Abstract
Solvent access to the protein interior plays an important role in the function of many proteins. Phytochromes contain a specific structural feature, a hairpin extension that appears to relay structural information from the chromophore to the rest of the protein. The extension interacts with amino acids near the chromophore, and hence shields the chromophore from the surrounding solvent. We envision that the detachment of the extension from the protein surface allows solvent exchange reactions in the vicinity of the chromophore. This can facilitate for example, proton transfer processes between solvent and the protein interior. To test this hypothesis, the kinetics of the protonation state of the biliverdin chromophore from Deinococcus radiodurans bacteriophytchrome, and thus, the pH of the surrounding solution, is determined. The observed absorbance changes are related to the solvent access of the chromophore binding pocket, gated by the hairpin extension. We therefore propose a model with an “open” (solvent-exposed, deprotonation-active on a (sub)second time-scale) state and a “closed” (solvent-gated, deprotonation inactive) state, where the hairpin fluctuates slowly between these conformations thereby controlling the deprotonation process of the chromophore on a minute time scale. When the connection between the hairpin and the biliverdin surroundings is destabilized by a point mutation, the amplitude of the deprotonation phase increases considerably. In the absence of the extension, the chromophore deprotonates essentially without any “gating”. Hence, we introduce a straightforward method to study the stability and fluctuation of the phytochrome hairpin in its photostationary state. This approach can be extended to other chromophore-protein systems where absorption changes reflect dynamic processes of the protein.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The view of proteins as static structures obtained from X-ray crystallography, has evolved to a more biologically relevant understanding of proteins as dynamic molecules in solution [1,2,3]. This is in part due to advances in experimental techniques, which are able to detect the sparsely populated, transitory states. Such methods include nuclear magnetic resonance (NMR) [4], hydrogen–deuterium exchange (H/D) [5], single molecule fluorescence methods [6], and more recently time-resolved crystallography [7] and time-resolved serial femtosecond crystallography [8, 9]. Notwithstanding, elaborated molecular dynamics simulations have often indicated the role of the dynamics of proteins in their function [2].
A complete overview of protein motion requires experimentation on a range of timescales covering over ten orders of magnitude: from picosecond side-chain rotations, to larger amplitude collective motions taking place on timescales from nanosecond to milliseconds, as well as global or subglobal unfolding events with time constants of seconds to hours. The goal is to determine the relationship between these motions and the function of the protein [10]. For example, the exact role dynamics plays in enzyme catalysis is still not fully understood or agreed upon [11,12,13]. There are numerous examples of enzymes where the active site is protected by loops that open to let substrate in and close to protect the reaction from solvent [14, 15].
Often, solvent dynamics can be linked to the dynamic nature of proteins [16,17,18]. Furthermore, solvent exchange reactions involving water from bulk solvent to the protein interior influence the activity of the enzymes.
Recent studies have reported a link between a decrease in bilin-specific absorption and the deprotonation of the positively charged chromophore, which is embedded within a binding pocket of the phytochrome [19, 20]. Rather than using the above-mentioned extensive methods, we describe here a straightforward measurement approach where the deprotonation process of the biliverdin is linked to a subglobal protein motion, and further, solvent accessibility to the protein interior.
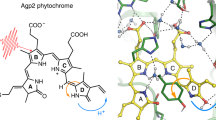
Phytochromes are red light-sensing, multidomain proteins, which generally consist of an N-terminal photosensory module and a C-terminal output module. Here we study the photosensory module of a bacteriophytochrome from Deinococcus radiodurans (DrBphP). This CBD-PHY fragment consists of a chromophore binding domain (CBD) and a phytochrome-specific GAF-related domain (PHY). In the dark, CBD-PHY maintains the structural features characteristic of red-light absorbing state (Pr) shown in ribbon representation in Fig. 1a. The biliverdin (BV) chromophore is covalently bound in a pocket of the CBD, and shown as ball and sticks in Fig. 1a. Absorption of red light in Pr causes isomerization of the BV D-ring which leads to formation of the far red-light absorbing state (Pfr) (Fig. S1), which is accompanied by large-scale structural changes within the CBD-PHY [21]. This photo-conversion acts as a control switch for activity of the output module, like enzymatic activity in two-component signaling [22]. Through these structural processes, phytochromes convert the ambient light cues to the host of biological processes within the cell.
