Abstract
Antimicrobial resistance is emerging as a global health challenge that requires immediate and concerted attention. Accordingly, the WHO has issued alerts urging to continue developing antibiotics with novel mechanisms of action toward clinically important pathogens, including Acinetobacter baumannii. In this context, fungi have played a crucial role in the discovery and development of antibiotics. Therefore, in this work, three fungal strains were prioritized based on their metabolic profiles and antibacterial activity against a pan-resistant isolate of A. baumannii, to identify potential antibiotic molecules. Chemical investigation of the selected fungi (mangrove endophytes) led to the isolation of asperazine (1), aurasperone B (2), aurasperone F (3), TMC-256A1 (4), fonsecin B (5), dianhydroaurasperone C (6), aurasperone A (7), pyrophen (8), and penicillide (9). Moreover, an in vitro assay to detect ligands of the filamentous temperature-sensitive mutant Z enzyme of A. baumannii (AbFtsZ), a GTPase that plays a central role in bacterial division, was developed to correlate the antibacterial properties of the isolated molecules to a mechanism of action. Compounds 1–4 and 9 inhibited the growth of A. baumannii. Interestingly, compounds 2, 3, and 5–9 interacted with AbFtsZ1-412, increasing its GTPase activity. Conversely, compound 4 exhibited an outstanding ability to act as an inhibitor of both the enzymatic activity and the growth of the strain under study.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Antimicrobial resistance (AMR) has emerged as a major health challenge of the twenty-first century and is considered the most serious infectious syndrome in developing countries (Murray et al. 2022). In 2019, infectious diseases caused by bacterial organisms resulted in 4.95 million deaths, of which 1.27 million were directly attributable to resistant bacterial strains (Murray et al. 2022). The top six pathogens responsible for AMR-related deaths are Escherichia coli, Staphylococcus aureus, Klebsiella pneumoniae, Streptococcus pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa (WHO 2017; Murray et al. 2022). In Mexico, antimicrobial resistance is already a significant healthcare challenge. Major infections are caused by pandrug-resistant A. baumannii, penicillin-resistant E. coli, and third-generation cephalosporins-resistant K. pneumoniae and P. aeruginosa (Founou et al. 2017; Sosa-Hernández et al. 2020). Considering the ever-growing rate of resistant infections, the World Health Organization (WHO) has called for increased efforts in the discovery and development of antibiotics with novel mechanisms of action to combat AMR. If current trends continue, AMR is projected to become the leading cause of death by 2050, resulting in approximately 10 million deaths annually worldwide (O'Neill 2016; Founou et al. 2017; Tacconelli et al. 2018).
One strategy for preventing bacterial growth is to inhibit cell division, which is a fundamental process in the spread of infections. In bacteria, the protein FtsZ (a GTPase enzyme) plays a key role in cell division. It is the first protein to be located at the dividing site (Adams and Errington 2009). Through autopolymerization processes, it assembles a ring-like structure (the Z-ring) that facilitates the generation of two new cells from one. FtsZ consists of two domains connected by an α-helix (Tripathy and Sahu 2019). The N-terminal domain contains a parallel β-strand surrounded by two and three helices and holds the nucleotide binding site. In addition, the C-terminal domain is a central β-strand supported by two helices, and it harbors the T7 loop, which is essential for the formation of the catalytic site (Kusuma et al. 2019). This enzyme offers several advantages as a molecular target for antibiotic development: (i) it is indispensable for bacterial growth (Carro 2019); (ii) it is highly conserved in prokaryotes and absent in eukaryotic animal cells (Carro 2019); (iii) ligands that inhibit or enhance the activity of this protein can disrupt the Z-ring structure, preventing bacterial division (Carro 2019); and (iv) currently there are no approved drugs targeting this protein, even less AbFtsZ ligands (Hurley et al. 2016; Carro 2019).
Based on these considerations, in this study, a multi-informative approach joining in bioassay-guided methodologies was used to find fungal antimicrobial metabolites against a pandrug-resistant A. baumannii strain (A564); to establish a putative molecular target for the isolated molecules, an in vitro assay to monitor the binding properties of candidate molecules toward A. baumannii FtsZ (AbFtsZ) was developed. Chemical investigation of three selected mangrove endophytes identified as Aspergillus sp. IQ-503, Aspergillus sp. IQ-548, and Talaromyces sp. I-567 led to the isolation and characterization of asperazine (1), aurasperone B (2), aurasperone F (3), TMC-256A1 (4), fonsecin B (5), dianhydroaurasperone C (6), aurasperone A (7), pyrophen (8), and penicillide (9). Among these metabolites, compounds 1–4 exhibited bacterial growth inhibition with IC50 values ranging from 6.9 ± 0.7 to 9.9 ± 0.8 µg/ml, while compounds 2, 3, and 5–9 increased the activity of AbFtsZ. In addition, compound 4 acted as the sole inhibitor of the activity of AbFtsZ while inhibiting bacterial growth.
Material and Methods
Fungal Isolation
Biological samples for fungal isolation were collected from lagoons in Guerrero (Tecomate: 16° 41′ 29.6″N 99° 19′ 42.6″W; Chautengo: 16° 36′ 51.5″N 99° 07′ 17.5″W) and Oaxaca (Corralero: 16° 13′ 23.9″N 98° 11′ 07.8″W, Chacahua: 16° 00′ 00.3″N 97° 44′ 16.0″W; Ventanilla: 15° 40′ 20.5″N 96° 34′ 47.5″W), Mexico, by the Rivera-Chávez Natural Products Research Laboratory in February 2020. Samples collected included mangrove soil, leaf, and root samples (from Rhizophora mangle and Laguncularia racemosa). Fungal microorganisms were isolated from soil samples using a serial dilution protocol, starting with a suspension of 1 g of soil in 9 ml of water. From this suspension, four serial dilutions (concentrations of 10−2, 10−3, 10−4, and 10−5 g/ml) were prepared. Then, 100 μl of each solution was plated on potato dextrose agar (PDA) supplemented with amoxicillin (500 mg/l). Mangrove leaves and roots were sampled in small pieces and placed on PDA plates containing amoxicillin. The plates were then incubated at room temperature for 48–72 h until clear growth was observed. Emerging microorganisms were then transferred to new plates. This procedure resulted in a collection of 250 axenic cultures (Jiménez-Arreola et al. 2020).
Antibacterial Activity
Test Microorganisms
The multidrug-resistant clinical bacterial strain of Acinetobacter baumannii (A564) was supplied by the National Institute of Pediatrics, Mexico City. It was cultured in Müller-Hinton broth at 37 °C and then stored in Müller-Hinton broth and sterile glycerol (4:1) at − 80 °C.
Antibacterial Assay
This assay was performed according to the method described in CLSI M07-A10 guidelines (microdilution assay) (CLSI 2015). The bacterial strain was suspended in Müller-Hinton broth at 37 °C for 12 h and the turbidity was adjusted to 0.5 McFarland standard units (108 CFU/ml). Molecules were evaluated at 100 µg/ml and extracts and fractions were evaluated at 250 µg/ml in DMSO. The microdilution setup consisted of 10 µl bacterial suspension, 2 µl sample (extracts/fractions/molecules), and 88 µl Müller-Hinton broth. The optical density of the plate was then measured at 600 nm (OD600) using a plate reader. The plate was then incubated at 37 °C for 24 h, and the measurement was repeated to calculate the inhibition percentage and growth rate. Gentamicin was used as a positive control in the assay (100 µg/ml). Inhibition values were calculated by nonlinear regression using GraphPad Prism 8.0 software.
where S.OD is sample optical density, and C.OD is negative control optical density.
