstendorf, H., Lamparter, T., Hughes, J., Gartner, W., & Siebert, F. (2000). The photoreactions of recombinant phytochrome from the cyanobacterium Synechocystis: a low-temperature UV-Vis and FT-IR spectroscopic study. Photochemistry and Photobiology, 71, 655–661.
Foerstendorf, H., et al. (2001). FTIR studies of phytochrome photoreactions reveal the C=O bands of the chromophore: Consequences for its protonation states, conformation, and protein interaction. Biochemistry, 40, 14952–14959.
Kim, P. W., Rockwell, N. C., Martin, S. S., Lagarias, J. C., & Larsen, D. S. (2014). Heterogeneous photodynamics of the P-fr State in the Cyanobacteriai phytochrome Cph1. Biochemistry, 53, 4601–4611.
Muller, M. G., Lindner, I., Martin, I., Gartner, W., & Holzwarth, A. R. (2008). Femtosecond kinetics of photoconversion of the higher plant photoreceptor phytochrome carrying native and modified chromophores. Biophysical Journal, 94, 4370–4382.
Schumann, C., et al. (2008). Subpicosecond midinfrared spectroscopy of the P-fr reaction of phytochrome Agp1 from Agrobacterium tumefaciens. Biophysical Journal, 94, 3189–3197. https://doi.org/10.1529/biophysj.107.119297
Wang, D., et al. (2019). Elucidating the molecular mechanism of ultrafast Pfr state photoisomerization in Bathy Bacteriophytochrome PaBphP. Journal of Physical Chemistry Letters, 10, 6197–6201.
Escobar, F. V., et al. (2015). Conformational heterogeneity of the Pfr chromophore in plant and cyanobacterial phytochromes. Frontiers in Molecular Biosciences. https://doi.org/10.3389/fmolb.2015.00037
Fischer, T., et al. (2022). Influence of the PHY domain on the ms-photoconversion dynamics of a knotless phytochrome. Photochemical and Photobiological Sciences. https://doi.org/10.1007/s43630-022-00245-9
Fischer, T., et al. (2021). Ultrafast photoconversion dynamics of the knotless phytochrome SynCph2. International Journal of Molecular Sciences. https://doi.org/10.3390/ijms221910690
Song, C., et al. (2018). 3D Structures of plant phytochrome A as Pr and Pfr from solid-state NMR: implications for molecular function. Frontiers in Plant Science. https://doi.org/10.3389/fpls.2018.00498
Legris, M., Ince, Y. C., & Fankhauser, C. (2019). Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nature Communications. https://doi.org/10.1038/s41467-019-13045-0
Anders, K., Gutt, A., Gartner, W., & Essen, L. O. (2014). Phototransformation of the red light sensor cyanobacterial Phytochrome 2 from Synechocystis species depends on its tongue motifs. Journal of Biological Chemistry, 289, 25590–25600.
Bischoff, M., Hermann, G., Rentsch, S., & Strehlow, D. (2001). First steps in the phytochrome phototransformation: a comparative femtosecond study on the forward (Pr -> Pfr) and back reaction (Pfr -> Pr). Biochemistry, 40, 181–186.
Essen, L. O., Mailliet, J., & Hughes, J. (2008). The structure of a complete phytochrome sensory module in the Pr ground state. Proceedings of the National Academy of Sciences USA, 105, 14709–14714. https://doi.org/10.1073/pnas.0806477105
Nagano, S., et al. (2020). Structural insights into photoactivation and signalling in plant phytochromes. Nature Plants. https://doi.org/10.1038/s41477-020-0638-y
Takala, H., et al. (2014). Signal amplification and transduction in phytochrome photosensors. Nature, 509, 245–248.
Yang, X., Kuk, J., & Moffat, K. (2008). Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: photoconversion and signal transduction. Proceedings of the National Academy of Sciences USA, 105, 14715–14720. https://doi.org/10.1073/pnas.0806718105
Stensitzki, T., et al. (2017). Influence of heterogeneity on the ultrafast photoisomerization dynamics of Pfr in Cph1 phytochrome. Photochemistry and Photobiology, 93, 703–712.
Yang, Y., Heyne, K., Mathies, R. A., & Dasgupta, J. (2016). Non-bonded interactions drive the sub-picosecond bilin photoisomerization in the P-fr state of phytochrome Cph1. ChemPhysChem, 17, 369–374.
Inomata, K., et al. (2005). Sterically locked synthetic bilin derivatives and phytochrome Agp1 from Agrobacterium tumefaciens form photoinsensitive Pr- and Pfr-like adducts. Journal of Biological Chemistry, 280, 24491–24497.
