Abstract
The Coronavirus Disease 2019 (COVID-19) which first emerged in Wuhan, China in late December, 2019, has now spread to all the countries in the world. Conventional testing methods such as the antigen test, serology tests, and polymerase chain reaction tests are widely used. However, the test results can take anything from a few hours to a few days to reach the patient. Chest CT scan images have been used as alternatives for the detection of COVID-19 infection. Use of CT scan images alone might have limited capabilities, which calls attention to incorporating clinical features. In this paper, deep learning algorithms have been utilized to integrate the chest CT scan images obtained from patients with their clinical characteristics for fast and accurate diagnosis of COVID-19 patients. The framework uses an ANN to obtain the probability of the patient being infected with COVID-19 using their clinical information. Beyond a certain threshold, the chest CT scan of the patient is classified using a deep learning model which has been trained to classify the CT scan with 99% accuracy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The spread of COVID-19 has been rapid. The pandemic has resulted in over 30 million cases worldwide, with over 1 million fatalities. The symptoms include respiratory diseases like cough, fever, and shortness of breath. Since no absolute cure is available, the only alternative to control the spread of the disease is to determine the infection as early as possible and take appropriate steps. Early recognition of the disease can save a person’s life by rapid application of treatment options before the virus infects major portions of the lung. Early detection is also effective to curb widespread transmission by placing the patient under quarantine before the patient infects more people.
The current scenario depicts burden on the healthcare resources of a nation. Widespread testing is hampered by lack of test kits. Conventional testing methods have several limitations associated with them. They usually do not have good accuracy, which leads to multiple tests, increasing the financial burden on the patient. The test results are not produced in a quick manner, and sometimes take days to reach the patient because of backlog of the test center or location of the patient [1]. Without rapid detection of the virus, patients who are suspected to be infected might transmit the virus to others if proper steps are not taken to isolate them.
Chest X-ray and CT scan imaging have been widely used in medical research [2]. Early detection of COVID-19 will be more accurate by using CT scan imaging instead of X-Ray imaging. This is because chest X-rays might remain normal even after a few days, whereas CT scan can show an opacity pattern [3, 4]. Chest CT scan imaging has proved to be valuable in the diagnosis of a COVID-19 patient. Although it provides good results, CT images can sometimes be normal for a patient who is COVID-19 positive, which can produce false-negative results. The proposed classification framework joins the CT scan data along with the corresponding clinical characteristics of the patient to get more accurate results. This paper also predicts the severity of infection in the patient. The severity score can be helpful for pathologists to determine the stage of infection and extent of the lesions formed inside the lung. This can help to determine whether more aggressive treatment options might be required. The proposed framework plays an important part in the rapid identification and detection of COVID-19, thus decreasing the burden on the health institutions of a country.
The clinical characteristics and corresponding chest CT scan images have been obtained from various hospitals in India. The raw clinical information data set has been cleaned to remove the null values. This was then joined with the corresponding patient ID. This final data set was used for the training process. Clinical data from 3000 patients were analyzed. The raw data set obtained from various hospitals is not publicly available to protect patient privacy and integrity.
In this paper, our objective is to overcome the challenges presented by various deep learning models developed to correctly identify presence of infection within a person. We have established that analyzing chest CT scan images are better than conventional testing methods but can still be erroneous. The motivation behind this work is providing more accurate test results in a swift manner. Since we take into account the clinical characteristics of the person before training the CT scan images, we can drastically reduce the probability of false negatives and false positives. The parameters used in this paper are to classify the patients into their respective categories, the deep learning architecture used, and coming back to the clinical parameters of said patient to rank the severity of infection is novel.
This paper has been divided into five sections. "Introduction" gives the introduction and background of the paper. Section 2 provides a brief literature survey and short description of previous results for COVID-19 diagnosis. Section 3 provides the detailed explanation of the proposed framework, starting from handling of the data set to prediction of the severity of infection in a patient. Section 4 provides a discussion of the results achieved by the paper. Section 5 provides the concluding remarks of this paper.
