Abstract
The low-expression level of lactoferrin (LF) in the production process poses a significant challenge. This study aimed to efficiently express bovine lactoferrin (BLF) using Pichia pastoris GS115 as the expression host and PIC9K as the recombinant vector. Optimization strategies included codon usage, promoter selection, and fermentation conditions. The blf gene was optimized for P. pastoris GS115 bias, resulting in the construction of the recombinant vector pPIC9K-UBLF1-3 controlled by the AOX1 promoter. SDS-PAGE analysis revealed soluble and efficient expression of ublf3 in P. pastoris GS115, with a molecular mass of approximately 76 kDa. The transformant P. pastoris GS115/pGAP9K-UBLF3-4 resistant at 4 mg·mL−1 G418, exhibited a ublf3 gene copy number of 5.88 through high-copy screening. Optimal expression conditions of recombinant UBLF were determined as 24℃, pH 5.0 and 220 r·min−1 through fermentation condition optimization. Under these conditions, recombinant UBLF production reached 40.62 mg·L−1. The yield of recombinant UBLF was reached 824.93 mg·L−1 through high-density fermentation. Antibacterial assay demonstrated the efficacy of recombinant UBLF against Escherichia coli JM109 and Staphylococcus aureus CGMCC 1.282. This study successfully achieved the efficient heterologous expression of recombinant UBLF in P. pastoris GS115, providing valuable insight for industrial production and the potential development of natural antibacterial agents.
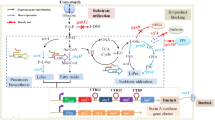
Graphical abstract








Similar content being viewed by others
Availability of data and materials
The datasets supporting the conclusions in this article are included in the manuscript.
References
Susana AG-C, Sigifredo A-G, Quintín R-C. Lactoferrin: structure, function and applications. Int J Antimicrob Agents. 2009;33(4):1–8.
Séverin S, Wenshui X. Milk biologically active components as nutraceuticals: review. Crit Rev Food Sci Nutr. 2005;45(7–8):645–56.
Masson PL, Heremans JF, Dive CH. An iron-binding protein common to many external secretions. Clin Chim Acta. 1966;14(6):735–9.
Wang X, Hao Y, Wang J, Teng D, Wang X. Research and development on lactoferrin and its derivatives in China from 2011–2015. Biochem Cell Biol. 2017;95(1):162–70.
Seok J-H, Hee-Gyun P, Sang-Hyeon L, Soo-Wan N, Sung-Jong J. High-level secretory expression of recombinant beta-agarase from zobellia galactanivorans in Pichia pastoris. Korean J Microbiol Biotechnol. 2010;38(1):40–5.
Takase M. Antimicrobial properties of lactoferrin in milk. J Ped Dermatol. 1996;15:1–5.
Jenssen H, Hancock REW. Antimicrobial properties of lactoferrin. Biochimie. 2009;91(1):19–29.
Wang B, Timilsena YP, Blanch E, Adhikari B. Lactoferrin: structure, function, denaturation and digestion. Crit Rev Food Sci Nutr. 2019;59(4):580–96.
Cutone A, Rosa L, Ianiro G, Lepanto MS, Musci G. Lactoferrin’s anti-cancer properties: safety, selectivity, and wide range of action. Biomolecules. 2020;10(3):456.
Blanca IF, Norberto VG, Tania S-C, Sugey S-G, Sigifredo A-G, Quintín R-C. High-level expression of recombinant bovine Lactoferrin in Pichia pastoris with antimicrobial activity. Int J Mol Sci. 2016;17(6):902.
Elzoghby AO, Abdelmoneem MA, Hassanin IA, Abd Elwakil MM, Elnaggar MA, Mokhtar S, Elkhodairy KA. Lactoferrin, a multi-functional glycoprotein: active therapeutic, drug nanocarrier and targeting ligand. Biomaterials. 2020;263:120355.
Sorensen M, Sorensen SPL. The proteins in whey. Comptes Rendus Des Travaux Du Laboratoire Carlsberg Ser Chimique. 1941;23:120355.
Groves ML. The isolation of a red protein from Milk2. J Am Chem Soc. 1960;82(13):3345–50.
Bai X, Teng D, Tian Z, Zhu Y, Yang Y, Wang J. Contribution of bovine lactoferrin inter-lobe region to iron binding stability and antimicrobial activity against Staphylococcus aureus. Biometals Int J Role Metal Ions Biol Biochem Med. 2010;23(3):431–9.
Conesa C, Sánchez L, Rota C, Pérez M-D, Calvo M, Farnaud S, Evans RW. Isolation of lactoferrin from milk of different species: calorimetric and antimicrobial studies. Compar Biochem Physiol Part B Biochem Mol Biol. 2008;150(1):131–9.
Jin L, Zhang RZ, Zhou L, Li L, Li J. Improving expression of bovine lactoferrin N-lobe by promoter optimization and codon engineering in Bacillus subtilis and its antibacterial activity. J Agric Food Chem. 2019;67(35):9749–56.
Kehoe SI, Jayarao BM, Heinrichs AJ. A survey of bovine colostrum composition and colostrum management practices on pennsylvania dairy farms. J Dairy Sci. 2007;9(9):4108–16.
Powell MJ, Ogden JE. Nucleotide sequence of human lactoferrin cDNA. Nucl Acids Res. 1990;18(13):4013.
Garcia-Montoya I, Salazar-Martinez J, Arevalo-Gallegos S, Sinagawa-Garcia S, Rascon-Cruz Q. Expression and characterization of recombinant bovine lactoferrin in E. coli. BioMetals Int J Role Metal Ions Biol Biochem Med. 2013;26(1):113–22.