Gross, R., et al. (2009). Ultrafast protein conformational alterations in bacteriorhodopsin and its locked analogue BR5.12. Journal of Physical Chemistry B, 113, 7851–7860.
Linke, M., et al. (2013). Electronic transitions and heterogeneity of the bacteriophytochrome Pr absorption band: An angle balanced polarization resolved femtosecond VIS pump-IR probe study. Biophysical Journal, 105, 1756–1766.
Fan, G. B., Siebert, F., Sheves, M., & Vogel, R. (2002). Rhodopsin with 11-cis-locked chromophore is capable of forming an active state photoproduct. Journal of Biological Chemistry, 277, 40229–40234.
Zhong, Q., et al. (1996). Reexamining the primary light-induced events in bacteriorhodopsin using a synthetic C-13=C-14-locked chromophore. Journal of the American Chemical Society, 118, 12828–12829.
Foster, K. W., Saranak, J., Krane, S., Johnson, R. L., & Nakanishi, K. (2011). Evidence from Chlamydomonas on the photoactivation of rhodopsins without isomerization of their chromophore. Chemistry and Biology, 18, 733–742. https://doi.org/10.1016/j.chembiol.2011.04.009
Salem, L., & Bruckmann, P. (1975). Conversion of a photon to an electrical signal by sudden polarisation in the N-retinylidene visual chromophore. Nature, 258, 526–528. https://doi.org/10.1038/258526a0
Mathies, R., & Stryer, L. (1976). Retinal has a highly dipolar vertically excited singlet state: implications for vision. Proceedings of the National Academy of Sciences USA, 73, 2169–2173. https://doi.org/10.1073/pnas.73.7.2169
Lewis, A. (1978). The molecular mechanism of excitation in visual transduction and bacteriorhodopsin. Proceedings of the National Academy of Sciences USA, 75, 549–553. https://doi.org/10.1073/pnas.75.2.549
Neumann-Verhoefen, M. K., et al. (2013). Ultrafast infrared spectroscopy on channel rhodopsin-2 reveals efficient energy transfer from the retinal chromophore to the protein. Journal of the American Chemical Society, 135, 6968–6976. https://doi.org/10.1021/ja400554y
Das, I., Pushkarev, A., & Sheves, M. (2021). Light-induced conformational alterations in heliorhodopsin triggered by the retinal excited state. The Journal of Physical Chemistry B, 125, 8797–8804. https://doi.org/10.1021/acs.jpcb.1c04551
Tahara, S., Kuramochi, H., Takeuchi, S., & Tahara, T. (2019). Protein dynamics preceding photoisomerization of the retinal chromophore in bacteriorhodopsin revealed by deep-UV femtosecond stimulated Raman spectroscopy. Journal of Physical Chemistry Letters, 10, 5422–5427.
Stensitzki, T., Adam, S., Schlesinger, R., Schapiro, I., & Heyne, K. (2020). Ultrafast backbone protonation in channel rhodopsin-1 captured by polarization resolved Fs Vis-pump-IR-Probe spectroscopy and computational methods. Molecules. https://doi.org/10.3390/molecules25040848
Yang, Y., et al. (2022). Ultrafast proton-coupled isomerization in the phototransformation of phytochrome. Nature Chemistry, 14, 823–830.
Heyne, K. (2022). Impact of ultrafast electric field changes on photoreceptor protein dynamics. The Journal of Physical Chemistry B, 126, 581–587.
Claesson, E., et al. (2020). The primary structural photoresponse of phytochrome proteins captured by a femtosecond X-ray laser. eLife. https://doi.org/10.7554/eLife.53514
Nogly, P., et al. (2018). Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser. Science. https://doi.org/10.1126/science.aat0094
Kovacs, G. N., et al. (2019). Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin. Nature Communications. https://doi.org/10.1038/s41467-019-10758-0
Yang, Y., et al. (2012). Real-time tracking of phytochrome’s orientational changes during Pr photoisomerization. Journal of the American Chemical Society, 134, 1408–1411.
Yang, Y., et al. (2014). Active and silent chromophore isoforms for phytochrome Pr photoisomerization: an alternative evolutionary strategy to optimize photoreaction quantum yields. Structural Dynamics. https://doi.org/10.1063/1.4865233
Hahn, J., Strauss, H. M., & Schmieder, P. (2008). Heteronuclear NMR investigation on the structure and dynamics of the chromophore binding pocket of the cyanobacterial phytochrome Cph1. Journal of the American Chemical Society, 130, 11170–11178. https://doi.org/10.1021/Ja8031086
Roben, M., et al. (2010). NMR Spectroscopic investigation of mobility and hydrogen bonding of the chromophore in the binding pocket of phytochrome proteins. ChemPhysChem, 11, 1248–1257. https://doi.org/10.1002/cphc.200900897
Schmieder, P., Strauss, H. M., & Hughes, J. (2005). Heteronuclear solution-state NMR studies of the chromophore in cyanobacterial phytochrome Cph1. Biochemistry, 44, 8244–8250. https://doi.org/10.1021/Bi050457r
Song, C., et al. (2011). Two ground state isoforms and a chromophore D-ring photoflip triggering extensive intramolecular changes in a canonical phytochrome. Proceedings of the National Academy of Sciences USA, 108, 3842–3847.