Literature Survey
This section gives a brief description on various studies conducted for COVID-19. The studies have been analyzed so as to gain useful insight into the methods used in the preparation of the framework. Many results have been published in the field of medical imaging [5,6,7]. These studies have confirmed that using deep learning and AI techniques help to achieve superior results. This is in comparison to traditional machine learning techniques, hand crafted, texture analysis and manual selection methods [8, 9]. Deep learning has had many wide ranging applications. It has achieved immensely successful results in medical image classification tasks, disease prediction and medical segmentation tasks. CNN (convolution neural networks) have been widely used in the prediction, diagnosis and treatment of many diseases like Alzheimer’s disease, breast cancer detection, brain tumor detection, etc [10,11,12].
Chest CT scans are a valuable tool in early detection and diagnosis of suspected COVID-19 patients. In early investigations, scientists and researchers found specific common patterns in the chest CT images [13,14,15]. It was found that patients who have tested positive for COVID-19 have some common abnormalities in their CT scans [16,17,18,19]. AI and deep learning models have been widely used to recognize these patterns to and perform automated classification on CT images. Recent studies have confirmed the presence of few abnormalities in the absence of COVID-19 infection [20]. This is the reason to suggest the use of clinical characteristics in conjunction with CT scan imaging. This decreases the probability of false results and increases the reliability of the model. Different levels of severity of infection lead to different types of symptoms and medical conditions, and have different effects on people. These symptoms have been observed in various different COVID-19 patients and several treatment options have been suggested [21,22,23].
An initial analysis of the patients from the epicentre of the virus, Wuhan was completed. Its purpose was to improve diagnosis by describing the clinical findings and the chest CT imaging. This study used demographics data of 234 patients, high-resolution CT signs, ABG analysis and CT scores of the patients. Since it was conducted during the initial days of the pandemic, fever and cough were the main symptoms [24]. The clinical and CT scan findings of COVID-19-infected patients were compared between those who survived and died. The SpO2 and OI decreased during the fourth stage, while the CT score of fibrosis and air trapping was low in the first stage. An analysis using clinical features and CT images of COVID-19 patients using AI was conducted. This study confirmed the presence of various blood results such as WBC count, and lymphocyte counts were observed [25]. These studies provide the knowledge base to rank patients by their severity in terms of what factors should be used. A more recent analysis combined the CT images with clinical data to aid in the rapid diagnosis of COVID-19 [26]. This study achieved acquired data from Chinese hospitals and confirmed their results with expert radiologists. Their model achieved higher AUC score than senior radiologists. According to their framework, they have appended the clinical data set with the flattened CT image feature vector to train the model simultaneously. This is the main limitation of their work, because the combination of clinical data and CT scan image data means that we are combining clinical characteristics with pixel values of an image. In our paper, we use the clinical characteristics data as a filter rather than appending it with pixel values before passing it to a neural network.
Numerous results have been published which demonstrate the use of transfer learning for medical imaging purposes. Results have been published which utilize various individual architectures trained using pre-trained weights. Fine tuning architectures on these weights have given promising results and have been proven to be computationally fast. This paper utilizes the concept of transfer learning for classification tasks. The proposed architecture is a combination of features extracted from three different pre-trained networks, namely, DenseNet-201, MobileNetV2 and NASNet-Mobile. This has been done to overcome challenges presented by each of the individual networks and utilizing the power of transfer learning to reduce computational time and power in an attempt to increase model efficiency.
Results have been published utilizing various individual architectures which were trained using the pre-trained weights [27]. They achieved their best accuracy of 0.96 using MobileNetV2 model. Since the model was pretty primitive and had small training data and the depth was shallow, it cannot be reliable in testing. They have acknowledged the need to use CT scan images instead of X-ray images along with patient data to provide more accurate results. K-Fold and transfer learning have been used to provide results with the highest accuracy at 0.98 [28]. The K-Fold model is used better in a clustering algorithm while doing segmentation rather than classification. This is why their model will not be able to work on diverse test data, because it is over fitting on the training data.
A model named Deep-COVID achieved a sensitivity rate of 0.97 (with 0.05 tolerance) and specificity rate of 0.9 with a 0.96 accuracy using transfer learning, by fine tuning popular architectures [29]. Their model was a concatenation of three models; however, the use of chest X-ray images has brought down their specificity values. As mentioned in their model, the SqueezeNet architecture provides poor results. The COVID-Net [30] model gave an accuracy of 0.933. It performed comparative analysis with other models like ResNet and VGG16. Their process can be reliable if their data set had more amount of COVID-19 data instead of pneumonia CT images. However, they used only single architecture which has its own disadvantages and use of another architecture to counter them provides more reliable results.