Structure of DrBphP constructs showing solvent protection of the BV binding pocket by the \(\beta\)-stranded hairpin extension A) Ribbon structure of the CBD-PHY dimer with one monomer coloured grey and the other coloured according to domains: the PHY domain is coloured purple and domains that make up the CBD, Per/ARNT/Sim (PAS) and cGMP phosphor diesterase/adenyl cyclase/FhlA (GAF), are coloured green and blue, respectively. The BV chromophore is represented as sticks with carbons coloured grey, nitrogens blue and oxygens red. The pyrrole water is shown as a yellow sphere. The hairpin is circled and tyrosine 263 is shown in yellow in both monomers. B) The coloured monomer subunit in A is shown in space fill representation with an \(\sim\) 90\(^{\circ }\) rotation out of the page along the vertical axis. The BV is protected from solvent by the hairpin extension with only the carbonyl oxygen from the BV A-ring and the pyrrole water visible. C) The CBD is shown in the same orientation as B. Since the PHY domain is absent, the BV binding pocket is more exposed to solvent in this truncated construct. The exposed atoms include oxygen from the BV A-ring (red), the pyrrole water (yellow), an ordered water (pink), as well as the oxygens of the D207 side chain. These atoms are also indicated in the magnification of BV above. The figure was prepared using PDB 4q0j [23] and VMD [24]
A peculiar structural feature of phytochromes is a hairpin extension (circled in Fig. 1a) that initiates from the PHY domain and extends to form close contacts with the CBD [25]. Along the photoactivation of the protein, this extension has been shown to change its fold from \(\beta\)-sheet structure in Pr, to \(\alpha\)-helix in Pfr, with each fold forming unique interactions with the CBD [21]. In Pr, these interactions are extensive enough to completely cover the binding pocket and protect BV from solvent [25] as shown in Fig. 1b. In the truncated CBD construct, shown in Fig. 1c, the BV is solvent-exposed due to the absence of the hairpin extension. Here, we link the movement of the hairpin extension in the dark state to the (de)protonation process of the BV-molecule under basic conditions. This is based on the assumption that the solvent access to the CBD pocket facilitates the proton transfer reaction (Fig. 2). Thus, by inducing a rapid pH change of (sub-)second time-scale and detecting the spectral changes of the biliverdin chromophore due to the changes of the BV protonation state, we are able to indirectly detect slow time-scale fluctuation of the hairpin extension and its effect on the solvent exposure to the CBD domain in the phytochrome in its Pr state.
Cartoon model of BV deprotonation in CBD-PHY in its Pr state. The opening and closing of the hairpin extension is governed by the rate constants k\(_{op}\) and k\(_{cl}\) respectively. When the hairpin extension is open, BV is exposed to solvent and is deprotonated with a rate constant k\(_{dep}\). In this model, it is assumed that in the closed state (red CBD-PHY) BV deprotonation does not occur
2 Results and discussion
The rate of BV deprotonation in the Pr state was determined for three different DrBphP samples, wild-type CBD-PHY (WT), CBD-PHY Y263F variant, and CBD. From previous studies it was shown that a decrease in absorbance at 700 nm with a concomitant increase at 600 nm can be associated with BV deprotonation in DrBphP [20]. Here, a jump to pH 10.8, near the pH titration midpoint for all three samples (Fig. S2), produces a clearly observable change of absorption at 700 nm, describing the decreased population of the protonated form (BV-H\(_4\))\(^+\), while maintaining the functionality of the protein. The time it takes to equilibrate from the “fully protonated” to the “deprotonated” state (BV-H\(_3\)) at pH 10.8 gives information about the solvent access to the binding site.
To confirm the functionality under the experimental conditions, the protein was left at pH 10.8 for 2.5 h and then illuminated with red light (Fig. S3). Consequently, the absorbance decreased at 700 nm and increased at 750 nm, as expected due to photoisomerization of the BV from Pr to Pfr [26]. This transition is reversible: the absorbance spectrum returned to the original state after illumination with far-red light (Fig. S3). However, very long incubation times (four hours or more) tend to lead to protein degradation (Fig. S4). By switching using red and far-red light illuminations, transitions between Pr and Pfr at pH 10.8, with final far-red illumination, can produce the endpoint absorbance values for the Pr state as well (Figs. 3 and S4). In this case, the switching between the states causes unfolding/folding of the hairpin which opens the hairpin extension and allows solvent access to the BV leading to fast deprotonation.
