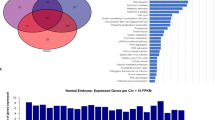
Abstract
Embryos are diagnosed as mosaic if their chromosomal copy number falls between euploid and aneuploid. The purpose of this study was to investigate the impact of mosaicism on global gene expression. This study included 42 blastocysts that underwent preimplantation genetic testing for aneuploidy (PGT-A) and were donated for IRB approved research. Fourteen blastocysts were diagnosed as mosaic with Next-generation Sequencing (NGS). Three NGS diagnosed euploid embryos, and 25 aneuploid embryos (9 NGS, 14 array Comparative Genomic Hybridization, 2 Single Nucleotide Polymorphism array) were used as comparisons. RNA-sequencing was performed on all of the blastocysts. Differentially expressed genes (DEGs) were calculated using DESeq2/3.5 (R Bioconductor Package) with p < 0.05 considered significantly differentially expressed. Pathway analysis was performed on mosaic embryos using EnrichR with p < 0.05 considered significant. With euploid embryo gene expression used as a control, 12 of 14 mosaic embryos had fewer DEGs compared to aneuploid embryos involving the same chromosome. On principal component analysis (PCA), mosaic embryos mapped separately from aneuploid embryos. Pathways involving cell proliferation, differentiation, and apoptosis were the most disrupted within mosaic embryos. Mosaic embryos have decreased disruption of global gene expression compared to aneuploid embryos. This study was limited by the small sample size, lack of replicate samples for each mosaic abnormality, and use of multiple different PGT-A platforms for the diagnosis of aneuploid embryos.




Similar content being viewed by others
Data Availability
Raw data, sample descriptions, and processed data (gene normalized counts) for all samples are publically available on GEO (Gene Expression Omnibus), accession number GSE151219.
Code Availability
Bioinformatics was performed using the following software: STAR/2.5 aligner (to align sequence reads to the hg19 ensemble reference transcriptome), featureCounts/1.5.3 (gene counting), DESeq2/3.5 using the R Bioconductor Package (Differential Gene Expression), EnrichR (pathway enrichment analysis).
References
Harton GL, Munne S, Surrey M, Grifo J, Kaplan B, McCulloh DH, Griffin DK, Wells D. Diminished effect of maternal age on implantation after preimplantation genetic diagnosis with array comparative genomic hybridization. Fertil Steril. 2013;100:1695–703.
Hassold T, Chiu D. Maternal age-specific rates of numerical chromosome abnormalities with special reference to trisomy. Hum Genet. 1985;70:11–7.
Hassold T, Chen N, Funkhouser J, Jooss T, Manuel B, Matsuura J, Matsuyama A, Wilson C, Yamane JA, Jacobs PA. A cytogenetic study of 1000 spontaneous abortions. Ann Hum Genet. 1980;44:151–78.
Werner M, Reh A, Grifo J, Perle MA. Characteristics of chromosomal abnormalities diagnosed after spontaneous abortions in an infertile population. J Assist Reprod Genet. 2012;29:817–20.
Forman EJ, Hong KH, Ferry KM, Tao X, Taylor D, Levy B, Treff NR, Scott RT Jr. In vitro fertilization with single euploid blastocyst transfer: a randomized controlled trial. Fertil Steril. 2013;100:100–7.
Forman EJ, Tao X, Ferry KM, Taylor D, Treff NR, Scott RT Jr. Single embryo transfer with comprehensive chromosome screening results in improved ongoing pregnancy rates and decreased miscarriage rates. Hum Reprod. 2012;27:1217–22.
Scott RT Jr, Upham KM, Forman EJ, Hong KH, Scott KL, Taylor D, Tao X, Treff NR. Blastocyst biopsy with comprehensive chromosome screening and fresh embryo transfer significantly increases in vitro fertilization implantation and delivery rates: a randomized controlled trial. Fertil Steril. 2013;100:697–703.
Yang Z, Liu J, Collins GS, Salem SA, Liu X, Lyle SS, Peck AC, Sills ES, Salem RD. Selection of single blastocysts for fresh transfer via standard morphology assessment alone and with array CGH for good prognosis IVF patients: results from a randomized pilot study. Mol Cytogenet. 2012;5:24.
Munne S, Kaplan B, Frattarelli JL, Child T, Nakhuda G, Nicholas Shamma F, Silverberg K, Kalista T, Handyside AH, Katz-Jaffe M, Wells D, Gordon T, Stock-Myer S, William S. Preimplantation genetic testing for aneuploidy versus morphology as selection criteria for single frozen-thawed embryo transfer in good-prognosis patients: a multicenter randomized clinical trial. Fertil Steril. 2019;112(6):1071–9.
Grifo JA, Hodes-Wertz B, Lee HL, Amperloquio E, Clarke-Williams M, Adler A. Single thawed euploid embryo transfer improves IVF pregnancy, miscarriage, and multiple gestation outcomes and has similar implantation rates as egg donation. J Assist Reprod Genet. 2013;30:259–64.
Wells D, Kaur K, Grifo J, Glassner M, Taylor JC, Fragouli E, Munne S. Clinical utilisation of a rapid low-pass whole genome sequencing technique for the diagnosis of aneuploidy in human embryos prior to implantation. J Med Genet. 2014;51:553–62.
Zheng H, Jin H, Liu L, Liu J, Wang WH. Application of next-generation sequencing for 24-chromosome aneuploidy screening of human preimplantation embryos. Mol Cytogenet. 2015;8:38.