Equation 2 was used to calculate IC50 values using Graph Pad Prism 8.0 software.
where X is the concentration and S is the Hill slope of each curve.
Molecular Network Analysis
Of the initial 250 isolates, 222 were discarded based on the antibacterial activity of their extracts (Fig. S1). The remaining 28 microorganisms were grown on a larger scale to increase the yield of organic extracts, and these extracts were analyzed through molecular networking. The complexity of the extract was then analyzed using UPLC-MS/MS. Chromatographic analyses were performed using an Agilent 1260 Infinity HPLC system coupled to an Agilent G6530BA MS-(ESI+)-SQ-TOF mass spectrometer, and a Gemini NX-C18 reverse phase column (3 µm, 2.0 × 75 mm; Phenomenex, Torrance, CA, USA) was used for separation, with a binary elution system of MeCN-H2O (0.1% formic acid). The gradient started at a ratio of 15:85 and reached 100:0 over 8 min, followed by isocratic hold for 1.5 min at a flow rate of 0.4 ml/min. The parameters for the mass spectrometric analysis were as follows: positive ionization mode in the range of 100–2500 m/z, selecting the two most abundant ions per cycle for fragmentation in automatic mode (Auto MS2). The mass spectra in .d format were converted to .mzML format using the ProteoWizard tool MsConvert (version 3.0.20239). The resulting files were analyzed on the Global Natural Products Social (GNPS) platform (https://gnps.ucsd.edu) using the methodology described by Aron et al. (2020). The data was filtered by removing all MS/MS fragment ions within ± 17 Da of the precursor m/z. MS/MS spectra were window filtered by choosing only the top six fragment ions in the ± 50-Da window throughout the spectrum. The precursor ion mass tolerance was set to 2.0 Da and a MS/MS fragment ion tolerance of 0.5 Da. A network was then created where edges were filtered to have a cosine score above 0.7 and more than six matched peaks. Further, edges between two nodes were kept in the network if and only if each of the nodes appeared in each other’s respective top 10 most similar nodes. Finally, the maximum size of a molecular family was set to 100, and the lowest scoring edges were removed from molecular families until the molecular family size was below this threshold. The spectra in the network were then searched against GNPS spectral libraries. The library spectra were filtered in the same manner as the input data. All matches kept between network spectra and library spectra were required to have a score above 0.7 and at least 6 matched peaks. The molecular networks generated on GNPS were dereplicated using the Derreplicator+ and MolNetEnhancer algorithms (Shannon et al. 2003; Van Der Hooft et al. 2016; Mohimani et al. 2018; Ernst et al. 2019). The criteria considered for a reliable tentative identification were an error < 10 ppm (Δ ppm) and a cosine score > 0.7 (Table S1-S2). Each of the identified compounds was compared with the databases on the GNPS platform (MIADB, CASMI, MASSBANK, and NIST) (Aron, et al. 2020). Molecular networks were visualized using Cytoscape 3.8.1. (Shannon et al. 2003).
Fermentation and Extraction
Seed cultures for each fungus were prepared in centrifuge tubes containing 15 ml of potato dextrose broth (PDB). After 5 days of agitation, the seed cultures were poured onto 10 g of commercially available Cheerios® cereal (× 10). The cultures were allowed to grow and undergo fermentation for a span of 30 days, under regular light and dark cycles and at room temperature. After fermentation, each organic extract was obtained by maceration and liquid-liquid partition. Initially, 80 ml of a 9:1 mixture of AcOEt-Me2CO was added to each replicate. The biomass underwent fragmentation with stirring for 24 h. Following this period, the cultures were vacuum filtered, and the filtrate was concentrated. The extract was reconstituted using a 1:1 mixture of aqueous MeOH (4:1):CH2Cl2. The resulting two-phase system was transferred to a separation funnel. The organic layer was further concentrated in vacuo. Finally, the organic extract was reconstituted with a mixture of MeOH:MeCN (1:1) and hexane. The two-phase mixture was transferred to a separation funnel and the MeOH:MeCN phase was concentrated and stored for further analysis (Jiménez-Arreola et al. 2020).
Analytical Techniques
NMR experiments were performed in CDCl3 or MeOD-d4. Spectra were acquired using a BRUKER-AVANCE 700 spectroscope operating at 700 MHz for 1H and 175 MHz for 13C. All chemical shifts (δ) were referenced to the deuterated solvent peak. The exact masses of compounds were determined using either a JEOL AccuTOF JMS-T100LC spectrometer (HR-DART-MS) with PEG-600 as an internal standard or HPLC (Agilent 1260 Infinity) coupled with MS-(ESI+)-SQ-TOF (Agilent 1260 Infinity-Agilent G6530BA).
HPLC separations were performed on a Waters system (2535 quaternary pump) connected to an autosampler and equipped with PDA (2998) and ELSD (2424) detectors. Data acquisition and analysis were performed using Empower 3 software (Waters). Analytical and semi-preparative HPLC was performed on C18 Gemini-NX columns (5 µm) (4.6 × 250 mm for analytical and 10.0 × 250 mm for semi-preparative; Phenomenex, Torrance, CA, USA) (Rivera-Chávez et al. 2019a, b, c).
Isolation and Purification
The fungal extracts IQ-503 (0.5 g), IQ-548 (1.1 g), and IQ-567 (1.2 g) underwent molecular exclusion chromatography using Sephadex LH-20 as the stationary phase and MeOH as the mobile phase at a flow rate of 2 ml/min. These procedures were performed on a Buchi Pure C-810 instrument equipped with a PDA detector (190–800 nm), resulting in the acquisition of 10 fractions (F1-F10) for IQ-548 fungal extract, 26 fractions (FI-FXXVI) for IQ-503, and 14 fractions (FA-FN) for IQ-567 (Rivera-Chávez et al. 2019a, b, c).
For all the selected fungal extracts, the fractions showing promising antibacterial activity (> 40%, at 250 µg/ml) and good yield (> 10 mg) were selected to proceed with the isolation of the active metabolites. From the extract of the microorganism IQ-548, fractions F4–F7 were selected; for IQ-503, fraction FX was selected; and finally for the extract of fungus IQ-567, fraction FD was chosen. Metabolite isolations were performed by semi-preparative HPLC using a Gemini NX-C18 column, 5 μm (10.0 × 250 mm; Phenomenex, Torrance, CA, USA) as the stationary phase and an elution gradient specific for each fraction (solvent A, MeCN; solvent B, H20:0.1% formic acid). The resolution of fraction F4 was performed using a gradient of 30–50% solvent A for 30 min and led to the isolation of compounds 1 and 2 (tR = 12.9 min, 4 mg; tR = 28.0 min, 1.5 mg, respectively). Resolution of fraction F5 was performed using a gradient of 35–60% solvent A for 30 min and led to the isolation of 7 (tR = 34.0 min, 1.5 mg). Resolution of fraction F6 was performed using a gradient of 30–60% solvent A for 30 min and led to the isolation of 4 (tR = 13.6 min, 6.0 mg) and 5 (tR = 16.5 min, 4.3 mg). Fraction F7 was resolved on the same column using a gradient of 30–50% solvent A for 30 min and led to the isolation of compounds 3 (tR = 25.2 min, 6.0 mg) and 6 (tR = 33.7 min, 4.3 mg). Fraction FX was resolved on the same column using a gradient of 30–70% solvent A for 30 min and led to the isolation of 8 (tR = 12.1 min, 2.4 mg). FD fraction was eluted on the same column using a gradient of 35–65% solvent A for 30 min and led to the isolation of compound 9 (tR = 19.2 min, 8.0 mg).
