Abstract
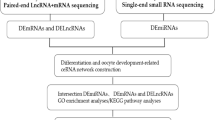
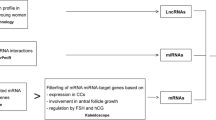
To explore the expression profiles of mRNAs, long-noncoding RNAs (lncRNAs), circular RNAs (circRNAs) and construct the competitive endogenous RNA networks in granulosa cells (GCs) of infertile women with ovarian endometriosis. RNA sequencing was conducted for RNA expression profiling from GCs of five women with ovarian endometriosis and five with tubal factor infertility. The differential expression of mRNAs, lncRNAs and circRNAs was compared. Then, the lncRNA-miRNA-mRNA and circRNA-miRNA-mRNA networks were constructed. Finally, the Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway were performed to determine the role of the differential expression of mRNA. A total of 12,498 mRNAs, 724 lncRNAs and 2269 circRNAs were identified in ovarian endometriosis and controls. 37 mRNAs, 51 lncRNAs and 101 circRNAs were detected to be differentially expressed in women with ovarian endometriosis. Ten lncRNAs and 22 differentially expressed mRNAs were selected to build the lncRNA–miRNA–mRNA network, while 12 circRNAs and four differentially expressed mRNAs were selected to build the circRNA–miRNA–mRNA network. GO analysis suggested that the differentially expressed mRNAs were mainly involved in regulation of cell differentiation, cell cycle while KEGG pathway analysis showed that pathways involved in the MAPK signaling pathway and FoxO signaling pathway were enriched with differentially upregulated mRNAs. We generated mRNAs, lncRNAs and circRNAs expression profiles and identified differentially expressed RNAs of GCs in infertile women with ovarian endometriosis. These findings provide a basis for further understanding of the underlying etiology of endometriosis-related infertility.





Similar content being viewed by others
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code Availability
Not applicable.
Change history
18 July 2022
This article was updated to fix the tagging of the equal contribution note.
References
Chapron C, Marcellin L, Borghese B, Santulli P. Rethinking mechanisms, diagnosis and management of endometriosis. Nat Rev Endocrinol. 2019;15(11):666–82. https://doi.org/10.1038/s41574-019-0245-z.
El-Toukhy T. Prevalence of endometriosis: how close are we to the truth? BJOG. 2021;128(4):666. https://doi.org/10.1111/1471-0528.16466.
Bulletti C, Coccia ME, Battistoni S, Borini A. Endometriosis and infertility. J Assist Reprod Genet. 2010;27(8):441–7. https://doi.org/10.1007/s10815-010-9436-1.
Saunders PTK, Horne AW. Endometriosis: Etiology, pathobiology, and therapeutic prospects. Cell. 2021;184(11):2807–24. https://doi.org/10.1016/j.cell.2021.04.041.
Peiris AN, Chaljub E, Medlock D. Endometriosis Jama. 2018;320(24):2608. https://doi.org/10.1001/jama.2018.17953.
Vercellini P, Viganò P, Somigliana E, Fedele L. Endometriosis: pathogenesis and treatment. Nat Rev Endocrinol. 2014;10(5):261–75. https://doi.org/10.1038/nrendo.2013.255.
Eisenberg VH, Weil C, Chodick G, Shalev V. Epidemiology of endometriosis: a large population-based database study from a healthcare provider with 2 million members. BJOG. 2018;125(1):55–62. https://doi.org/10.1111/1471-0528.14711.
Tanbo T, Fedorcsak P. Endometriosis-associated infertility: aspects of pathophysiological mechanisms and treatment options. Acta Obstet Gynecol Scand. 2017;96(6):659–67. https://doi.org/10.1111/aogs.13082.
Dumesic DA, Meldrum DR, Katz-Jaffe MG, Krisher RL, Schoolcraft WB. Oocyte environment: follicular fluid and cumulus cells are critical for oocyte health. Fertil Steril. 2015;103(2):303–16. https://doi.org/10.1016/j.fertnstert.2014.11.015.
Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21(1):22–36. https://doi.org/10.1038/s41568-020-00306-0.
Song G, Yang Z, Guo J, Zheng Y, Su X, Wang X. Interactions Among lncRNAs/circRNAs, miRNAs, and mRNAs in Neuropathic Pain. Neurotherapeutics. 2020;17(3):917–31. https://doi.org/10.1007/s13311-020-00881-y.