Munne S, Blazek J, Large M, Martinez-Ortiz PA, Nisson H, Liu E, Tarozzi N, Borini A, Becker A, Zhang J, et al. Detailed investigation into the cytogenetic constitution and pregnancy outcome of replacing mosaic blastocysts detected with the use of high-resolution next-generation sequencing. Fertil Steril. 2017;108:62–71.
Fragouli E, Alfarawati S, Spath K, Babariya D, Tarozzi N, Borini A, Wells D. Analysis of implantation and ongoing pregnancy rates following the transfer of mosaic diploid-aneuploid blastocysts. Hum Genet. 2017;136:805–19.
Victor AR, Tyndall JC, Brake AJ, Lepkowsky LT, Murphy AE, Griffin DK, McCoy RC, Barnes FL, Zouves CG, Viotti M. One hundred mosaic embryos transferred prospectively in a single clinic: exploring when and why they result in healthy pregnancies. Fertil Steril. 2019;111(2):280–93.
Munné S, Spinella F, Grifo J, Zhang J, Parriego Beltran M, Fragouli E, Fiorentino F. Clinical outcomes after the transfer of blastocysts characterized as mosaic by high resolution Next Generation Sequencing-further insights. European J of Med Genet. 2020;63:103741.
Viotti M, Victor AR, Barnes FL, Zouves CG, Besser AG, Grifo JA, Cheng E, Lee M, Horcajadas JA, Corti L, Fiorentino F, Spinella F, Minasi MG, Greco E, Munné S. Using outcome data from one thousand mosaic embryo transfers to formulate an embryo ranking system for clinical use. Fertil Steril. 2021;115(5):1212–24.
Maxwell SM, Colls P, Hodes-Wertz B, McCulloh DH, McCaffrey C, Wells D, Munne S, Grifo JA. Why do euploid embryos miscarry? A case-control study comparing the rate of aneuploidy within presumed euploid embryos that resulted in miscarriage or live birth using next-generation sequencing. Fertil Steril. 2016;106:1414–9.
Friedenthal J, Maxwell SM, Munne S, Kramer Y, McCulloh DH, McCaffrey C, Grifo JA. Next generation sequencing for preimplantation genetic screening improves pregnancy outcomes compared with array comparative genomic hybridization in single thawed euploid embryo transfer cycles. Fertil Steril. 2018;109:627–32.
Munch EM, Sparks AE, Gonzalez Bosquet J, Christenson LK, Devor EJ, Van Voorhis BJ. Differentially expressed genes in preimplantation human embryos: potential candidate genes for blastocyst formation and implantation. J Assist Reprod Genet. 2016;33:1017–25.
Licciardi F, Lhakhang T, Kramer YG, Zhang Y, Heguy A, Tsirigos A. Human blastocysts of normal and abnormal karyotypes display distinct transcriptome profiles. Sci Rep. 2018;8:14906.
Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–8.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Tiegs AW, Tao X, Zhan Y, Whitehead C, Kim J, Hanson B, Osman E, Kim TJ, Patounakis G, Gutmann J, Castelbaum A, Seli E, Jalas C, Scott RT Jr. A multicenter, prospective, blinded, nonselection study evaluating the predictive value of an aneuploid diagnosis using a targeted next-generation sequencing-based preimplantation genetic testing for aneuploidy assay and impact of biopsy. Fertil Steril. 2021;115(3):627–37.
Sachdev NM, McCulloh DH, Kramer Y, Keefe D, Grifo JA. The reproducibility of trophectoderm biopsies in euploid, aneuploid, and mosaic embryos using independently verified next-generation sequencing (NGS): a pilot study. J Assist Reprod Genet. 2020;37(3):559–71.
Bolton H, Graham SJ, Van der Aa N, Kumar P, Theunis K, Fernandez Gallardo E, Voet T, Zernicka-Goetz M. Mouse model of chromosome mosaicism reveals lineage-specific depletion of aneuploid cells and normal developmental potential. Nat Commun. 2016;7:11165.
Capalbo A, Wright G, Elliott T, Ubaldi FM, Rienzi L, Nagy ZP. FISH reanalysis of inner cell mass and trophectoderm samples of previously array-CGH screened blastocysts shows high accuracy of diagnosis and no major diagnostic impact of mosaicism at the blastocyst stage. Hum Reprod. 2013;28(8):2298–307.
Macaulay IC, Teng MJ, Haerty W, Kumar P, Ponting CP, Voet T. Separation and parallel sequencing of the genomes and transcriptomes of single cells using G&T-seq. Nat Protoc. 2016;11:2081–103.
Acknowledgements
We would like to thank Bevin Kaplan for supporting this research as well as the Genome Technology Center and the Applied Bioinformatics Laboratories departments for their collaboration.
Author information
Authors and Affiliations
Contributions
All authors contributed to study concept and design. Sample preparation and acquisition of data was performed by Susan M. Maxwell, Yael G. Kramer, Yutong Zhang, and Fang Wang. Analysis and interpretation of data was performed by all authors. The manuscript was drafted by Susan M. Maxwell, and all authors participated in manuscript revision and approval of the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
This study was non-human subject research utilizing de-identified human embryos donated for research. Patients signed a consent for the disposal of their embryos for research purposes. The collection and use of discarded embryos for research was approved by the IRB at the New York University School of Medicine, study number i16-00154.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Susan M. Maxwell and Tenzin C. Lhakhang should be regarded as joint First Authors.
Rights and permissions
About this article
Cite this article
Maxwell, S.M., Lhakhang, T.C., Lin, Z. et al. Investigation of Global Gene Expression of Human Blastocysts Diagnosed as Mosaic using Next-generation Sequencing. Reprod. Sci. 29, 1597–1607 (2022). https://doi.org/10.1007/s43032-022-00899-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43032-022-00899-x