A model was proposed using transfer learning which gave very promising results. This model achieved an accuracy of 0.98 [31]. However, the use of small number of training data and shallow network structure means that the model over fits on training data and will not be able to identify test data images. In [32], a study showing the comparison of transfer learning techniques has been presented which gives the results of training chest X-ray images on various individual architectures. Classification of COVID-19 patients using DenseNet-201 and Transfer learning [33] gave a test accuracy of 0.96. They have used a pretty deep architecture and have made good use of the power of dense neural networks, but have used a single architecture. Use of shallow model can be justified if we use a semi supervised model [34]. However, using semi supervised shallow architecture results in longer time to train the model, thus opposing the need for a faster method to test patients for infection. The proposed method of classification aims to overcome the challenges associated with each of the individual methods used as well as combine the strengths of the individual models and give the most accurate results.
Proposed Framework
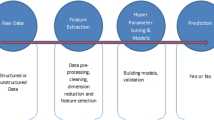
This section will describe in detail the initial processing done on the data set, the algorithm used to extract features from the CT scan images, image processing technique applied to the CT images and the model used to classify the training CT images and image features. The algorithm of the framework is illustrated in Fig. 1. The clinical information data are first processed and cleaned. The clinical features and laboratory findings are passed to an ANN model which computes the probability of patient being infected based on the observed symptoms and observations. A threshold probability of 0.25 has been set below which the patient is considered not being infected with the virus. A threshold of 0.5 has been used [35] in the previous research which has several limitations like using imperfect ground truth images for lung localization as well as the fact there all data used in said research came from a single hospital. In this paper, we started with the value of 0.5, but the results obtained mismatched with the actual values by a large amount, and thus, after repeating the process, we have set our threshold at 0.25. The framework does not outright make a classification of the patient only based on clinical factors. This is because a large number of patients may have symptoms on a particular day which might not be COVID-19 related. The laboratory findings also have overlapping values for each class, which cannot be accurately clustered into two separate groups, as shown in Table 1. If the probability of infection is more than 0.25, the CT scan images of the patient has to be taken into consideration.
This paper uses a feature extraction architecture built on the backbone of recent deep neural network architectures, DenseNet-201, MobileNetV2 and NasNet-Mobile [36,37,38]. Transfer learning concept will be applied to make the computation faster. This is done in an attempt to increase the accuracy of the model, overcome limitations of the individual models as well as to achieve the foremost objective which was rapid detection. The combined features are passed to a classification model to predict the probability of infection. This model has the ability to compute the probability of a new patient being infected with COVID-19 based on the chest CT scan.
This paper proposes four separate thresholds which have been set up to obtain the severity of infection. If the probability of infection is less than 0.30, the patient is classified as not infected with COVID-19. The other ranges of probability of infection are between (0.3, 0.65), (0.65, 0.9) and probability greater than 0.9. The severity of infection has been determined by taking into account the age groups of various patients as well as their blood oxygen levels (SpO2).
The ANN model and the CNN model which compute the probability of infection based on clinical parameters and chest CT images have been trained beforehand. After observing the classification results, they have been incorporated into the framework. The ANN used for clinical information classification is made of three layers of three layers of dense hidden neural networks consisting of 64, 128 and 256 neurons, respectively, and dropout regularization layers, in between the input and output layers. The architecture of the CNN model has been described in Section 3.3 in detail. The architectures, advantages and limitations of each of the individual models have also been described in brief.
DenseNet-201 The DenseNet model was developed to simplify the pattern of connections between various layers. In DenseNet, each layer is connected to every other layer. The problems previously associated with deep neural architectures were the redundancy issues in the deeper layers. DenseNet sought to rectify those issues. The training time for similar architectures like ResNet and Inception is very high in comparison to DenseNet. This model does not have the problem of vanishing gradient, since there is no need to learn redundant feature maps. Each layer has access to gradients and loss values. Various models of DenseNet differ in the number of convolutional and pooling blocks but have the same basic structure.
MobileNetV2 The MobileNet model has achieved very good performance on models across varying sizes and tasks. This is used in mobile and resource constrained environments. This model has an inverted residual network with a linear bottleneck. This structure makes the MobileNet model more accurate than other state-of-the-art models. This model is widely used for object detection and provides the similar accuracy as conventional frameworks like SSD and YoloV2, with less computational power.