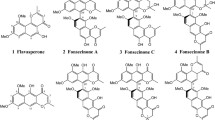
The chemical structure of each compound was determined by comparison of spectroscopic (1D and 2D NMR) and spectrometric (HR-DART-MS or MS-SQ-TOF) data with those reported in the literature: asperazine (1) (Varoglu et al. 1997; Loach et al. 2016), aurasperone B (2) (Priestap 1984; Siriwardane et al. 2015), aurasperone F (3) (Bouras et al. 2005; Antonov et al. 2021), TMC-256A1 (4) (Sakurai et al. 2002), fonsecin B (5) (Priestap 1986), dianhydroaurasperone C (6) (Pilevneli et al. 2021), aurasperone A (7) (Campos et al. 2005), pyrophen (8) (Zhang et al. 2010), and penicillide (9) (Suzuki et al. 1991).
Expression and Purification AbFtsZ1-412
The codon-optimized sequence of the protein filament temperature-sensitive mutant FtsZ1-412 of Acinetobacter baumannii (ID GenBank CU459141.1) was cloned into expression vector pET-28-HT (Addgene), which thrombin site has been changed for TEV site. The construction was amplified by PCR using the specific oligonucleotide FtsZ Fwd 5′-CCGCATATGGCCTCATTTGAATTTATAGAAG-3′ and FtsZ Rev 5′-TTCAAGCTTACTTACGTTGCTGATTTTTC-3′ which introduce 5′-NdeI and 3′-HindIII restriction sites. The resulting vector AbFtsZ1-412 was verified by DNA sequencing (Laragen, Inc) (Chen et al. 2005).
To express the recombinant protein, 20 ml of LB media containing kanamycin (50 µl/ml) and chloramphenicol (30 µl/ml) was inoculated with one colony of previously transformed E. coli Rosetta (DE3) pLysS strain (Novagen, Madison, WI, USA) with AbFtsZ1-412 vector and grown overnight at 37 °C with continuous shaking. The next day, the culture was diluted at 1:20 in fresh LB media with antibiotics and incubated at 37 °C until it reached an OD600 of 0.6. Protein expression was induced by addition 0.5 mM IPTG (Goldbio, USA) at 30 °C for 6 h. The cell pellet was harvested by centrifugation (5500 × g, 10 min, 4 °C) and kept at − 20 °C until required for protein purification.
Purification of FtsZ (A. baumannii) was carried out following the procedure described by Jiménez-Arreola et al. (2020) and Chen et al. (2005) with some modifications. Briefly, the cell pellet from a 500-ml culture was thawed on ice and suspended in 40 ml of lysis buffer (300 mM KCl, 50 mM Tris, and 10% glycerol at pH 8). Lysis was carried out by sonication (Misonix 3000) with an output of 750 W, by intervals of 5 s on and 30 s off for a total of 4 min of total sonication time. The soluble fraction was clarified by centrifugation (45 min at 30,000 × g, 4 °C), after filtration with a PVDF membrane (pore size of 45 µm). AbFtsZ1-412 enzyme was purified by affinity chromatography (His-Trap). The affinity column was equilibrated with lysis buffer. The elution consisted of lysis buffer and imidazole (50 mM). The protein was used immediately or stored at − 80 °C in the same lysis buffer.
Enzymatic Assay of AbFtsZ1-412
This assay was performed according to the methodology reported by Quan and Robinson (2005), Martín-García et al. (2012), and Baykov et al. (1988). Briefly, the assay was completed in 96-well plates with a final volume of 100 μl. Each well contained final concentrations of AbFtsZ1-412 at 4 μM, GTP at 50 μM, test compounds at 100 µg/ml, and berberine at 186 µg/ml (or 500 µM). Assays were performed at pH 7 in assay buffer (300 mM KCl, 50 mM Tris, 2.5 mM MgCl2). The negative control (used for percent inhibition) was prepared by mixing the enzyme (4 μM), GTP (50 μM), assay buffer, and DMSO (sample volume). The plate was then incubated at 37 °C for 30 min, followed by the addition of 20 μl of a final malachite green solution. The plate was further incubated at 37 °C with agitation for 5 min. Finally, the absorbance of each well was read at 630 nm. The percentage of enzyme activity activation was calculated as a percentage relative to DMSO according to Eq. 3. The entire assay procedure up to incubation was performed in a cold room at 4 °C. The final malachite green solution was prepared by combining 500 μl of a stock solution of malachite green (1.22 mg/ml malachite green reagent in sulfuric acid (20% v/v)) with the addition of 125 μl ammonium molybdate (7.5% w/v in deionized water) and 10 μl Tween 20 (11% v/v).
All experiments were performed in quadruple. Data are presented as mean ± standard deviation.
Results and Discussion
This study began with the isolation of fungi associated to Mexican wetlands along the Pacific coast (Guerrero and Oaxaca, Mexico). The outcome of this procedure was a collection of 250 axenic isolates. Then, each microorganism was fermented in a solid medium (PDA). After the incubation period, the organic extract for each fungal strain was prepared and evaluated against multidrug-resistant A. baumannii (Fig. S1). Through this process, the 28 most promising fungi (inhibition greater than 40%: preselected fungi) were selected for up-scaling in a solid medium (Cheerios® cereal), taxonomic identification, and subsequent antibacterial evaluation. This procedure selected strains IQ-503 (Aspergillus sp.), IQ-548 (Aspergillus sp.), and IQ-567 (Talaromyces sp.), from which nine active molecules were isolated (Fig. S1-S2).
Diversity of Mangrove Fungi
Mexico is ranked in the sixth position in mangrove abundance worldwide (Lacerda et al. 1993). Mangroves are highly biodiverse ecosystems that harbor a wide range of species, especially heterotrophic microorganisms such as fungi, with ecological, chemical, and economic importance (Sosa-Rodríguez et al. 2009). These microorganisms perform various functions in this environment, such as decomposition of wood or organic matter from sediments, fragmentation of leaves, and as endophytes in symbiotic processes with mangrove roots and leaves (Sosa-Rodríguez et al. 2009). A review by Devadatha et al. (2021) indicates that approximately 850 mangrove fungi have been identified by 2020, being Xylaria spp., Aspergillus spp., Penicillium spp., Trichoderma spp., and Fusarium spp. the most represented, in agreement with outcomes from this work. In this scenario, endophytic fungi isolated from Mexican mangroves were studied looking for molecules with promising biological activity.
The taxonomic identification of 24/28 preselected microorganisms was performed using molecular methods. For this purpose, the ITS region of the ribosomal DNA was amplified and sequenced. The data are available in GenBank. To show the evolutionary relationships among the organisms, the sequences were then aligned and placed in a phylogenetic tree constructed using the maximum likelihood method (Fig. 1). In this study, 24 nucleotide sequences from fungi associated with Mexican Pacific wetlands and 38 reference sequences were included in the analysis. All identified fungi belong to the orders Botryosphaeriales, Eurotiales, and Hypocreales of the phylum Ascomycota. Within the Eurotiales, the genera Talaromyces, Aspergillus, and Penicillium were the most abundant, while the genera Phyllosticta and Lasiodiplodia, Botryosphaeriales, were less copious.