Stoll L, Sobel J, Rodriguez-Trejo A, Guay C, Lee K, Venø MT, et al. Circular RNAs as novel regulators of β-cell functions in normal and disease conditions. Mol Metab. 2018;9:69–83. https://doi.org/10.1016/j.molmet.2018.01.010.
Savelli L. Transvaginal sonography for the assessment of ovarian and pelvic endometriosis: how deep is our understanding? Ultrasound Obstet Gynecol. 2009;33(5):497–501. https://doi.org/10.1002/uog.6392.
Quinn MC, McGregor SB, Stanton JL, Hessian PA, Gillett WR, Green DP. Purification of granulosa cells from human ovarian follicular fluid using granulosa cell aggregates. Reprod Fertil Dev. 2006;18(5):501–8. https://doi.org/10.1071/rd05051.
Geng Y, Sui C, Xun Y, Lai Q, Jin L. MiRNA-99a can regulate proliferation and apoptosis of human granulosa cells via targeting IGF-1R in polycystic ovary syndrome. J Assist Reprod Genet. 2019;36(2):211–21. https://doi.org/10.1007/s10815-018-1335-x.
Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–15. https://doi.org/10.1038/s41587-019-0201-4.
Frazee AC, Pertea G, Jaffe AE, Langmead B, Salzberg SL, Leek JT. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat Biotechnol. 2015;33(3):243–6. https://doi.org/10.1038/nbt.3172.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. https://doi.org/10.1093/bioinformatics/bts635.
Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666–70. https://doi.org/10.1261/rna.043687.113.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. https://doi.org/10.1093/bioinformatics/btp616.
Ru Y, Kechris KJ, Tabakoff B, Hoffman P, Radcliffe RA, Bowler R, et al. The multiMiR R package and database: integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res. 2014;42(17): e133. https://doi.org/10.1093/nar/gku631.
Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39(Database issue):D163-9. https://doi.org/10.1093/nar/gkq1107.
Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v.20: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92-7. https://doi.org/10.1093/nar/gkt1248.
Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5(1):R1. https://doi.org/10.1186/gb-2003-5-1-r1.
Meuleman C, Vandenabeele B, Fieuws S, Spiessens C, Timmerman D, D’Hooghe T. High prevalence of endometriosis in infertile women with normal ovulation and normospermic partners. Fertil Steril. 2009;92(1):68–74. https://doi.org/10.1016/j.fertnstert.2008.04.056.
Bouet PE, Chao de la Barca JM, El Hachem H, Descamps P, Legendre G, Reynier P, et al. Metabolomics shows no impairment of the microenvironment of the cumulus-oocyte complex in women with isolated endometriosis. Reprod Biomed Online. 2019;39(6):885–92. https://doi.org/10.1016/j.rbmo.2019.08.001.
Albertini DF, Combelles CM, Benecchi E, Carabatsos MJ. Cellular basis for paracrine regulation of ovarian follicle development. Reproduction. 2001;121(5):647–53. https://doi.org/10.1530/rep.0.1210647.
Da Broi MG, Navarro PA. Oxidative stress and oocyte quality: ethiopathogenic mechanisms of minimal/mild endometriosis-related infertility. Cell Tissue Res. 2016;364(1):1–7. https://doi.org/10.1007/s00441-015-2339-9.
Xu B, Guo N, Zhang XM, Shi W, Tong XH, Iqbal F, et al. Oocyte quality is decreased in women with minimal or mild endometriosis. Sci Rep. 2015;5:10779. https://doi.org/10.1038/srep10779.
Lin X, Dai Y, Tong X, Xu W, Huang Q, Jin X, et al. Excessive oxidative stress in cumulus granulosa cells induced cell senescence contributes to endometriosis-associated infertility. Redox Biol. 2020;30: 101431. https://doi.org/10.1016/j.redox.2020.101431.
Orazov MR, Radzinsky VY, Ivanov II, Khamoshina MB, Shustova VB. Oocyte quality in women with infertility associated endometriosis. Gynecol Endocrinol. 2019;35(sup1):24–6. https://doi.org/10.1080/09513590.2019.1632088.
Zondervan KT, Becker CM, Missmer SA. Endometriosis. N Engl J Med. 2020;382(13):1244–56. https://doi.org/10.1056/NEJMra1810764.