NASNet-Mobile In the NASNet model, the blocks are not predefined by the authors like other models. They are searched using reinforcement learning to optimize the configuration of the architectures. The initial filters are free parameters and are used for scaling. There are two types of cells called the Normal cell and Reduction cell. Normal cells are convolution cells which return a feature map of the same dimension, while reduction cells are the cells which return a feature map where the height and width is decreased by a factor of two. This model used a new regularization technique called ScheduledDropPath. This model achieved state of the art on the Image Net data set with a smaller model size and lower complexity ops.
Data Set Preparation and Processing
The data set was obtained from various different hospitals in India to obtain randomness and maintain data from a diverse group of patients. This was to prevent over generalization of the model to be applied to patients from a single geographical area or neighborhood. This data set contained various clinical factors including the age, gender, temperature, WBC count, Lymphocyte, Neutrophil count and SpO2 levels (blood oxygen content) as well as presence of clinical symptoms such as fever, cough, difficulty in breathing, chest pain along with some less common symptoms such as aches, sore throat, diarrhoea, etc. All the symptoms were not observed in every patient. The presence of these symptoms and patient status play a vital role in computing the severity of infection. The rows of clinical information with null values were removed and the data were normalized. Since the caseload in India is quite high, a large number of data still remained for training the artificial neural network (ANN) model after processing the data set and discarding unusable row data. The final data set contained the clinical information of 3000 patients, the characteristics of the entire data set along with all the parameters are provided in Table 1. The chest CT scan images were obtained from various open source and public data sets [39, 40]. These have been combined by different individual organizations from different parts of the world.
Clinical Data Set The parameters like age, temperature and laboratory findings containing ’±’ indicate mean ± standard deviation of all the values. The clinical symptoms give the number of patients who have the symptoms. The WBC, Lymphocyte and Neutrophil count are in the range of 10e9. The SpO2 levels indicate the blood oxygen saturation of the patient. The ideal blood oxygen levels in healthy persons are in the range of 92–100%. If the blood oxygen levels drop below 90%, the condition is called hypoxemia, which can lead to organ failure and cardiac arrest. Monitoring the blood oxygen levels has very important application in measuring the severity of infection in a patient. Table 1 shows that the average temperature of patient with COVID-19 is higher, while the average blood oxygen level is lower than that of uninfected patient.
Feature Extraction from Chest CT Images
After the data set is ready to be passed to the training model, all the features are extracted from the CT images. The model architecture used to extract the features is built by combining various deep neural networks. To make the process computationally fast, transfer learning has been applied. The images were first preprocessed to remove noise and probable irrelevant features. These images were passed to the CNN model as inputs. The features were extracted and mapped in multi-dimensional arrays, and flattened into a single dimension array.
Image Preprocessing and Augmentation Since the images were obtained from multiple sources independently, they were not of the same dimension. They needed to be processed, so that we could extract features from them. Preprocessing involves normalization of the images, resizing them to \(512\,\times \, 512\) dimensions. A portion of the images were white spaces or had very low intensities. Therefore, for efficient classification, these irrelevant parts of the CT image were removed without changing the shape of the image altogether. The images are smoothed using a Gaussian filter. The background noise is removed using a two-dimensional median filter. Contrast limited adaptive histogram equalization (CLAHE) algorithm has been used to enhance the contrast of the images. This is used, because medical images, especially CT images, have lot of contrast problems. The clip limit has been kept at 3. Finally, data augmentation was applied randomly on the images. This is very useful to improve the performance of our model by forming new and different examples to train data set. It adds randomness and diversification of the images, by application of various transformations like rotation, shearing, and horizontal and vertical flip throughout the data set without changing the image labels. This is useful, because it increases the performance of the model because of addition of diversified images without actually increasing the size of the data set. Data augmentation techniques enable machine learning models to be more robust by creating variations that the model may see in the real world.