Prioritization of Microorganisms
Activity Screening
The 28 preselected isolates were cultured in solid medium (Cheerios®) to increase yields. The antibacterial potential of each extract was assessed against A. baumannii to confirm its activity. Extracts with growth inhibition greater than or equal to 40% at 250 µg/ml toward A. baumannii A564 were selected. Some of the most outstanding extracts were (Fig. S3) IQ-503 (Aspergillus sp., 54.3 ± 5), IQ-512 (Aspergillus sp., 75.9 ± 3%), IQ-547 (Fusarium sp., 35.0 ± 3%), IQ-548 (Aspergillus sp. 60.2 ± 2), IQ-567 (Talaromyces sp, 75.0 ± 5), IQ-568 (Penicillium sp., 41.9 ± 6), IQ-573 (Penicillium sp., 42.7 ± 7), and IQ-574 (Talaromyces sp., 40.3 ± 8). Aspergillus and Penicillium correspond to the two strains with the highest reported number of naturally occurring, structurally diverse products, with 2607 and 2192 products, respectively (NPAtlas, 2023).
Untargeted Metabolomic Studies
Simultaneously, the chemical composition information obtained by LC-MS/MS for the 28 extracts was organized into molecular networks using the GNPS platform with the Derreplicator+ and MolNetEnhancer algorithms. The molecular network was constructed with 6123 mass spectra grouped into 930 nodes and categorized into 13 groups of composite families (Fig. 2). The molecular families (clusters) illustrated in the molecular networks are based on the similarity of the mass spectra. The largest group was that of benzoic acids with 108 nodes, whereas the pyrones and pyrans group with 13 nodes was remarkable. In contrast, the smallest group comprised benzodiazepines with only two nodes. Furthermore, this analysis facilitated the dereplication of 32 metabolites (with mass error < 10 ppm and cosine similarity > 0.7, Table S2), including some mycotoxins such as citrinin, harmane, meleagrin, and sterigmatocystin (Fig. 2). It is worth noting that out of the 32 identified molecules, nine have been reported to possess hepatotoxic (Flajs and Peraica 2009), neurotoxic (Lešić et al. 2019), nephrotoxic (Hamed et al. 2021), and/or mutagenic activities (Gupta et al. 2018) (Table S1). This analysis was performed to exclude extracts containing previously reported cytotoxic compounds (15 out of 28: Table S2) thus prioritizing the isolation of chemical entities with promising biological activity toward A. baumannii. Based on the results of antibacterial activity (inhibition > 40%, Fig. S1) and dereplication (Table S1), three extracts were prioritized. The selected microorganisms included Aspergillus sp. IQ-503, Aspergillus sp. IQ-548, and Talaromyces sp. IQ-567, all endophytic microorganisms from the species Rhizophora mangle (red mangrove), collected in Tecomate Lagoon, Guerrero, Mexico.
Chemical Study
The bioassay-guided chemical study of the bioactive extract of Aspergillus sp. IQ-548 led to the isolation of seven molecules characterized as asperazine (1) (Varoglu et al. 1997; Loach et al. 2016), aurasperone B (2) (Siriwardane et al. 2015; Priestap 1986), aurasperone F (3) (Antonov et al. 2021), TMC-256A1 (4) (Sakurai et al. 2002), fonsecin B (5) (Priestap 1986), dianhydroaurasperone C (6) (Pilevneli et al. 2021), and aurasperone A (7) (Campos et al. 2005). In parallel, investigation of Aspergillus sp. IQ-503 and Talaromyces sp. IQ-567 allowed the isolation of pyrophen (8) (Zhang et al. 2010) and penicillide (9) (Suzuki et al. 1991), respectively. All isolated molecules were identified by matching their spectroscopic and spectrometric data with those reported in the literature (Supporting Information).
Asperazine (1) belongs to the diketopiperazine group and inhibits the growth of human leukemia, murine, and human colon cell lines. It was isolated from a marine A. niger (Varoglu et al. 1997). Compounds 2–7 (aurasperone F, TMC-256A1, fonsecin B, dianhydroaurasperone C, and aurasperone A) are benzophenones derived from Aspergillus spp., principally A. niger; however, this kind of molecules and other benzophenones (e.g., penibenzophenones, tenellones, asperphenin) have also been isolated from Penicillium spp., Diaporthe spp., and Phomopsis spp. (Ibrahim et al. 2023). Pyrophen (8) is a pyrone that was first isolated from A. niger from maize in Puerto Rico (Barnes et al. 1990) and later in the species A. brasiliensis (Perrone et al. 2007). Finally, penicillide (9) is a benzodioxocin previously isolated from Penicillium sp., Pestalotiopsis sp., and various Talaromyces species with relevant biological activities (see below) (Salituro et al. 1993; Tao et al. 2017).

After the structural characterization, compounds 1–9 were manually annotated into the constructed molecular networking (Fig. S4). Asperazine (1) falls within the indole derivatives group and can be observed within a two-node cluster. Compounds 2–7 are found in small clusters. Compounds 2 and 3 exhibit a high degree of similarity in their molecular structure, differing only in small changes at positions 2′, 3′, and 8, specifically a dehydration and loss of methoxy group, respectively. Likewise, compounds 6 and 7 also formed a single cluster because these benzophenones have distinct substituents at C-8, hydroxy, and methoxy groups, respectively. However, although compounds 4 and 5 are related, both were found in different clusters. Compound 8 appears as a member of a cluster formed by 10 nodes, indicating that some other analogues of pyrophen are present in fungal extracts. Penicillide (9) was found only in a single node.
Antimicrobial Potential of Isolates
The antimicrobial potential of the isolated molecules was determined according to the CLSI guidelines (CLSI 2015). The results of this evaluation showed that compounds 1 (IC50 6.9 ± 0.7 µg/ml), 2 (IC50 9.9 ± 0.8 µg/ml), 3 (IC50 8.3 ± 0.9 µg/ml), 4 (IC50 7.4 ± 1 µg/ml), and 9 (40 ± 9% inhibition, at 100 µg/ml) inhibited the growth of multidrug-resistant A. baumannii A564 (Fig. 3A and Table S3) in a concentration-dependent manner, with IC50 values below 10 µg/ml for compounds 1–4. Compound 9 displayed an inhibition of 40 ± 9% at 100 µg/ml. In all cases, the inhibition of bacterial growth exceeded that of the positive control gentamicin (20 ± 1% at 100 µg/ml). These results highlight the potential of benzophenones 2–4 and asperazine (1) as suitable candidates for developing antibacterial agents against A. baumannii A564, an intrahospital isolate resistant to last-generation drugs, including ciprofloxacin, gentamicin, meropenem, imipenem, doripenem, cefepime, ceftriaxone, ceftazidime, piperacillin, and ampicillin. In addition, it shows moderate resistance to colistin, a last-generation antibiotic (García-Patiño et al. 2017; Raorane et al. 2019). Based on these findings, compounds 1–4 and 9 are proposed as possible references for developing antibiotics to help combat AMR, particularly against A. baumannii, as it has become one of the main causes of resistant infections worldwide.