For kinetic data, the absorbance of the samples was measured after 13 s and up to 120 mins after dilution (Fig. 3). Each time point was measured separately after dilution to avoid illumination artifacts. The deprotonation process was followed using the absorbance at 700 nm, which is plotted as function of incubation time after the pH jump (Fig. 3, lower panel). The absorbance kinetics of the CBD-PHY WT and the Y263F variant are best fit to two exponential phases while one is sufficient for CBD (Eq. 1, Table 1). The fitted end-point absorbance for each construct is very similar to corresponding values from pH titrations (Fig. S2), and photoisomerization at pH 10.8 (Fig. S4) confirming little to no measurement artifacts.
2.1 Fast and slow BV deprotonation is observed
For all samples, there is a significant absorbance decrease that occurs within the 13 s experimental dead time. An exponential fit gives a rate constant of about 10 min\(^{-1}\) for this phase (k\(_1\)), reflecting a process of about 6 s and therefore being beyond the time-resolution limit of the experiment. A second, slower phase (k\(_2\)) with a rate constant of 0.013 min\(^{-1}\) corresponding to a time constant of about 76 min (Table 1), exists for the CBD-PHY WT and Y263F samples.
In addition to the rate constants, the amplitudes of the phases give important information. Since each protein has different BV absorbance properties and slightly different pK\(_a\) values, the total absorbance change from pH 8.0 to 10.8 is variable (Fig. 3). For easier comparison, the kinetics are plotted as a function of total amplitude change in Fig. 4. From this and Table 1, the fast phase amplitude A\(_1\) for CBD-PHY WT and Y263F is 47 and 60 % of the total absorbance change. For CBD, only a fast phase is observed and therefore makes up 100% of the amplitude change.
Absorbance-monitored BV deprotonation Top row: absorbance spectra of A CBD-PHY, B CBD-PHY Y263 and C CBD in the Pr state monitored after a jump from pH 8 (thick lines) to pH 10.8 (thin lines). Spectra are normalized to the absorbance at 700 nm of the protein in pH 8.0 buffer. The representative spectra correspond to the time points of the filled circles in the plots below. The equilibrium absorbance at pH 10.8 determined by pH titration experiments (Fig. S2) is indicated by an arrow to the right-hand side axes. Bottom row: The normalized absorbance value at 700 nm is plotted against time (circles), and according to Eq. 1, a free parameter fit to a double or single exponential (red line) is presented. The 700 nm absorbance expected at equilibrium was determined by a jump to pH 10.8, followed by immediate illumination with red light (down arrow) and then driving the sample back to Pr with far red-light (up arrow)(Fig. S4)
Kinetics of BV deprotonation scaled according to the fraction of total amplitude and modelled as in Fig. 2. Absorbance values (filled circles) have been converted to fraction of total amplitude of the exponential fits for easier comparison. At the top, 0 represents no change and 1 represents 100% of the total change. The black (CBD-PHY WT) and blue (CBD-PHY Y263F) solid lines were generated by substituting the corresponding rate constants for k\(_{op}\), k\(_{cl}\) and k\(_{dep}\) in Table 1 into Eqs. S4 and S5.
2.2 Two-state model for BV deprotonation
Although the pH titrations for CBD and CBD-PHY are similar (Fig. S2), CBD deprotonates within seconds while CBD-PHY takes over 2 h to reach the expected endpoint. This is attributed to gating of solvent access to the BV binding site by the extension as outlined in Fig. 2. In theory, the hairpin may assume many conformations and multiple deprotonation pathways; however, the model assumes two general cases: closed and open, which prevents and allows deprotonation, respectively. At neutral pH, the positively charged BV is highly favoured and there is no net change in the protonation state. If the pH is increased above the pK\(_{a}\) of BV, the deprotonation reaction becomes significant. The amplitude of the fast phase reflects the distribution of states at the initial conditions [27]; therefore, the K\(_{eq}\) is reflective of the open and closed populations at pH 8.0.
For CBD, all of the molecules are considered to be in the open state. Hence, the observed single exponential rate constant is taken as a limiting value for k\(_{dep}\). For PHY-containing proteins, the observed deprotonation kinetics are a combination of the microscopic rate constants that define each step in Fig. 2. Since this deprotonation model is analogous to the Linderstrom-Lang model used for H/D exchange (Eq. 3), we can use equations derived for H/D to analyse the deprotonation to obtain k\(_{op}\) and k\(_{cl}\) from the observed kinetics (reviewed in [5, 28]).