Kim WS, Shimazaki KI, Tamura T. Expression of bovine lactoferrin C-lobe in Rhodococcus erythropolis and its purification and characterization. Biosci Biotechnol Biochem. 2006;70(11):2641–5.
Jin L, Lihong Z, Lixian Z, Rongzhen X, Yan LJ. Improving expression of bovine lactoferrin N-lobe by promoter optimization and codon engineering in Bacillus subtilis and its antibacterial activity. J Agric Food Chem. 2019;67(35):9749–56.
Macauley-Patrick S, Fazenda ML, Mcneil B, Harvey LM. Heterologous protein production using the Pichia pastoris expression system. Yeast. 2005;22(4):249–70.
Sreekrishna K, Brankamp RG, Kropp KE, Blankenship DT, Tsay JT, Smith PL, Wierschke JD, Subramaniam A, Birkenberger LA. Strategies for optimal synthesis and secretion of heterologous proteins in the methylotrophic yeast Pichia pastoris. Gene. 1997;190(1):55–62.
Guiziou S, Sauveplane V, Chang HJ, Clerté C, Bonnet J. A part toolbox to tune genetic expression in Bacillus subtilis. Nucl Acids Res. 2016;44(15):7495–508.
Qin X, Qian J, Yao G, Zhuang Y, Zhang S, Chu J. GAP promoter library for fine-tuning of gene expression in Pichia pastoris. Appl Environ Microbiol. 2011;77(11):3600–8.
Xuemei L, Jie W, Xiaobao J, Jiayong Z. High-level expression of a novel liver-targeting fusion interferon with preferred Escherichia coli codon preference and its anti-hepatitis B virus activity in vivo. BMC Biotechnol. 2015.
Scorer CA, Clare JJ, Mccombie WR, Romanos MA, Sreekrishna K. Rapid selection using G418 of high copy number Transformants of Pichia pastoris for high–level foreign gene expression. Bio/Technolgy. 1994;12(2):181–4.
Yadav D, Ranjan B, Mchunu N, Le Roes-Hill M, Kudanga T. Enhancing the expression of recombinant small laccase in Pichia pastoris by a double promoter system and application in antibiotics degradation. Folia Microbiol. 2021;66(6):917–30.
Rahmasari R, Raekiansyah M, Azallea SN, Nethania M, Bilqisthy N, Rozaliyani A, Bowolaksono A, Sauriasari R. Low Cost SYBR Green-Based RT-qPCR for Detecting SARS-CoV-2 in Indonesia setting using WHO-recommended primers. Social Science Electronic Publishing.
Waterham HR, Digan ME, Koutz PJ, Lair SV, Cregg JM. Isolation of the Pichia pastoris glyceraldehyde-3-phosphate dehydrogenase gene and regulation and use of its promoter. Gene. 1997;186(1):37–44.
Karaoglan M, Fidan EK. Effect of codon optimization and promoter choice on recombinant endo-polygalacturonase production in Pichia pastoris. Enzyme Microb Technol. 2020;139(1):109589.
Unver Y, Gün BE, Acar M, Yildiz S. Heterologous expression of azurin from Pseudomonas aeruginosa in the yeast Pichia pastoris. Preparat Biochem Biotechnol. 2021;51(7):723–30.
En-Peng Z, Zi-Li LV, Tian D, Ai-Zhen G, Liang W. Structural design of bovine lactoferricin-derived peptide and its activity assay after expressed in Pichia pastoris. Sci Technol Food Ind. 2019.
Liu C, Gong J-S, Su C, Li H, Qin J, Xu Z-H, Shi J-S. Increasing gene dosage and chaperones co-expression facilitate the efficient dextranase expression in Pichia pastoris. LWT. 2023;181: 114753.
Mohd FYNB, Naoki T, Yuki IK, Hiroshi T, Yukio K. The unfolded protein response in Pichia pastoris without external stressing stimuli. FEMS Yeast Res. 2020;20(7):7.
Sun J, Jiang J, Zhai X, Zhu S, Wei C. Coexpression of Kex2 endoproteinase and Hac1 transcription factor to improve the secretory expression of bovine lactoferrin in Pichia pastoris. Biotechnol Bioprocess Eng. 2019;24(6):934–41.
Acknowledgements
We are grateful to the Lab of Brewing Microbiology and Applied Enzymology, School of Biotechnology and Key Laboratory of Industrial Biotechnology of Ministry of Education, Jiangnan University in encouraging us for the studies.
Funding
This project was supported by the National Key Research and Development Program of China (2023YFA0914500), the National Science Foundation of China (32271487), the National First-class Discipline Program of Light Industry Technology and Engineering (LITE2018-12), and the Program of Introducing Talents of Discipline to Universities (111-2-06).
Author information
Authors and Affiliations
Contributions
XY conducted experiments, analyzed and interpreted data, and wrote the draft. ZW and HT helped with the data analysis and performed the calculation. RZ led the project and revised the manuscript. YX and WC revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
Yan Xu as the Editor-in-Chief of SMAB, the authors declare a conflict of interest for this paper. Yan Xu has been excluded from the peer review and decision-making process to ensure unbiased evaluation and strict adherence to ethical guidelines.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, X., Xi, Z., Zhao, H. et al. Efficient heterologous expression of bovine lactoferrin in Pichia pastoris and characterization of antibacterial activity. Syst Microbiol and Biomanuf (2024). https://doi.org/10.1007/s43393-024-00266-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s43393-024-00266-8