In 2018, Zulqarnain et al. (2020) reported the antifungal activity of asperazine (1) against Fusarium oxysporum (at a minimum inhibitory concentration of 60 µg/ml). Furthermore, a more recent study in 2021 showed that 1 inhibited the proliferation of cervical cancer cells (Abdou et al. 2021). To date, this is the first report of this molecule as a growth inhibitor of a multidrug-resistant A. baumannii. Compounds 2 and 3, aurasperones B and F, are benzophenones with antioxidant (Leutou et al. 2016; Zhang et al. 2007), antifungal (Zhang et al. 2007), anticancer (Antonov et al. 2021; Fang et al. 2016), and antimicrobial activities against B. subtilis and E. coli (Bouras et al. 2005; Song et al. 2004). However, there are no reports of these molecules as inhibitors of multidrug-resistant A. baumannii strains. Compound 4 belongs to the naphthopyrone group and has been reported as an antioxidant (Leutou et al. 2016); furthermore, it exhibits anticancer activity against human cervical (Sakurai et al. 2002), liver (Huang et al. 2011), brain (Huang et al. 2011), and mama (Huang et al. 2011) cells and showed inhibition of IgE antibody activity, leading to its proposal as an antiallergic agent (Sakurai et al. 2002); to date, this is the first report of compound 4 as a bacterial inhibitor. Penicillide (9) has been reported to be an anticholesterolemic (Zeng et al. 2022), anticancer (Tao et al. 2017), and antimicrobial agent against strains such as S. aureus, S. albus, Vibrio alginolyticus, and V. parahaemolyticus (Song et al. 2022). This is the first report of compounds 1, 2, 3, 4, and 9 as growth inhibitors of A. baumannii.
AbFtsZ1-412 as a Putative Mechanism of Action
To establish a possible mechanism of action for the isolated molecules with antimicrobial potential, the enzyme AbFtsZ1-412 was selected as a candidate. This selection was based on the fact that molecules with similar nuclei to compounds 2–7, such as mycopyranone and viriditoxin (binaphthopyranones), interact with FtsZ proteins of S. aureus and E. coli (Rivera-Chávez et al. 2019a, b, c), suggesting this protein as a putative molecular target. To test this hypothesis, AbFtsZ1-412 was cloned and expressed in a heterologous host (Supporting Information).
Enzymatic Assay
Once the enzyme was obtained, a photometric assay was developed in a 96-well plate format at 37 °C to screen pure molecules as protein ligands. For this assay AbFtsZ1-412 (4 μM) and GTP (50 μM) were used in a final volume of 100 μl at pH 7. Assay conditions were adapted from the literature with minor modifications (Quan and Robinson 2005, Baykov et al. 1988, Martín-García et al. 2012). Malachite green was used to monitor enzyme activity and inhibition at 630 nm. To validate the ability of the assay to detect molecules capable of altering the GTPase activity of AbFtsZ1-412, berberine was used as a positive control. This alkaloid has previously been reported as an inhibitor of the FtsZ enzyme in S. aureus (Domadia et al. 2008). In this study, berberine was found to inhibit AbFtsZ1-412 activity by no more than 50% at a concentration of 186 µg/ml (500 μM) (Fig. S6).
Compounds as AbFtsZ Binders
Compounds 1–9 were evaluated against AbFtsZ1-412 at 100 µg/ml. Compound 4 showed inhibition of AbFtsZ1-412 activity, while compounds 2–3 and 5–9 showed interaction with the enzyme and enhanced its activity (Fig. 3B, Table S3). In addition, compound 4 also inhibited bacterial growth of A. baumannii A564 strain, suggesting AbFtsZ as a possible molecular target. In 2008, Rai et al. (2008) found that curcumin activates the GTPase activity of the FtsZ enzyme in E. coli. Similarly, in 2012, Ma et al. (2013) reported a group of trisubstituted benzimidazoles that activate GTPase activity of FtsZ in M. tuberculosis. In addition, vitamin K increases the GTPase activity of Streptococcus pneumonia FtsZ enzyme (Pushpakaran et al. 2022). In these reports, the authors discussed that increasing the enzyme activity raises the concentration of GDP in the bacterial cell, leading to depolymerization of the Z-ring and subsequently inhibiting cell division, resulting in enlargement and finally death. None of the FtsZ binders (2–9) identified in this study has previously been reported as ligands of any FtsZ enzyme.
Compound 5, fonsecin B, has been previously reported (Carboué et al. 2019) to possess antioxidant capacity. Lee et al. (2010) demonstrated that fonsecin B has the ability to affect the expression of specific genes involved in the production of immunoglobulins in germinal cells. Compound 6, dianhydroaurasperone C, is also a naphthopyrone with activity against skin cancer. This compound increases the intracellular concentration of vinblastine in KB-8-5 cancer cells and prevents its efflux, potentially enhancing the efficacy of vinblastine in the treatment of drug-resistant cancer (Ikeda et al. 1990). Compound 7, aurasperone A, has been reported to have antibacterial activity against B. subtilis, E. coli, and P. fluorescence, and antifungal activity against Candida albicans and Trichophyton rubrum (Lu et al. 2014). No inhibitory activity against A. baumannii was detected in this study, but it acts as a ligand for the enzyme AbFtsZ1-412. Moreover, pyrophen (8) and penicillide (9) showed significant biological activity as AbFtsZ ligands.
In prior studies, pyrophen (8) was found to hinder the growth of Aeromonas hydrophila (Agrawal et al. 2020), Micrococcus luteus (Agrawal et al. 2020), and Listeria innocua (Agrawal et al. 2020). Additionally, penicillide (9) exhibited inhibition against the microorganisms S. aureus (Song et al. 2004), C. albicans (Song et al. 2004), and E. coli (Song et al. 2004). This is the initial report of these compounds (2–9) with activity against FtsZ of A. baumannii.
Based on these considerations, it is proposed that compounds 2–9 have the potential to be FtsZ-targetting antimicrobials. The bacterial growth inhibition observed in A. baumannii by compounds 2–4 and 9 supports this hypothesis. Despite being activators of the AbFtsZ protein, compounds 5–8 did not demonstrate bacterial growth inhibition against A. baumannii, suggesting the activation of varied resistance mechanisms in A. baumannii, including changes in membrane permeability that inhibit the entry of molecules, as well as effluence of xenobiotics mediated by efflux pumps (García-Patiño et al. 2017; Raorane et al. 2019; Kyriakidis et al. 2021). Based on a structural analysis of compounds 2 and 3 in comparison to 6 and 7, it can be inferred that having a hydroxy group at position C-2 in molecules 2 and 3 affects their antibacterial activity, probably due to the hybridization (sp3) of C-2 and in consequence, the geometry of itself. In contrast, compounds 6 and 7 lacking this moiety displayed no inhibition against multi-resistant A. baumannii. There is currently no other documented fungal inhibitor AbFtsZ1-412 enzyme that simultaneously inhibits bacterial growth. Therefore, compound 4 presents a potential opportunity to develop antibiotics targeting a novel molecular pathway.
Conclusions
Natural products from fungi have often been an important source for antimicrobial drug development. In this study, 250 axenic fungal extracts from Mexican mangroves were screened against a multidrug-resistant strain of Acinetobacter baumannii. Bioassay-guided studies combined with untargeted metabolomic, taxonomic analyses, and dereplication tools enabled the prioritization of the most promising strains for isolating molecules with antimicrobial potential. By applying these criteria, the chemical study of three prioritized fungal isolates resulted in the isolation of nine molecules (1–9) with biological activity, either as inhibitors of bacterial growth or as ligands of the A. baumannii FtsZ enzyme.
Compound 4 (TMC-256A1) is the first reported fungal inhibitor of the AbFtsZ1-412 enzyme. Furthermore, this molecule inhibited bacterial growth of the multi-resistant clinical strain A. baumannii A564. Conversely, compounds 2, 3, and 9 increased FtsZ enzyme activity and showed antibacterial properties. Compounds 1–3 showed antibacterial activity only against A. baumannii A564, whereas products 5–8 interacted with AbFtsZ1-412 and enhanced its enzymatic activity. In brief, this study highlights the importance of studying the mycobiota of under-explored ecosystems in Mexico to obtain and uncover molecular scaffolds to develop antibiotics with different structural classes and novel targets that can contribute to the fight against antimicrobial resistance.
