Abstract
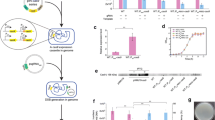
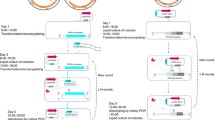
The traditional homologous recombination (HR) gene-editing method faces problems such as low editing efficiency and absence of marker genes. CRISPR–Cas9-editing efficiency is high and has been widely used in bacteria and yeast. In comparison with CRISPR–Cas9, CRISPR–Cas12a has many outstanding advantages. Here, we report an Acidaminococcus sp. BV3L6 (As) Cas12a-based genome-editing method used for Starmerella bombicola. To demonstrate the high efficiency of the CCMGE system, we verified a counter-selectable marker in S. bombicola, orotidine 5’-phosphate decarboxylase (100% for URA3). We also tested the common gene UDP-glucosyltransferase (100% for UGTA) using a 300 bp donor containing hygromycin expression cassette. This toolkit was further extended to simultaneously edit two genes (18% for UGTA and leu) and three genes (13.8% for UGTA, leu and URA3). The system greatly reduces the screening time for such multi-site editing. Based on the CCMGE system, the PHA (polyhydroxyalkanoate)-producing strain was constructed by increasing the copy number of the PHA synthase (PHAC). The PHA content and DCW reached 11.8% and 30.1 g/L, respectively. The yield of PHA was about three times higher than that of the single-copy strain using the same fermentation method.







Similar content being viewed by others
Availability of data and materials
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
Not applicable.
References
Pekin G, Vardar-Sukan F, Kosaric N. Production of sophorolipids from Candida bombicola ATCC 22214 using Turkish corn oil and honey. Eng Life Sci. 2005;5(4):357–62.
Daniel HJ, Reuss M, Syldatk C. Production of sophorolipids in high concentration from deproteinized whey and rapeseed oil in a two stage fed batch process using Candida bombicola ATCC 22214 and Cryptococcus curvatus ATCC 20509. Biotech Lett. 1998;20(12):1153–6.
Jiang Y, et al. CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum. Nat Commun. 2017;8:15179.
Wang H, et al. Starmerella bombicola: recent advances on sophorolipid production and prospects of waste stream utilization. J Chem Technol Biotechnol. 2019;94(4):999–1007.
Anna Y, et al. Production of D-lactic acid containing polyhydroxyalkanoate polymers in yeast Saccharomyces cerevisiae. J Ind Microbiol Biotechnol. 2021;5–6:5–6.
Vyas VK, Barrasa MI, Fink GR. A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Advance. 2015;1(3):e1500248.
Yu Z, Jefd B. CRISPR–Cas12a system in fission yeast for multiplex genomic editing and CRISPR interference. Nucleic Acids Res. 2020;48(10):5788–98.
Chen JS, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):791.
Moreno-Mateos MA, et al. CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nat Commun. 2017;8(1):2024.
Yang Z, Edwards H, Xu P. CRISPR-Cas12a/Cpf1-assisted precise, efficient and multiplexed genome-editing in Yarrowia lipolytica. Metab Eng Commun. 2020;10:e00112. https://doi.org/10.1016/j.mec.2019.e00112
Zhang L. A CRISPR–Cas9 system for multiple genome editing and pathway assembly in Candida tropicalis. Biotechnol Bioeng. 2019;117(2):531–42.
Jakounas T, et al. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng. 2015;28:213–22.
Zetsche B, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–71.
Zhao Y, Boeke JD. CRISPR-Cas12a system in fission yeast for multiplex genomic editing and CRISPR interference. Nucleic Acids Res. 2020;48(10):5788–98. https://doi.org/10.1093/nar/gkaa329.
Su BM, et al. Recent advances in the CRISPR genome editing tool set. Exp Mol Med. 2019;51(11):1–11.
Zetsche B, et al. Multiplex gene editing by CRISPR–Cpf1 using a single crRNA array. Nat Biotechnol. 2016;35(1):31.
Ouedraogo J-P, Tsang A. CRISPR_Cas systems for fungal research. Fungal Biol Rev. 2020;34(4):189–201. https://doi.org/10.1016/j.fbr.2020.10.002.
Malizia A, Monmany-Garzia AC. Terrestrial ecologists should stop ignoring plastic pollution in the Anthropocene time. Sci Total Environ. 2019;668(JUN.10):1025–9.
Haddouche R, et al. Engineering polyhydroxyalkanoate content and monomer composition in the oleaginous yeast Yarrowia lipolytica by modifying the -oxidation multifunctional protein. Appl Microbiol Biotechnol. 2011;91(5):1327–40.