The two phases observed for PHY-containing proteins are taken to reflect the relative population of the closed and open states that exist at time zero [27]. This is expected when k\(_{cl}\) and k\(_{op}\) are of similar magnitude so that both the closed and open states are significantly populated [27]. The simplified equations typically used for H/D are therefore not applicable (see Supplementary Information); deprotonation kinetics require the full equation for exchange, described in detail by Qian and Chan [27] and outlined in Supplementary Information in Eqs. S4 and S5.
For CBD-PHY WT and Y263F, the fractional amplitude change of the fast phase is 0.47 and 0.6, respectively. Since these values indicate the fraction of molecules that exist in solution, the values can be used to calculate the equilibrium constant (K\(_{eq}\)) for the transition from open to closed states as shown in Table 1. The remaining slow phase amplitude is due to the population of PHY domain-containing proteins in the closed state. The rate of deprotonation in this case is limited by k\(_{op}\) and can, therefore, be assigned to the rate constant measured for this phase (see Table 1) [27]. This condition is considered the EX1 limit of exchange as in Eq. S3. Using this value for k\(_{op}\) and K\(_{eq}\), the value for k\(_{cl}\) can be calculated as shown in Table 1. These rate constants were substituted into Eqs. S4 and S5 to generate the lines for CBD-PHY WT and Y263F in Fig. 4, demonstrating the applicably of this model. These equations have also used to simulate deprotonation time courses to better understand how the microscopic rate constants affect the observed kinetics (Fig. S6).
Comparison of fAmp\(_1\) in WT and Y263F CBD-PHY suggests more molecules are in the open state in solution for the mutant. Since the slow rate constant is the same for WT and Y263F, this results in a decreased value of k\(_{cl}\) for the mutant. An example of the effect, an even larger selective decrease of k\(_{cl}\) has on deprotonation is shown in the simulated black and red solid lines in Fig. S6.
The results of Y263F are in line with our previous study which shows that the conformation of the hairpin becomes partially uncoupled from the Pr/Pfr transition of the biliverdin chromophore [29]. Specifically, the extension in Y263F is more likely to adopt Pfr-like \(\alpha\)-helical architecture even when BV remains in the Pr state. According to the crystal structure, the hairpin in the Pfr conformation does not protect BV from solvent (Fig. S5) which is observed experimentally as fast deprotonation when CBD-PHY is in the Pfr conformation (Fig. S4). The “closed” hairpin in Y263F likely adopts conformations that behave as open ones, granting solvent access to the binding site.
2.3 Dynamics of the extension
The minute timescales of opening and closing of the extension determined using this method are much slower than typically observed in protein dynamics and for loops in particular [30]. It is not particularly surprising that the dynamics of the extension differs from other mobile loops presented so far: it is formed by 37 residues (Pro443 to His484) and is, therefore, much longer than most loops. Further, it contains clear secondary structures and thus does not fall neatly into any one category of loop architecture [31]. In light of this, it can be even considered as a separate subdomain. Indeed the dynamic behaviour is more like subglobal unfolding; for example, unfolding of the lysozyme \(\beta\)-domain occurs on the second timescale in the naturally occurring mutants I156T and D67H [32]. The opening and closing of the \(\Omega\)-loop in cytosolic phosphoenolpyruvate carboxykinase [33] and in cytochrome c [34] have also been described as folding and unfolding transitions as opposed to active site loops that remain rigid like a swinging gate [14, 15]. To note, dynamic events in the hairpin extension in the dark-adapted phytochromes have been reported by means of NMR (internal dynamics on the \(\mu\)s to ms timescale) [35] and by H/D exchange studies (on the second time-scale) [36].
A folding transition of the extension is known to occur as a part of the function of phytochrome: it readily adopts an \(\alpha\)-helical structure in the light-activated Pfr state shown in Fig. S1 [21, 37]. The idea that proteins have pre-existing pathways of structural organization has gained attention in the last decade [2, 38]. Specifically, the alternate conformations adopted after ligand binding or other modifiers are always accessible to the protein as dynamic substates, even in the absence of the triggering event. It is therefore conceivable that conformational dynamics of the extension occurs in the absence of any light induced BV isomerization. Especially extension residues 472–477 that form a solvent exposed \(\beta\)-strand in the crystal structure [21, 23, 25] but are actually predicted to form an \(\alpha\)-helix based on the primary sequence (Fig. S1).