Data Availability
All the data generated in this work are available on request through the corresponding author.
References
Abdou R, Alqahtani AM, Attia GH (2021) Anticancer natural products from Aspergillus neoniger, an endophyte of Ficus carica. Bull Natl Res Cent 4:74. https://doi.org/10.1186/s42269-021-00536-8
Adams DW, Errington J (2009) Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol 7:642–653. https://doi.org/10.1038/nrmicro2198
Agrawal S, Deshmukh SK, Reddy MS, Prasad R, Goel M (2020) Endolichenic fungi: a hidden source of bioactive metabolites. S Afr J Bot 134:163–186. https://doi.org/10.1016/j.sajb.2019.12.008
Antonov AS, Leshchenko EV, Zhuravleva OI, Dyshlovoy SA, Von Amsberg G, Popov RS, Denisenko VA, Kirichuk NN, Afiyatullov SS (2021) Naphto-Γ-pyrones from the marine-derived fungus Aspergillus foetidus. Nat Prod Res 35:131–134. https://doi.org/10.1080/14786419.2019.1610954
Aron AT, Gentry EC, McPhail KL, Nothias LF, Nothias-Esposito M, Bouslimani A, Petras D, Gauglitz JM, Sikora N, Vargas F (2020) Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat Protoc 15:1954–1991. https://doi.org/10.1038/s41596-020-0317-5
Barnes CL, Steiner JR, Torres E, Pacheco R, Marquez H (1990) Structure and absolute configuration of pyrophen, a novel pryrone derivative of L-phenylalanine from Aspergillus niger. Int J Pept Protein Res 36:292–296. https://doi.org/10.1111/j.1399-3011.1990.tb00981.x
Baykov A, Evtushenko O, Avaeva S (1988) A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal Biochem 171:266–270. https://doi.org/10.1016/0003-2697(88)90484-8
Bouras N, Mathieu F, Coppel Y, Lebrihi A (2005) Aurasperone F–a new member of the naphtho-gamma-pyrone class isolated from a cultured microfungus, Aspergillus niger C-433. Nat Prod Res 19:653–659. https://doi.org/10.1080/14786410412331286955
Breda A, Valadares NF, de Souza ON, Garratt RC (2006) Protein structure, modelling and applications. In: Gruber A, Durham AM, Huynh C et al (eds) Bioinformatics in tropical disease research: a practical and case-study approach, Chapter A06. National Center for Biotechnology Information, US, pp 1–3
Campos FR, Barison A, Daolio C, Ferreira AG, Rodrigues-Fo E (2005) Complete 1H and 13C NMR assignments of aurasperone A and fonsecinone A, two bis-naphthopyrones produced by Aspergillus aculeatus. Magn Reson Chem 43:962–965. https://doi.org/10.1002/mrc.1654
Carboué Q, Maresca M, Herbette G, Roussos S, Hamrouni R, Bombarda I (2019) Naphtho-gamma-pyrones produced by Aspergillus tubingensis G131: new source of natural nontoxic antioxidants. Biomolecules 10:29. https://doi.org/10.3390/biom10010029
Carro L (2019) Recent progress in the development of small-molecule FtsZ inhibitors as chemical tools for the development of novel antibiotics. Antibiotics 8:217. https://doi.org/10.3390/antibiotics8040217
Chen Y, Bjornson K, Redick SD, Erickson HP (2005) A rapid fluorescence assay for FtsZ assembly indicates cooperative assembly with a dimer nucleus. Biophys J 88:505–514. https://doi.org/10.1529/biophysj.104.044149
CLSI (2015) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically M07-A10 Standard. Clinical Laboratory Standards Institute 32:24-29
Detlefsen N, Hauberg S, Boomsma W (2022) Learning meaningful representations of protein sequences. Nat Commun 13:1914. https://doi.org/10.1038/s41467-022-29443-w
Devadatha B, Jones E, Pang K, Abdel-Wahab M, Hyde K, Sakayaroj J, Bahkali A, Calabon M, Sarma V, Sutreong S (2021) Occurrence and geographical distribution of mangrove fungi. FUngal Diversity 106:137–227. https://doi.org/10.1007/s13225-020-00468-0
Domadia PN, Bhunia A, Sivaraman J, Swarup S, Dasgupta D (2008) Berberine targets assembly of Escherichia coli cell division protein FtsZ. Biochemistry 47:3225–3234. https://doi.org/10.1021/bi7018546
Ernst M, Kang KB, Caraballo-Rodríguez AM, Nothias LF, Wandy J, Chen C, Wang M, Rogers S, Medema MH, Dorrestein PC (2019) MolNetEnhancer: enhanced molecular networks by integrating metabolome mining and annotation tools. Metabolites 9:144. https://doi.org/10.3390/metabo9070144
Fang W, Lin X, Wang J, Liu Y, Tao H, Zhou X (2016) Asperpyrone-type bis-naphtho-γ-pyrones with COX-2–inhibitory activities from marine-derived fungus Aspergillus niger. Molecules 21:941. https://doi.org/10.3390/molecules21070941
Flajs D, Peraica M (2009) Toxicological properties of citrinin. Arh Hig Rada Toksikol 60:457. https://doi.org/10.2478/10004-1254-60-2009-1992
Founou RC, Founou LL, Essack SY (2017) Clinical and economic impact of antibiotic resistance in developing countries: a systematic review and meta-analysis. PLoS ONE 12:e0189621. https://doi.org/10.1371/journal.pone.0189621
García-Patiño MG, García-Contreras R, Licona-Limón P (2017) The immune response against Acinetobacter baumannii, an emerging pathogen in nosocomial infections. Front Immunol 8:441. https://doi.org/10.3389/fimmu.2017.00441
Gupta RC, Srivastava A, Lall R (2018) Ochratoxins and citrinin. In: Gupta RC. Veterinary toxicology, Chapter 72, Academic Press, pp 1019–1027. https://doi.org/10.1016/B978-0-12-811410-0.00072-6
Hamed A, Abdel-Razek AS, Araby M, Abu-Elghait M, El-Hosari DG, Frese M, Soliman HS, Stammler HG, Sewald N, Shaaban M (2021) Meleagrin from marine fungus Emericella dentata Nq45: crystal structure and diverse biological activity studies. Nat Prod Res 35:3830–3838. https://doi.org/10.1080/14786419.2020.1741583
Huang HB, Xiao ZE, Feng XJ, Huang CH, Zhu X, Ju JH, Li MF, Lin YC, Liu L, She ZG (2011) Cytotoxic naphtho-γ-pyrones from the mangrove endophytic fungus Aspergillus tubingensis (GX1-5E). Helv Chim Acta 94:1732–1740. https://doi.org/10.1002/hlca.201100050
Hurley KA, Santos TM, Nepomuceno GM, Huynh V, Shaw JT, Weibel DB (2016) Targeting the bacterial division protein FtsZ. J Med Chem 59:6975–6998. https://doi.org/10.1021/acs.jmedchem.5b01098
Ibrahim SR, ALsiyud DF, Alfaeq AY, Mohamed SG, Mohamed GA (2023) Benzophenones-natural metabolites with great Hopes in drug discovery: structures, occurrence, bioactivities, and biosynthesis. RSC Adv 13:23472-23498. https://doi.org/10.1039/D3RA02788K
Ikeda SI, Sugita M, Yoshimura A, Sumizawa T, Douzono H, Nagata Y, Akiyama SI (1990) Aspergillus species strain m39 produces two naphtho-γ-pyrones that reverse drug resistance in human KB cells. Int J Cancer 45:508–513. https://doi.org/10.1002/ijc.2910450323