Classification of Chest CT Images
The CNN model proposed in this paper is extremely effective and efficient for feature extraction process. This architecture was first trained separately to work on the CT images alone, independent of the proposed framework. The processed chest CT images are input to the model which diverges into three separate individual architectures. They are the DenseNet-201, MobileNetV2 and NASNet-Mobile models. The pre-trained Image Net weights for each architecture are loaded into their corresponding models instead of training the models from scratch. This is done to use the results of a previous classification task as the starting weights of the training process, instead of randomly applied weights. This has a major effect on computational power required to train the model. When each of the individual models are initialized independently with the same images as input, they extract independent features. These features are passed into the Global Average Pooling Layer in two dimensions. This is done to decrease the dimensionality of the image feature map, decrease in the number of parameters involved and increase in computational efficiency. The features extracted from each of the individual models are combined. Each of the models individually are not completely efficient and miss out various relevant features. Combination of features is done to overcome obstacles presented by any of the individual models, using the advantages of each of the individual models to strengthen the final feature matrix by returning the maximum number of relevant features. The features are then flattened to form a one-dimensional feature matrix, because the model accepts data in this format.
This actually marks the end of the feature extraction process and begins the classification process. The images are classified using an ANN architecture. This consists of a batch normalization layer, multiple fully connected layers with 256, 512 and 1024 neurons each. Dropout layers were added in between them for regularization to prevent over fitting on the training data set. Finally, the dense layer consisting of two neurons with the sigmoid activation function was added to provide the probability of output classes. The model classified a patient as COVID-19 positive or negative based on the larger of the output probabilities of each predicted class. During the training process, binary cross-entropy loss function was used with the Adam optimization function. Due to application of transfer learning, this process is quite fast in comparison to training the model using random weights. Due to the very accurate results obtained, which can be seen in the classification report in Table 2, as well as the confusion matrix and ROC curve in the next section, this architecture has been incorporated into the framework.
As mentioned, the CNN model which has been incorporated into the framework to classify the chest CT images, outright classifies them into two classes based on their probabilities. Since in this framework, the CT scan image of a new patient is being used only after it is classified as being possibly infected with COVID-19 based on the clinical information beforehand, the probabilities of infection are used to calculate the severity of the infection in the patient.
Determination of Severity of Infection
As shown in Figs. 1 and 2, after the patient’s clinical characteristics pass through the ANN model and if the probability of infection is above the threshold value, the CT scan of the patient is passed through the CNN model. The features are extracted, flattened and passed through the fully connected layer to obtain the probability of infection. This probability is input to the severity framework, along with the age and SpO2 level of the patient. These two factors are important parameters in deciding the severity of infection of the patient. If the probability of infection from the CNN model is less than 0.3, the patient is classified as COVID-19 negative. If the probability is more than 0.3, it is again divided into three different levels, [0.3, 0.65], [0.65, 0.9] and greater than 0.9. These categories have been made in accordance with the clinical severity factors decided by the Government of India and can thus be justified. These groups are subdivided into different branches of decision making based on the patient’s age and blood oxygen level. The patient’s age has been taken into account, because it is known that the health of older patients worsens quickly in comparison to the health of younger patients. The blood oxygen level is an important factor, because deficiency of oxygen in different cells of the body can lead to adverse effects on various organ systems which in turn prohibits the body from functioning normally. Figure 3 illustrates the CNN model used for the feature extraction process.
Results and Discussion
This section demonstrates the results achieved by this paper. It proves the superiority of the proposed model in terms of accuracy comparison with the previous architectures. Table 2 illustrates the classification report for the task of classifying the chest X-ray images. This report contains important evaluation metrics for a classification task. The accuracy score obtained using the proposed model for classification of chest X-ray images to detect the presence of COVID-19 patients is 99%, which can be confirmed by the following table. Recall gives the total results correctly classified which are relevant, whereas precision refers to the percentage of relevant results. The support column gives the number of images for the class. As mentioned in the support column of Table 2, there are a total of 688 images in the test set, out of which 360 are COVID negative, while 328 are COVID positive. F1-score is the harmonic mean of precision and recall. Theoretically, it is not possible to maximize both precision and recall at the same time, so a trade-off between the two metrics is needed. The aim of a classification job is to increase the value of precision and recall to as high as possible.