The stability of the extension in the context of the Pr state as a whole may only be slightly tipped in favour of \(\beta\)-sheet structure. This coincides with the determined K\(_{eq}\), indicating the closed and open states have similar energy levels (Table 1). For Y263F, which disrupts an interaction between the extension and the CBD, the balance seems to be further tipped away from retaining the \(\beta\)-sheet conformation. Although the closed and open states may be close in energy, the slow interconversion suggests a high energy barrier between them. If pre-existing pathways are considered, the extension needs to break contacts with the CBD, unfold, fold to an \(\alpha\)-helix, unfold, and then fold back to a \(\beta\)-sheet reforming contacts. The k\(_{op}\) and k\(_{cl}\) includes a rate constant for all these unfolding and refolding processes.
The ability to act as a solvent gate to the BV binding site is similar to other loops that gate solvent and substrate access to the active site, like in triosephosphate isomerase [39] and lactate dehydrogenase [40]. Presently, a clear functional purpose for this type of gate is missing, although (dynamic) interactions between water molecules and the biliverdin have been indicated, both in the resting state as well as in the intermediate states of the photocycle [41,42,43].
3 Conclusions
Here, we show a method that uses the intrinsic pH dependence of BV absorbance to monitor the equilibrium and kinetic properties of the PHY domain hairpin extension; an interesting and unusual structural feature of phytochromes for which the function has yet to be established.
The method assumes that access to bulk solvent is required for BV deprotonation to proceed and that this access is gated by the extension (Fig. 2). The observed deprotonation kinetics, monitored by absorbance at 700 nm after a jump to high pH, reflect the energetic transition of the hairpin extension from a closed solvent protected, to an open solvent exposed state. To demonstrate that the effect observed in this study originates from the stability of the hairpin extension, we use a truncated construct CBD which lacks the PHY domain, and a CBD-PHY Y263F mutant, which has been shown to drive the equilibrium population of the extension from \(\beta\)-strand to \(\alpha\)-helical structure [21, 29]. For CBD, deprotonation is complete within seconds, while the Y263F mutation increases the amplitude of this fast phase relative to CBD-PHY WT. The rate of BV deprotonation is, therefore, dependent on the presence/stability of the PHY domain.
Energetic parameters were extracted from the observed kinetics using Eqs. S5 and S4 (Table 1). They indicate that the closed and open states are similar in energy but have a high energy barrier between them. This interpretation derives from the fast and slow phases of CBD-PHY WT and Y263F deprotonation: the fast phase coinciding with the significant population of open state at equilibrium, and the slow phase due to rate limiting opening of the extension. We note that with this approach, we are unable to access the mechanism of the proton transfer reaction from the chromophore to the solvent, or the proton exchange reactions within the chromophore binding pocket. To expand the knowledge about these proton transfer events, faster detection methods and additional mutations at the chromophore vicinity would be required.
4 Experimental
4.1 Protein purification
The expression constructs, DrBphP coding domains CBD (residues 1–321) and CBD-PHY (residues 1–502) were kindly provided by the laboratories of Prof. R. D. Vierstra and Prof. K. T. Forest. Cloning of the CBD-PHY Y263F mutant is described elsewhere [29]. For all constructs, the expression vector pET21b(+) containing a C-terminal His6-tag (Novagen) was used. Proteins were expressed and purified as previously outlined [21, 44]. Purified proteins from the final size exclusion chromatography step were concentrated to 30 mg mL\(^{-1}\) in 30 mM Tris pH 8, flash-frozen and stored at \(-80^{\circ }\)C.
4.2 UV–vis spectroscopy
The absorbance measurements were carried out using a Cary 8454 UV–vis spectrometer (Agilent Technologies). For pH jump experiments on a given day, a stock of phytochrome protein was made in 3 mM Tris pH 8.0 with an absorbance value at 700 nm of 0.5. The stock was mixed 1+1 with either 60 mM Tris pH 8.0 or 60 mM glycine pH 10.8 giving a final buffer concentration of 31.5 mM Tris pH 8.0 or 1.5 mM Tris and 30 mM glycine pH 10.8 (confirmed with a pH meter). The final concentration of phytochrome after mixing gave an absorbance at 700 nm of 0.25. The absorption spectrum at pH 8.0 was used as a zero time point and for normalization. For kinetic data, absorbance spectra were measured at specific time intervals after the dilution into pH 10.8 buffer. Using this manual mixing set up, the smallest possible time interval between dilution and measurement is 13 s and is referred to as the experimental dead-time. The longest time interval was 250 min. As each measurement exposes the phytochrome to UV and visible light, repeated measurements were not taken to avoid illumination artifacts; instead, a separate dilution was made for every time interval.