Jiménez-Arreola BS, Aguilar-Ramírez E, Cano-Sánchez P, Morales-Jiménez J, González-Andrade M, Medina-Franco JL, Rivera-Chávez J (2020) Dimeric phenalenones from Talaromyces sp. (IQ-313) inhibit hPTP1B1-400: insights into mechanistic kinetics from in vitro and in silico studies. Bioorg Chem 101:103893. https://doi.org/10.1016/j.bioorg.2020.103893
Kusuma KD, Payne M, Ung AT, Bottomley AL, Harry EJ (2019) FtsZ as an antibacterial target: status and guidelines for progressing this avenue. ACS Infect Dis 5:1279–1294. https://doi.org/10.1021/acsinfecdis.9b00055
Kyriakidis I, Vasileiou E, Pana ZD, Tragiannidis A (2021) Acinetobacter baumannii antibiotic resistance mechanisms. Pathogens 10:373. https://doi.org/10.3390/pathogens10030373
Lacerda L, Carvalho CEV, Tanizaki K, Ovalle ARC, Rezende CE (1993) The biogeochemistry and trace metals distribution of mangrove rhizospheres. Biotropica 25:252–257. https://doi.org/10.2307/2388783
Lee HM, Chan DSH, Yang F, Lam HY, Yan SC, Che CM, Ma DL, Leung CH (2010) Identification of natural product fonsecin B as a stabilizing ligand of c-myc G-quadruplex DNA by high-throughput virtual screening. Chem Commun 46:4680–4682. https://doi.org/10.1039/B926359D
Lešić T, Kmetič I, Kiš M, Vulić A, Kudumija N, Zadravec M, Murati T, Pleadin J (2019) Sterigmatocistin–prekursor aflatoksina B1 u hrani i hrani za životinje. Croatian J Food Technol Biotechnol Nutr 14:105–112. https://doi.org/10.31895/hcptbn.14.3-4.2
Leutou AS, Yun K, Son BW (2016) Induced production of 6, 9-dibromoflavasperone, a new radical scavenging naphthopyranone in the marine-mudflat-derived fungus Aspergillus niger. Arch Pharmacal Res 39:806–810. https://doi.org/10.1007/s12272-016-0764-2
Loach RP, Fenton OS, Movassaghi M (2016) Concise total synthesis of (+)-asperazine,(+)-pestalazine a, and (+)-iso-pestalazine A. Structure revision of (+)-pestalazine a. J Am Chem Soc 138:1057–1064. https://doi.org/10.1021/jacs.5b12392
Lu S, Tian J, Sun W, Meng J, Wang X, Fu X, Wang A, Lai D, Liu Y, Zhou L (2014) Bis-naphtho-γ-pyrones from fungi and their bioactivities. Molecules 19:7169–7188. https://doi.org/10.3390/molecules19067169
Ma S, Cong C, Meng X, Cao S, Yang H, Guo Y, Lu X, Ma S (2013) Synthesis and on-target antibacterial activity of novel 3-elongated arylalkoxybenzamide derivatives as inhibitors of the bacterial cell division protein FtsZ. Bioorg Med Chem Lett 23:4076–4079. https://doi.org/10.1016/j.bmcl.2013.05.056
Martín-García F, Salvarelli E, Mendieta-Moreno JI, Vicente M, Mingorance J, Mendieta J, Gómez-Puertas P (2012) Molecular dynamics simulation of GTPase activity in polymers of the cell division protein FtsZ. FEBS Lett 586:1236–1239. https://doi.org/10.1016/j.febslet.2012.03.042
Mohimani H, Gurevich A, Shlemov A, Mikheenko A, Korobeynikov A, Cao L, Shcherbin E, Nothias LF, Dorrestein PC, Pevzner PA (2018) Dereplication of microbial metabolites through database search of mass spectra. Nat Commun 9:4035. https://doi.org/10.1038/s41467-018-06082-8
Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Aguilar GR, Gray A, Han C, Bisignano C, Rao P, Wool E (2022) Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399:629–655. https://doi.org/10.1016/S0140-6736(21)02724-0
O'Neill J (2016) Tackling drug-resistant infections globally: final report and recommendations. Government of the United Kingdom, https://apo.org.au/node/63983. Accessed 20 May 2023
Perrone G, Susca A, Cozzi G, Ehrlich K, Varga J, Frisvad J, Meijer M, Noonim P, Mahakarnchanakul W, Samson R (2007) Biodiversity of Aspergillus species in some important agricultural products. Stud Mycol 59:53–66. https://doi.org/10.3114/sim.2007.59.07
Pilevneli AD, Ebada SS, Kaşkatepe B, Konuklugil B (2021) Penicacids H-J, three new mycophenolic acid derivatives from the marine-derived fungus Rhizopus oryzae. RSC Adv 11:34938–34944. https://doi.org/10.1039/D1RA07196C
Priestap HA (1984) New naphthopyrones from aspergillus fonsecaeus. Tetrahedron 40:3617–3624. https://doi.org/10.1016/S0040-4020(01)88792-5
Priestap HA (1986) 13C NMR spectroscopy of naphtho-γ-pyrones. Magn Reson Chem 24:875–878. https://doi.org/10.1002/mrc.1260241006
Pushpakaran A, Battaje RR, Panda D (2022) Vitamin K3 inhibits FtsZ assembly, disrupts the Z-ring in Streptococcus pneumoniae and displays anti-pneumococcal activity. Biochem J 479:1543–1558. https://doi.org/10.1042/BCJ20220077
Quan A, Robinson PJ (2005) Rapid purification of native dynamin I and colorimetric GTPase assay. Methods Enzymol 404:556–569. https://doi.org/10.1016/S0076-6879(05)04049-8
Rai D, Singh JK, Roy N, Panda D (2008) Curcumin inhibits FtsZ assembly: an attractive mechanism for its antibacterial activity. Biochem J 410:147–155. https://doi.org/10.1042/BJ20070891
Raorane CJ, Lee JH, Kim YG, Rajasekharan SK, García-Contreras R, Lee J (2019) Antibiofilm and antivirulence efficacies of flavonoids and curcumin against Acinetobacter baumannii. Front Microbiol 10:990. https://doi.org/10.3389/fmicb.2019.00990
Rivera-Chávez J, Caesar LK, Garcia-Salazar JJ, Raja HA, Cech NB, Pearce CJ, Oberlies NH (2019a) Mycopyranone: a 8,8ˈ-binaphthopyranone with potent anti-MRSA activity from the fungus Phialemoniopsis sp. Tetrahedron Lett 60:594–597. https://doi.org/10.1016/j.tetlet.2019.01.029
Rivera-Chávez J, El-Elimat T, Gallagher JM, Graf TN, Fournier J, Panigrahi GK, Deep G, Bunch RL, Raja HA, Oberlies NH (2019b) Delitpyrones: α-pyrone derivatives from a freshwater Delitschia sp. Planta Med 85:62–71. https://doi.org/10.1055/a-0654-5850
Rivera-Chávez J, Zacatenco-Abarca J, Morales-Jiménez J, Martinez-Avina B, Hernández-Ortega SN, Aguilar-Ramírez E (2019c) Cuautepestalorin, a 7,8-dihydrochromene–oxoisochromane adduct bearing a hexacyclic scaffold from Pestalotiopsis sp. IQ-011. Org Lett 21:3558–3562. https://doi.org/10.1021/acs.orglett.9b00962
Sakurai M, Kohno J, Yamamoto K, Okuda T, Nishio M, Kawano K, Ohnuki T (2002) TMC-256A1 and C1, new inhibitors of IL-4 signal transduction produced by Aspergillus niger var niger TC 1629. J Antibiotics 55:685–692. https://doi.org/10.7164/antibiotics.55.685
Salituro GM, Pettibone DJ, Clineschmidt BV, Williamson JM, Zink DL (1993) Potent, non-peptidic oxytocin receptor antagonists from a natural source. Bioorg Med Chem Lett 3:337–340. https://doi.org/10.1016/S0960-894X(01)80905-7