Two important performance measures in a classification task are the confusion matrix and the receiver-operating characteristics (ROC) curve. The confusion matrix gives a clear understanding of the model performance. It provides the number of images which are classified correctly and incorrectly. The ROC curve gives the diagnostic ability of the model. It gives the ability of the model to distinguish between the two classes. The area under the ROC curve otherwise known as the area under curve (AUC) should be maximized. Figure 4 illustrates the confusion matrix and Fig. 5 illustrates the ROC curve. As demonstrated in the ROC curve, the AUC score obtained in this paper is 0.986. The ROC curve gives the plot of the true positive rate V/S the true negative rate. In the confusion matrix, the number of true positive (TP) results is 326 and the number of true negative (TN) results is 357. This means that from the test set, 326 images were correctly predicted as COVID-19 positive, whereas 357 images were correctly predicted as negative. Only five images were wrongly predicted. Two images were predicted as COVID-19 negative, while their actual label was positive. Three images were predicted as COVID-19 positive, while their actual labels were negative. This means that there are only three false positives (FP) and two false negatives (FN) in the predicted values of the test set. Table 4 clearly demonstrates the superiority of the proposed model by providing a comparative analysis of the results achieved by our model against the results achieved by other authors, in terms of model accuracy. The sensitivity and specificity of the model are 99.39% and 99.16%, respectively. They are obtained using Eqs. 1 and 2
The output classes obtained by the CNN model are nothing but probabilities of prediction belonging to each output class where the threshold has been set at 0.5. The framework uses this CNN architecture, and instead of calculating the output class, the output probabilities are calculated which are in effect divided into four different classes. This technique works quite well, because during the training of the CNN model, very few incorrect results were observed for the completely unbiased test set. This meant that any new image had equal probability of belonging to either class. When incorporated into the framework, the model received a new image to classify after the corresponding patient’s clinical information was classified using an ANN. This meant that the patient had a higher probability of being infected with COVID-19 than otherwise. This is the reason to decrease the threshold probability of being COVID-19 negative to 0.30.
The proposed severity framework acts on the patients who have a probability of infection from COVID-19 more than 0.3 have been classified into three different levels of severity, mild, severe and critical. These levels of severity can give an idea of the process of treatment to be undertaken. The patients classified as mild are usually expected to recover quickly, without the need for further hospitalization. This is only possible by early and rapid detection of COVID-19; otherwise, the severity of infection is likely to increase. Common symptoms include fever, fatigue, and cough, and less common symptoms may be present which include sore throat, aches, etc. These patients might not need medical attention and should stay in quarantine indoors with less contact with other people. The patients who have been classified as severe may develop inflammation of the lungs. Common symptoms include high fever, breathing difficulty, chest pains, etc. These usually occur in patients with pre-existing medical conditions. These patients will require medical attention at some point, without which the infection will only grow inside the lungs. The patients who have been classified as critical develop severe life-threatening conditions. They usually have weak pulse, coldness in limbs and might need life supporting medical equipment so prevent organ failure, sepsis or respiratory failure. This condition is mostly seen in patients of older ages, but every age group of the population is susceptible unless early treatment is provided. Certain medical conditions like heart diseases, diabetes, hypertension and high blood pressure increase the risk of death.
We have demonstrated the correlation between visual features and clinical features by showing some CT scan images of different patients and have provided the Grad-Cam (Gradient-weighted Class Activation Mapping) visualizations to highlight the regions of the input CT scan which are important to the prediction. We have shown the CT scan images of three COVID-19-positive patients with their heat map visualization which give the region of interest in the prediction process. These are illustrated in Figs. 6, 7, and 8. As illustrated in these images, the severity of infection in each of the patients is mild, severe and critical. In Fig. 6, we can see that the lesions are quite small and sparsely populated in contrast to Fig. 7 where the lesions are small but more dense toward the start of the bronchi. In Fig. 8, we can see that the lesions are quite large and concentrated in both lungs, and therefore, the severity has been characterized as critical. Table 4 gives the respective clinical descriptions of these three patients. As we can see in Table 3, the severity classification of all three patients can be justified on the basis of Age and SpO2 levels more than other laboratory findings like lymphocyte, WBC, and neutrophil, because their values are not as distinct as they should be discern between them. As we can see, the factors like Fever, Cough and other symptoms present a weak classification, and that is why, they are used before incorporating the CNN architecture.
The challenges involved in the successful completion of this research is the difficulty involved in obtaining diverse patient data from different hospitals in different geographical regions due to various laws protecting patient privacy. Following that the different factors which actually determine how severe the infection is have to be studied. Since there are many deep neural architectures, it is important to choose the architectures which will diminish each other’s faults and prevent an over fitted model.