4.3 Data processing and analysis
Absorbance scans were normalized to the absorbance at 700 nm of the time zero pH 8.0 spectra measured on the same day. These absorbance values were plotted as a function of time and fit to the exponential Eq. 1
where A\(_{measured}\) is the normalized absorbance measured at 700 nm, A\(_i\) and k\(_i\) are the amplitude and rate constant for the i\(^{th}\) phase, respectively, t is time and A\(_{endpoint}\) is the final absorbance value.
The equation for BV deprotonation demonstrated in Fig. 2 is written below:
where [(BV-H\(_4\))\(^+\)]\(_{c}\) represents the fully protonated BV in the solvent protected form with the extension in close contact with the GAF domain and [(BV-H\(_4\))\(^+\)]\(_{o}\) represents the solvent exposed form when the extension is separated from GAF, k\(_{op}\) and k\(_{cl}\) are the rate constants for opening and closing of the extension and k\(_{dep}\) is the rate constant for deprotonation of the solvent exposed [(BV-H\(_4\))\(^+\)].
This approach is analogous to the Linderstrom-Lang model for amide exchange [45]:
in which closed represents the closed (protected), protonated state, open represents the open (unprotected) protonated state where exchange is possible and exchanged represents the deuterated state. The k\(_{op}\) and k\(_{cl}\) are the opening and closing rate constants for the structural event leading to exchange, k\(_{int}\) is the intrinsic rate constant for amide hydrogen-deuterium exchange in the open state [46].
References
Frauenfelder, H., Sligar, S., & Wolynes, P. (1991). The energy landscapes and motions of proteins. Science, 254(5038), 1598.
Bahar, I., Chennubhotla, C., & Tobi, D. (2007). Intrinsic dynamics of enzymes in the unbound state and relation to allosteric regulation. Current Opinion in Structural Biology, 17(6), 633.
Frauenfelder, H. (2017). Physical concepts in biology—past and future. Physical Biology, 14(1), 010402.
Peng, J. W. (2012). Exposing the moving parts of proteins with NMR spectroscopy. The Journal of Physical Chemistry Letters, 3(8), 1039.
Konermann, L., Pan, J., & Liu, Y. (2011). Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chemical Society Reviews, 40(3), 1224.
Schuler, B., Lipman, A., & Everett, W. E. (2002). SProbing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature, 419, 743.
Mehrabi, P., Schulz, E. C., Dsouza, R., Müller-Werkmeister, H. M., Tellkamp, F., Miller, R. J. D., & Pai, E. F. (2019). Time-resolved crystallography reveals allosteric communication aligned with molecular breathing. Science, 365, 1167.
Nogly, P., Weinert, T., James, D., Carbajo, S., Ozerov, D., Furrer, A., et al. (2018). Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser. Science, 361, eaat0094.
Claesson, E., Wahlgren, W. Y., Takala, H., Pandey, S., Castillon, L., Kuznetsova, V., Henry, L., Panman, M., Carrillo, M., Kübel, J., Nanekar, R., Isaksson, L., Nimmrich, A., Cellini, A., Morozov, D., Maj, M., Kurttila, M., Bosman, R., Nango, E., Tanaka, R., Tanaka, T., Fangjia, L., Iwata, S., Owada, S., Moffat, K., Groenhof, G., Stojković, E. A., Ihalainen, J. A., Schmidt, M., & Westenhoff, S. (2020). The primary structural photoresponse of phytochrome proteins captured by a femtosecond X-ray laser. eLife, 9, e53514.
Henzler-Wildman, K., Lei, M., Thai, V., Kerns, S., Karplus, M., & Kern, D. (2007). A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature, 450(7171), 913.
Kamerlin, S.C. & Warshel, A. (2010). At the dawn of the 21st century: Is dynamics the missing link for understanding enzyme catalysis?. Proteins, 78(6), 1339.
Olsson, M. H., Parson, W. W., & Warshel, A. (2006). Dynamical contributions to enzyme catalysis: Critical tests of a popular hypothesis. Chemical Reviews, 106(5), 1737.
Zhao, Q. (2017). On the indirect relationship between protein dynamics and enzyme activity. Progress in Biophysics and Molecular Biology, 125, 52.