Toya M, Saito H, Ohta N, Saito T, Kaneko T, Hiroi M. Moderate and severe endometriosis is associated with alterations in the cell cycle of granulosa cells in patients undergoing in vitro fertilization and embryo transfer. Fertil Steril. 2000;73(2):344–50. https://doi.org/10.1016/s0015-0282(99)00507-5.
Yue J, López JM. Understanding MAPK Signaling Pathways in Apoptosis. Int J Mol Sci. 2020;21(7). https://doi.org/10.3390/ijms21072346
Li Y, Liu YD, Chen SL, Chen X, Ye DS, Zhou XY, et al. Down-regulation of long non-coding RNA MALAT1 inhibits granulosa cell proliferation in endometriosis by up-regulating P21 via activation of the ERK/MAPK pathway. Mol Hum Reprod. 2019;25(1):17–29. https://doi.org/10.1093/molehr/gay045.
Brosens JJ, Wilson MS, Lam EW. FOXO transcription factors: from cell fate decisions to regulation of human female reproduction. Adv Exp Med Biol. 2009;665:227–41. https://doi.org/10.1007/978-1-4419-1599-3_17.
Cui C, Han S, Yin H, Luo B, Shen X, Yang F, et al. FOXO3 Is Expressed in Ovarian Tissues and Acts as an Apoptosis Initiator in Granulosa Cells of Chickens. Biomed Res Int. 2019;2019:6902906. https://doi.org/10.1155/2019/6902906.
Christian M, Lam EW, Wilson MS, Brosens JJ. FOXO transcription factors and their role in disorders of the female reproductive tract. Curr Drug Targets. 2011;12(9):1291–302. https://doi.org/10.2174/138945011796150253.
Li D, You Y, Bi FF, Zhang TN, Jiao J, Wang TR, et al. Autophagy is activated in the ovarian tissue of polycystic ovary syndrome. Reproduction. 2018;155(1):85–92. https://doi.org/10.1530/rep-17-0499.
Zhang JQ, Gao BW, Guo HX, Ren QL, Wang XW, Chen JF, et al. miR-181a promotes porcine granulosa cell apoptosis by targeting TGFBR1 via the activin signaling pathway. Mol Cell Endocrinol. 2020;499: 110603. https://doi.org/10.1016/j.mce.2019.110603.
Li Y, Xiang Y, Song Y, Wan L, Yu G, Tan L. Dysregulated miR-142, -33b and -423 in granulosa cells target TGFBR1 and SMAD7: a possible role in polycystic ovary syndrome. Mol Hum Reprod. 2019;25(10):638–46. https://doi.org/10.1093/molehr/gaz014.
Huo S, Qi H, Si Y, Li C, Du W. MicroRNA 26a targets Ezh2 to regulate apoptosis in mouse ovarian granulosa cells. Syst Biol Reprod Med. 2021;67(3):221–9. https://doi.org/10.1080/19396368.2021.1895362.
Acknowledgements
We thank Dr. Hongmin Chen for the help in data analysis. We also thank the patients who participated in this study.
Funding
This research was supported by the Basic and Applied Basic Research Foundation of Guangdong Province of China (No. 2020A1515110791).
Author information
Authors and Affiliations
Contributions
Jintao Peng and Xiaoyan Liang designed the experiments. Jiayi Guo performed the experiments and wrote the manuscript. Both Jiayi Guo and Haitao Zeng contributed equally to this study. Jintao Peng and Haitao Zeng performed data analysis and constructed the figures. Tingting Li assisted in the design of the experiments. Jintao Peng and Xiaoyan Liang revised the manuscript. All authors approved the final version of the manuscript for publication.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors have no conflicts of interest.
Ethics Approval
This study was approved by the Ethics Committee of the Six Affiliated Hospital of Sun Yat-sen University(2021ZSLYEC-299).
Consent for Publication
All patients obtained written informed consent.
Consent for Publication
Not applicable.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Guo, J., Zeng, H., Li, T. et al. mRNA, lncRNA and Circular RNA Expression Profiles in Granulosa Cells of Infertile Women with Ovarian Endometriosis. Reprod. Sci. 29, 2937–2946 (2022). https://doi.org/10.1007/s43032-022-00966-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43032-022-00966-3