Mroginski, M. A., Mark, F., Thiel, W., & Hildebrandt, P. (2007). Quantum mechanics/molecular mechanics calculation of the Raman spectra of the phycocyanobilin chromophore in alpha-c-phycocyanin. Biophysical Journal, 93, 1885–1894.
Mroginski, M. A., et al. (2011). Elucidating photoinduced structural changes in phytochromes by the combined application of resonance Raman spectroscopy and theoretical methods. Journal of Molecular Structure, 993, 15–25.
Gaussian09 (2009).
Levina, E. O., Lokshin, B. V., Mai, B. D., & Vener, M. V. (2016). Spectral features of guanidinium-carboxylate salt bridges. The combined ATR-IR and theoretical studies of aqueous solution of guanidinium acetate. Chemical Physics Letters, 659, 117–120.
Ihalainen, J. A., et al. (2018). Chromophore-protein interplay during the phytochrome photocycle revealed by step-scan FTIR spectroscopy. Journal of the American Chemical Society, 140, 12396–12404.
Braiman, M. S., Briercheck, D. M., & Kriger, K. M. (1999). Modeling vibrational spectra of amino acid side chains in proteins: effects of protonation state, counterion, and solvent on arginine C-N stretch frequencies. The Journal of Physical Chemistry B, 103, 4744–4750.
Lotze, S., & Bakker, H. J. (2015). Structure and dynamics of a salt-bridge model system in water and DMSO. The Journal of Chemical Physics. https://doi.org/10.1063/1.4918904
Escobar, F. V., et al. (2015). A protonation-coupled feedback mechanism controls the signalling process in bathy phytochromes. Nature Chemistry, 7, 423–430.
Theisen, M., et al. (2009). Femtosecond polarization resolved spectroscopy: A tool for determination of the three-dimensional orientation of electronic transition dipole moments and identification of configurational isomers. The Journal of Chemical Physics. https://doi.org/10.1063/1.3236804
Song, C., et al. (2013). Solid-state NMR spectroscopy to probe photoactivation in canonical phytochromes. Photochemistry and Photobiology, 89, 259–273. https://doi.org/10.1111/php.12029
Mroginski, M. A., et al. (2009). Chromophore structure of cyanobacterial phytochrome Cph1 in the Pr state: reconciling structural and spectroscopic data by QM/MM calculations. Biophysical Journal, 96, 4153–4163.
Toh, K. C., Stojkovic, E. A., van Stokkum, I. H. M., Moffat, K., & Kennis, J. T. M. (2010). Proton-transfer and hydrogen-bond interactions determine fluorescence quantum yield and photochemical efficiency of bacteriophytochrome. Proceedings of the National academy of Sciences of the United States of America, 107, 9170–9175.
Huerta-Viga, A., et al. (2015). The structure of salt bridges between Arg(+) and Glu(-) in peptides investigated with 2D-IR spectroscopy: Evidence for two distinct hydrogen-bond geometries. The Journal of Chemical Physics. https://doi.org/10.1063/1.4921064
Toh, K. C., et al. (2011). Primary Reactions of bacteriophytochrome observed with ultrafast mid-infrared spectroscopy. Journal of Physical Chemistry A, 115, 11985–11997.
Acknowledgements
The work was funded by the Deutsche Forschungsgemeinschaft (DFG) through project-ID 221545957−CRC 1078 ‘Protonation Dynamics in Protein Function’, subproject B07 (K.H., and J.H.), subprojects C02 (M.A.M.) and through DFG grant project HE 5206/3-2 (K.H.) and HU 702/12-1 (J.H.).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
This publication is dedicated to Prof. Silvia E. Braslavsky, a pioneer in photobiology and photobiophysics, on the occasion of her 80th birthday.
The original online version of this article was revised: the footnote on page 1 was missing.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, Y., Stensitzki, T., Lang, C. et al. Ultrafast protein response in the Pfr state of Cph1 phytochrome. Photochem Photobiol Sci 22, 919–930 (2023). https://doi.org/10.1007/s43630-023-00362-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43630-023-00362-z