Gerstein, M., Arthur, M., & Chothia, C. (1994). Structural mechanisms for domain movements in proteins. Biochemistry, 33(22), 6739.
Gora, A., Brezovsky, J., & Damborsky, J. (2013). Gates of enzymes. Chemical Reviews, 113(8), 5871.
Frauenfelder, H., Fenimore, P. W., & Young, R. D. (2007). Protein dynamics and function: Insights from the energy landscape and solvent slaving. IUBMB Life, 59(8–9), 506.
Frauenfelder, H., Fenimore, P., Chen, G., & McMahon, B. (2006). Protein folding is slaved to solvent motions. Proceedings of the National Academy of Sciences of the United States of America, 103(42), 15469.
Lewandowski, J. R., Halse, M. E., Blackledge, M., & Emsley, L. (2015). Direct observation of hierarchical protein dynamics. Science, 348(6234), 578.
Escobar, F. V., Lang, C., Takiden, A., Schneider, C., Balke, J., Hughes, J., et al. (2017). Protonation-dependent structural heterogeneity in the chromophore binding site of cyanobacterial phytochrome Cph1. Journal of Physical Chemistry B, 121(1), 47–57.
Rumfeldt, J. A., Takala, H., Liukkonen, A., & Ihalainen, J. A. (2019). UV-Vis spectroscopy reveals a correlation between Y263 and BV protonation states in bacteriophytochromes. Photochemistry Photobiology, 95(4), 969.
Takala, H., Björling, A., Berntsson, O., Lehtivuori, H., Niebling, S., Hoernke, M., Kosheleva, I., Henning, R., Menzel, A., Ihalainen, J. A., & Westenhoff, S. (2014). Signal amplification and transduction in phytochrome photosensors. Nature, 509(7499), 245.
Multamäki E., Nanekar R., Morozov D., Lievonen T., Golonka D., Wahlgren W. Y., Stucki-Buchli B., Rossi J., Hytönen V. P., Westenhoff S., Ihalainen J. A., Möglich A., Takala H. (2021). Comparative analysis of two paradigm bacteriophytochromes reveals opposite functionalities in two-component signaling. Nat Commun, 12(1), 4394.
Burgie, E. S., Wang, T., Bussell, A. N., Walker, J. M., Li, H., & Vierstra, R. D. (2014). Crystallographic and electron microscopic analyses of a bacterial phytochrome reveal local and global rearrangements during photoconversion. Journal of Biological Chemistry, 289(35), 24573.
Humphrey, W., Dalke, A., & Schulten, K. (1996). VMD: Visual molecular dynamics. Journal of Molecular Graphics, 14(1), 33.
Essen, L. O., Mailliet, J., & Hughes, J. (2008). The structure of a complete phytochrome sensory module in the Pr ground state. Proceedings of the National Academy of Sciences, 105(38), 14709.
Takala, H., Lehtivuori, H., Hammarén, H., Hytönen, V. P., & Ihalainen, J. A. (2014). Connection between absorption properties and conformational changes in Deinococcus radiodurans phytochrome. Biochemistry, 53(45), 7076.
Qian, H., & Chan, S. I. (1999). Hydrogen exchange kinetics of proteins in denaturants: A generalized two-process model. Journal of Molecular Biology, 286(2), 607.
Englander, S. W., Downer, N. W., & Teitelbaum, H. (1972). Hydrogen exchange. Annual Review of Biochemistry, 41(1), 903.
Takala, H., Lehtivuori, H. K., Berntsson, O., Hughes, A., Nanekar, R., Niebling, S., et al. (2018). On the (un)coupling of the chromophore, tongue interactions, and overall conformation in a bacterial phytochrome. Journal of Biological Chemistry, 293(21), 8161.
Fenwick, R., Esteban-Martín, S., & Salvatella, X. (2011). Understanding biomolecular motion, recognition, and allostery by use of conformational ensembles. European Biophysics Journal, 40(12), 1339.
Oliva, B., Bates, P. A., Querol, E., Avilés, F. X., & Sternberg, M. J. (1997). An automated classification of the structure of protein loops. Journal of Molecular Biology, 266(4), 814.
Dumoulin, M., Canet, D., Last, A. M., Pardon, E., Archer, D. B., Muyldermans, S., et al. (2005). Reduced global cooperativity is a common feature underlying the amyloidogenicity of pathogenic lysozyme mutations. Journal of Molecular Biology, 346(3), 773.