Conclusion
This paper proposes novel and efficient framework for the fast and automated classification of patients suspected as being infected with COVID-19 using clinical information, laboratory findings and chest CT scan images and calculation of the severity of infection in the patient. The framework has been divided into two steps where the clinical information is used to first filter out the patients who are possibly infected with COVID-19 and then use a CNN algorithm to confirm the infection and predict severity of infection. The framework makes use of two trained neural network architectures which have been tested and achieved promising results. The CNN algorithm makes use of transfer learning on three different models, namely DenseNet-201, MobileNetV2 and NASNet-Mobile. The models are trained using pre-trained weights as the starting point to save computational power and make the classification process faster. Each of the individual models generates a different feature map and none of the feature maps individually cover all the features. The three models are trained separately and the extracted features are combined. This is done in an attempt to increase the performance of the model. The model then classifies the chest CT scan images into COVID-19-negative and -positive images and achieves an accuracy of 99%. A comparison of the results achieved by previous studies has been done against the results achieved by our proposed model. The comparative analysis has been presented in a tabular form. This shows that the classification model which has been used achieved better results than previous studies in terms of accuracy and proves the superiority of our model. This model can make accurate and quick detection of COVID-19 infection in a patient and thus is incorporated into this framework. Since this method is automated, it decreases the need of physical contact with a doctor or clinician, further reducing the chances of transmission of the virus. The paper also proposes the algorithm of a framework to determine the severity of infection in the patient. This is determined on the basis of various factors which include the probability of patient being infected according to the CNN algorithm, the age of the patient and SpO2 (blood oxygen) level of the patient. Three levels of severity have been proposed, mild, severe and critical, which can influence the treatment options to be undertaken. The main limitation in this research is the unavailability of large raw data set. Although the patients are diverse in nature, the reliability of the model can be ensured if our training data had a larger number of images. Another limitation is that the model is yet to verify if any mutation in the COVID-19 virus can be identified.
References
Boger B, Fachi MM, Vilhena RO, et al. Systematic review with meta-analysis of the accuracy of diagnostic tests for COVID-19. Am J Infect Control. 2020;49(1):21–9.
Toussie D, Voutsinas N, Finkelstein M, et al. Clinical and chest radiography features determine patient outcomes in young and middle age adults with COVID-19. Radiology. 2020;297(1):197–206.
Phelan AL, Katz R, Gostin LO. The novel coronavirus originating in Wuhan, China: challenges for global health governance. JAMA. 2020;323(8):709–10.
Nishiura H, Jung S, Linton NM, et al. The extent of transmission of novel coronavirus in Wuhan, China, 2020. J Clin Med. 2020;9:330.
Suri JS, Puvvula A, Biswas M, et al. COVID-19 pathways for brain and heart injury in comorbidity patients: a role of medical imaging and artificial intelligence-based COVID severity classification: a review. Comput Biol Med Comput Biol Med. 2020:103960.
Kotian RP, Faujdar D, Kotian SP, D’souza B. Knowledge and understanding among medical imaging professionals in India during the rapid rise of the COVID-19 pandemic. Health Technol. 2020;10:1415–20.
Dai WC, Zhang HW, Yu J, et al. CT imaging and differential diagnosis of COVID-19. Can Assoc Radiol J. 2020;71(2):195–200.
Abdollahi-Arpanahi R, Gianola D, Penagaricano F. Deep learning versus parametric and ensemble methods for genomic prediction of complex phenotypes. Genet Sel Evol. 2020;52(1):1–15.
Nassif AB, Shahin I, Attili I, et al. Speech recognition using deep neural networks: a systematic review. IEEE Access. 2019;7:19143–65.
Khan SU, Islam N, Jan Z, et al. A novel deep learning based framework for the detection and classification of breast cancer using transfer learning. Pattern Recogn Lett. 2020;125:1–6.
Talo M, Yildirim O, Baloglu UB, et al. Convolutional neural networks for multi-class brain disease detection using MRI images. Comput Med Imaging Graph. 2019;78:101673.
Wen J, Thibeau-Sutre E, Diaz-Melo M, et al. Convolutional neural networks for classification of Alzheimer’s disease: overview and reproducible evaluation. Med Image Anal. 2020;63:101694.
Jacobi A, Chung M, Bernheim A, Eber C. Portable chest X-ray in coronavirus disease-19 (COVID-19): a pictorial review. Clin Imaging. 2020;64:35–42.
Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506.
Ng MY, Lee EY, Yang J, et al. Imaging profile of the COVID-19 infection: radiologic findings and literature review. Radiol Cardiothorac Imaging. 2020;2(1):200034.