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Siriwardane A, Kumar NS, Jayasinghe L, Fujimoto Y (2015) Chemical investigation of metabolites produced by an endophytic Aspergillus sp. isolated from Limonia acidissima. Nat Prod Res 29:1384–1387. https://doi.org/10.1080/14786419.2015.1025230
Song Y, Li H, Ye Y, Shan C, Yang Y, Tan R (2004) Endophytic naphthopyrone metabolites are co-inhibitors of xanthine oxidase, SW1116 cell and some microbial growths. FEMS Microbiol Lett 241:67–72. https://doi.org/10.1016/j.femsle.2004.10.005
Song F, Dong Y, Wei S, Zhang X, Zhang K, Xu X (2022) New antibacterial secondary metabolites from a marine-derived Talaromyces sp. strain BTBU20213036. Antibiotics 11:222. https://doi.org/10.3390/antibiotics11020222
Song F (2022) Antimicrobial natural products. Antibiotics 11:1765. https://doi.org/10.3390/antibiotics11121765
Sosa-Hernández O, Vázquez-Zamora C, Gutiérrez-Muñoz VH, Lugo-Zamudio GE, Cureño-Díaz MA (2020) Resultados del Programa de Uso Racional de Antimicrobianos en un hospital de México, 2013–2018. Rev Panam Salud Publica 44:e45. https://doi.org/10.26633/RPSP.2020.45
Sosa-Rodríguez T, Sánchez-Nieves J, Melgarejo LM (2009) Papel funcional de los hongos en ecosistemas de manglar. Bol Invest Mar Cost 38:38–57
Suzuki K, Nozawa K, Udagawa S, Nakajima S, Kawai K (1991) Penicillide and dehydroisopenicillide from Talaromyces derxii. Phytochemistry 30:2096–2098. https://doi.org/10.1016/0031-9422(91)85080-J
Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y (2018) Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. https://doi.org/10.1016/S1473-3099(17)30753-3
Tao H, Wei X, Lin X, Zhou X, Dong J, Yang B (2017) Penixanthones A and B, two new xanthone derivatives from fungus Penicillium sp. SYFz-1 derived of mangrove soil sample. Nat Prod Res 31:2218–2222. https://doi.org/10.1080/14786419.2017.1297442
Tripathy S, Sahu B (2019) FtsZ inhibitors as a new genera of antibacterial agents. Bioorg Chem 91:103169. https://doi.org/10.1016/j.bioorg.2019.103169
Van Der Hooft JJJ, Wandy J, Barrett MP, Burgess KE, Rogers S (2016) Topic modeling for untargeted substructure exploration in metabolomics. Proc Natl Acad Sci 113:13738–13743. https://doi.org/10.1073/pnas.1608041113
Varoglu M, Corbett TH, Valeriote FA, Crews P (1997) Asperazine, a selective cytotoxic alkaloid from a sponge-derived culture of Aspergillus niger. J Org Chem 62:7078–7079. https://doi.org/10.1021/jo970568z
WHO (2017) La OMS publica la lista de las bacterias para las que se necesitan urgentemente nuevos antibióticos. from https://www.who.int/es/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed. Accessed 7 Sept 2023
Zeng WN, Cai J, Wang B, Chen LY, Pan CX, Chen SJ, Huang GL, Zheng CJ (2022) A new bioactive isocoumarin from the mangrove-derived fungus Penicillium sp. TGM112. J Asian Nat Prod Res 24:679–684. https://doi.org/10.1080/10286020.2021.1952188
Zhang Y, Li XM, Wang BG (2007) Nigerasperones A~ C, new monomeric and dimeric naphtho-γ-pyrones from a marine alga-derived endophytic fungus Aspergillus niger EN-13. J Antibiot 60:204–210. https://doi.org/10.1038/ja.2007.24
Zhang Y, Li XM, Feng Y, Wang BG (2010) Phenethyl-α-pyrone derivatives and cyclodipeptides from a marine algous endophytic fungus Aspergillus niger EN–13. Nat Prod Res 24:1036–1043. https://doi.org/10.1080/14786410902940875
Zulqarnain ZI, Cox R, Anwar J, Ahmad N, Khan K, Iqbal M, Manzoor N, Khattak SU (2020) Antifungal activity of compounds isolated from Aspergillus niger and their molecular docking studies with tomatinase. Nat Prod Res 34:2642–2646. https://doi.org/10.1080/14786419.2018.1548447
Acknowledgements
We would like to thank Dr. Adriana Romo Pérez, Dr. María del Carmen García-González, MSc. Elizabeth Huerta (Instituto de Química, UNAM), and the LURMN laboratory for their assistance in the collection of spectroscopic and spectrometric data. We also thank Dr. Adela Rodríguez-Romero (Instituto de Química, UNAM) and Dr. Rodolfo García Contreras (Facultad de Medicina, UNAM) for their guidance and for providing bacterial strains. We also thank Dr. Armando Hernández García, Dr. Federico del Rio Portilla, and Dr. Corina Ceapa for providing the infrastructure to carry out the assays. MSc. Maricruz López López for handling and disposal of Hazardous Biological Infectious Waste material. Special thanks to MSc. Everardo Tapia for the acquisition of the MS/MS spectra. Special thanks to MSc. Enrique Aguilar for his guidance on preparative HPLC. KCJ is in debt to CONACyT/CONAHCyT for the fellowsip provided to pursue graduate studies (CVU 954406).
Funding
This research received financial support from CONACyT/CONAHCyT through Frontline Science project (“Programa Ciencia de Frontera”) CF-2019-263977. It also received partial support by the Dirección General de Asuntos del Personal Académico (DGAPA), Universidad Nacional Autónoma de México (UNAM) through the Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT: IA207422 and IA203220).
Author information
Authors and Affiliations
Contributions
All authors contributed to the conception and design of this study. Isolation of fungal microorganisms, extracts preparation, biodirected assay, isolation of compounds, and biological assays were performed by KCJ and FHS. Structural elucidation of the compounds and revision of the manuscript were done by CAFH and KCJ. Enzyme purification and enzyme assay were performed by PCS and KCJ. Identification of the fungal microorganisms was carried out by JMJ and JRC. BQG actively participated in the acquisition of the NMR spectra. The first draft of the manuscript was written by KCJ, and all authors commented on subsequent versions of the manuscript. All work was supervised by JRC. All authors read and approved the final manuscript.
Corresponding author
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Carrillo-Jaimes, K., Fajardo-Hernández, C.A., Hernández-Sedano, F. et al. Antibacterial Activity and AbFtsZ Binding Properties of Fungal Metabolites Isolated from Mexican Mangroves. Rev. Bras. Farmacogn. 34, 564–576 (2024). https://doi.org/10.1007/s43450-023-00507-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43450-023-00507-2