Cui, D. S., Broom, A., Mcleod, M. J., Meiering, E. M., & Holyoak, T. (2017). Asymmetric anchoring is required for efficient o-loop opening and closing in cytosolic phosphoenolpyruvate carboxykinase. Biochemistry, 56(15), 2106.
Krishna, M. M., Lin, Y., Rumbley, J. N., & Englander, S. W. (2003). Cooperative omega loops in cytochrome c: Role in folding and function. Journal of Molecular Biology, 331(1), 29.
Gustavsson, E., Isaksson, L., Persson, C., Mayzel, M., Brath, U., Vrhovac, L., et al. (2020). Modulation of structural heterogeneity controls phytochrome photoswitching. Biophysics of Journal, 118(2), 415.
Gourinchas, G., Etzl, S., Göbl, C., Vide, U., Madl, T., & Winkler, A. (2017). Long-range allosteric signaling in red light-regulated diguanylyl cyclases. Science Advances, 3, e1602498.
Burgie, E. S., Zhang, J., & Vierstra, R. D. (2016). Crystal structure of Deinococcus phytochrome in the photoactivated state reveals a cascade of structural rearrangements during photoconversion. Structure, 24(3), 448.
Haliloglu, T., & Bahar, I. (2015). Adaptability of protein structures to enable functional interactions and evolutionary implications. Current Opinion in Structural Biology, 35, 17.
Kursula, I., Salin, M., Sun, J., Norledge, B., Haapalainen, A., Sampson, N., & Wierenga, R. (2004). Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase. Protein Engineering, Design and Selection, 17(4), 375.
McClendon, S., Zhadin, N., & Callender, R. (2005). The approach to the Michaelis complex in lactate dehydrogenase: The substrate binding pathway. Biophysical Journal, 89(3), 2024.
Wagner, J. R., Brunzelle, J. S., Forest, K. T., & Vierstra, R. D. (2005). A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature, 438(7066), 325.
Lenngren, N., Edlund, P., Takala, H., Stucki-Buchli, B., Rumfeldt, J., Peshev, I., et al. (2018). Coordination of the biliverdin D-ring in bacteriophytochromes. Physical Chemistry Chemical Physics, 20(27), 18216.
Ihalainen, J. A., Gustavsson, E., Schroeder, L., Donnini, S., Lehtivuori, H., Isaksson, L., et al. (2018). Chromophore-protein interplay during the phytochrome photocycle revealed by step-scan FTIR spectroscopy. Journal of the American Chemical Society, 140(39), 12396.
Lehtivuori, H., Rissanen, I., Takala, H., Bamford, J., Tkachenko, N. V., & Ihalainen, J. A. (2013). Fluorescence properties of the chromophore-binding domain of bacteriophytochrome from Deinococcus radiodurans. The Journal of Physical Chemistry B, 117(38), 11049.
Hvidt, A., & Nielsen, S. O. (1966). Hydrogen exchange in proteins. Advances in Protein Chemistry, 21, 287.
Milne, B. Y. J., & Englander, L. M. (1993). Primary structure effects on peptide group hydrogen exchange. Proteins, 17(1), 75.
Drozdetskiy, A., Cole, C., & Procter, J. B. G. (2015). Pred4: A protein secondary structure prediction server. Nucleic Acids Research, 43(W1), W389.
Wildes, D., & Marqusee, S. (2004). Hydrogen-Exchange Strategies Applied to Energetics of Intermediate Processes in Protein Folding. Methods in Enzymology, 380, 328.
Clarke, J., & Itzhaki, L. S. (1998). Hydrogen exchange and protein folding. Current Opinion in Structural Biology, 8(1), 112.
Acknowledgements
The authors thank Alli Liukkonen and Heikki Häkkänen for help in protein purification procedures and in the spectroscopic experimental settings, respectively. The research was supported by the Academy of Finland (Grants 332742 for JAI and 330678 for HT).
Funding
Open access funding provided by University of Jyväskylä (JYU).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Pushing the limits of flash photolysis to unravel the secrets of biological electron and proton transfer—a topical issue in honour of Klaus Brettel.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rumfeldt, J., Kurttila, M., Takala, H. et al. The hairpin extension controls solvent access to the chromophore binding pocket in a bacterial phytochrome: a UV–vis absorption spectroscopy study. Photochem Photobiol Sci 20, 1173–1181 (2021). https://doi.org/10.1007/s43630-021-00090-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43630-021-00090-2