Bernheim A, Mei X, Huang M, et al. Chest CT findings in coronavirus disease 2019 (COVID-19): Relationship to duration of infection. Radiology. 2020;295.
Li L, Qin L, Xu Z, et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020.
Xu X, Jiang X, Ma C, et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6(10):1122–9.
Shiri I, Akhavanallaf A, Sanaat A, et al. Ultra-low-dose chest CT imaging of COVID-19 patients using a deep residual neural network. Eur Radiol. 2021;31(3):1420–31.
Bastos ML, Tavaziva G, Abidi SK, et al. Diagnostic accuracy of serological tests for COVID-19: systematic review and meta-analysis. BMJ. 2020;370.
Carfì A, Bernabei R, Landi F. Persistent symptoms in patients after acute COVID-19. JAMA. 2020;324(6):603–5.
Menni C, Valdes AM, Freidin MB, et al. Real-time tracking of self-reported symptoms to predict potential COVID-19. Nat Med. 2020;26(7):1037–40.
Jin X, Lian JS, Hu JH, et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69(6):1002–9.
Dai H, Zhang X, Xia J, et al. High-resolution chest CT features and clinical characteristics of patients infected with COVID-19 in Jiangsu, China. Int J Infect Dis. 2020;95:106–12.
Ren HW, Wu Y, Dong JH, et al. Analysis of clinical features and imaging signs of COVID-19 with the assistance of artificial intelligence. Eur Rev Med Pharmacol Sci. 2020;24(15):8210–8.
Mei X, Lee HC, Diao KY, et al. Artificial intelligence-enabled rapid diagnosis of patients with COVID-19. Nat Med. 2020;26(8):1224–8.
Apostolopoulos ID, Mpesiana TA. COVID-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43(2):635–40.
Razzak I, Naz S, Rehman A, et al. Improving coronavirus (COVID-19) diagnosis using deep transfer learning. medRxiv. 2020.
Minaee S, Kafieh R, Sonka M, et al. Deep-COVID: Predicting COVID-19 from chest x-ray images using deep transfer learning. Med Image Anal. 2020;65:101794.
Konar D, Panigrahi BK, Bhattacharyya S, et al. Auto-diagnosis of COVID-19 using lung CT images with semi-supervised shallow learning network. IEEE Access. 2021;9:28716–28.
Wang L, Lin ZQ, Wong A. COVID-net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-Ray images. Sci Rep. 2020;10(1):1–12.
Maghdid HS, Asaad AT, Ghafoor KZ, et al. Diagnosing COVID-19 pneumonia from X-ray and CT images using deep learning and transfer learning algorithms. In: Multimodal image exploitation and learning. International Society for Optics and Photonics; 2021.
Rahaman MM, Li C, Yao Y, et al. Identification of COVID-19 samples from chest X-Ray images using deep learning: A comparison of transfer learning approaches. J X-Ray Sci Technol. 2020;1–19.
Jaiswal A, Gianchandani N, Singh D, et al. Classification of the COVID-19 infected patients using DenseNet201 based deep transfer learning. J Biomol Struct Dyn. 2020:1–8.
Wang X, Deng X, Fu Q, et al. A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE Trans Med Imaging. 2020;39(8):2615–25.
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition; 2017. pp. 4700–8.
Sandler M, Howard A, Zhu M, Zhmoginov A, Chen LC. Mobilenetv2: Inverted residuals and linear bottlenecks. In: Proceedings of the IEEE conference on computer vision and pattern recognition; 2018. pp. 4510–20.(2018)
Zoph B, Vasudevan V, Shlens J, Le QV. Learning transferable architectures for scalable image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition; 2018. pp. 8697–710.
Zhao J, Zhang Y, He X, Xie P. COVID-CT-dataset: a CT scan dataset about COVID-19. 2020.
Rahimzadeh M, Attar A, Sakhaei SM. A fully automated deep learning-based network for detecting COVID-19 from a new and large lung CT scan dataset. Biomed Signal Process Control. 2021;68:102588.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mishra, S. Deep Transfer Learning-Based Framework for COVID-19 Diagnosis Using Chest CT Scans and Clinical Information. SN COMPUT. SCI. 2, 390 (2021). https://doi.org/10.1007/s42979-021-00785-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-021-00785-4