Poirier Y, Erard N, Petetot MC. Synthesis of polyhydroxyalkanoate in the peroxisome of Saccharomyces cerevisiae by using intermediates of fatty acid β-oxidation. Appl Environ Microbiol. 2001;67(11):5254–60.
Lang S. Production of native and modified sophorose lipids. Chim Oggi. 2000;18(10):76–9.
Roelants SLKW. Candida bombicola as a platform organism for the production of tailor-made biomolecules. Biotechnol Bioeng. 2013;110(9):2494–503.
Roelants SL, et al. Towards the industrialization of new biosurfactants: Biotechnological opportunities for the lactone esterase gene from Starmerella bombicola. Biotechnol Bioeng. 2016;113(3):550–9.
Saerens K, et al. Identification of the UDP-glucosyltransferase gene UGTA1, responsible for the first glucosylation step in the sophorolipid biosynthetic pathway of Candida bombicola ATCC 22214. FEMS Yeast Res. 2011;11(1):123–32.
Bogaert I, et al. Development of a transformation and selection system for the glycolipid-producing yeast Candida bombicola. Yeast. 2010;25(4):273–8.
Imkeller K, et al. gscreend: modelling asymmetric count ratios in CRISPR screens to decrease experiment size and improve phenotype detection. Genome Biol. 2020;21(1):53.
Huang FC, Peter A, Schwab W. Expression and Characterization of CYP52 Genes Involved in the Biosynthesis of Sophorolipid and Alkane Metabolism from Starmerella bombicola. Appl Environ Microbiol. 2014;80(2):766–76.
Saerens K, et al. Cloning and functional characterization of the UDP-glucosyltransferase UgtB1 involved in sophorolipid production by Candida bombicola and creation of a glucolipid-producing yeast strain. Yeast. 2011;28:279–92.
Gao C. Exploring medium-chain-length polyhydroxyalkanoates production in the engineered yeast Yarrowia lipolytica. J Ind Microbiol Biotechnol. 2015;42(9):1255.
Zetsche B, et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol. 2017;35(1):31–4. https://doi.org/10.1038/nbt.3737.
Malzahn AA, et al. Application of CRISPR-Cas12a temperature sensitivity for improved genome editing in rice, maize, and Arabidopsis. BMC Biol. 2019;17(1):9. https://doi.org/10.1186/s12915-019-0629-5.
Ciurkot K, et al. CRISPR/Cas12a multiplex genome editing of Saccharomyces cerevisiae and the creation of yeast pixel art. J Vis Exp. 2019;14:7. https://doi.org/10.3791/59350.
Moon SB, et al. Recent advances in the CRISPR genome editing tool set. Exp Mol Med. 2019;51(11):1–11. https://doi.org/10.1038/s12276-019-0339-7.
Yang Z, Blenner M. Genome editing systems across yeast species. Curr Opin Biotechnol. 2020;66:255–66.
Kim D, et al. Erratum: Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol. 2016;34(8):888.
Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36(8):765–71.
Song L, et al. Efficient genome editing using tRNA promoter-driven CRISPR/Cas9 gRNA in Aspergillus niger. PLoS ONE. 2018;13(8):e020868.
Min JK, et al. Practical guidance for the implementation of the CRISPR genome editing tool in filamentous fungi. Fungal Biol Biotechnol. 2019;6(1):15.
Malzahn AA, et al. Application of CRISPR-Cas12a temperature sensitivity for improved genome editing in rice, maize, and Arabidopsis. BMC Biol. 2019;17(1):1–9.
Van BINA, et al. Knocking out the MFE-2 gene of Candida bombicola leads to improved medium-chain sophorolipid production. FEMS Yeast Res. 2010;4:4.
Ramdane H, et al. Roles of multiple acyl-CoA oxidases in the routing of carbon flow towards beta-oxidation and polyhydroxyalkanoate biosynthesis in Yarrowia lipolytica. FEMS Yeast Res. 2010;10(7):917–27.
Poirier Y, et al. Peroxisomal beta-oxidation–a metabolic pathway with multiple functions. Biochim Biophys Acta. 2006;1763(12):1413–26.
Acknowledgements
The work was supported by the 111 Project (No. 111-2-06), National Natural Science Foundation of China (32001064), Key Research and Development Program of China (2021YFC2100102-03), China Postdoctoral Science Foundation (2020M671331), and Postgraduate Research and Practice Innovation Program of Jiangsu Province (No. KYCX20-1807).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Compliance with ethics requirements
This article does not contain any studies with human participants or animals performed by any of the authors.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, M., Shi, Y., Zhang, L. et al. A CRISPR–Cas12a system for multi-gene editing (CCMGE) and metabolic pathway assembly in Starmerella bombicola. Syst Microbiol and Biomanuf 2, 665–675 (2022). https://doi.org/10.1007/s43393-022-00093-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43393-022-00093-9