Abstract
Autophagy is a highly conserved self-degradation process of eukaryotic cells which is required for the effective elimination of damaged and unnecessary cytosolic constituents. Defects in the process can cause the intracellular accumulation of such damages, thereby leading to the senescence and subsequent loss of the affected cell. Defective autophagy hence is implicated in the development of various degenerative processes, including cancer, neurodegenerative diseases, diabetes, tissue atrophy and fibrosis, and immune deficiency, as well as in accelerated aging. The autophagic process is mediated by numerous autophagy-related (ATG) proteins, among which the ATG8/LC3/GABARAP (Microtubule-associated protein 1A/1B-light chain 3/Gammaaminobutyric acid receptor-associated protein) superfamily has a pivotal role in the formation and maturation of autophagosome, a key (macro) autophagic structure (the autophagosome sequesters parts of the cytoplasm which are destined for breakdown). While in the unicellular yeast there is only a single ATG8 protein, metazoan systems usually contain more ATG8 paralogs. ATG8 paralogs generally display tissue-specific expression patterns and their functions are not strictly restricted to autophagy. For example, GABARAP proteins also play a role in intracellular vesicle transport, and, in addition to autophagosome formation, ATG8 also functions in selective autophagy. In this review, we summarize the functional diversity of ATG8/LC3/GABARAP proteins, using tractable genetic models applied in autophagy research.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
During autophagy (cellular self-eating), damaged, unnecessary constituents of the cytoplasm are delivered to the lysosomal compartment for enzymatic degradation (Mizushima et al. 2008; Levine and Kroemer 2008; Vellai et al. 2009). Thus, autophagy plays a major role in maintaining cellular homeostasis. The mechanism of this catabolic process is highly conserved among diverse eukaryotic taxa (Takács-Vellai et al. 2006; Sigmond et al. 2008; Takács-Vellai and Vellai 2010; Kovács et al. 2017). In humans, defects in autophagy can lead to the intracellular accumulation of toxic proteins, thereby triggering the development of an organ dysfunction and age-associated disease. Such pathologies include various neurodegenerative diseases (among which the most prevalent ones involve Alzheimer’s, Parkinson’s and Huntington’s diseases, as well as Amyotrophic Lateral Sclerosis), cancer, muscle atrophy and diabetes (Mizushima et al. 2008; Deretic 2009; Tan et al. 2014). Hyperactivity of the process can also cause various age-related diseases (Gozuacik and Kimchi 2004). Based on the mechanism by which the autophagic cargo enters the lysosome, three major types of autophagy can be distinguished, chaperone-mediated autophagy, microautophagy and macroautophagy. Among these processes, quantitatively macroautophagy (hereafter referred to as autophagy) is the most significant one.
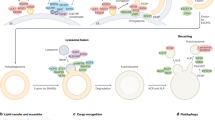
During autophagy, a primer isolation (double) membrane is formed, called phagophore, the growing and completion of which generate a vesicle called autophagosome. Autophagy proteins (ATG) are organized into different complexes that function in distinct steps of autophagosome biogenesis (Suzuki 2013). The induction of phagophore/PAS (pre-autophagosomal structure) depends on the operation of the Atg1/Atg13/Atg17 initiation complex (Wirth et al. 2013). The complex activates another complex, Atg6/Atg14/Vps34/Vps15 (Vacuolar protein sorting), which is required for the autophagic membrane nucleation. During vesicle nucleation, phosphatidylinositol (PI) is converted into phosphatidylinositol-3 phosphate (PI3P), and this reaction ensures that the generated membrane is committed to the autophagic way (Roberts and Ktistakis 2013). Atg18 binds to membranes labelled by PI3P and recruits ubiquitin-like complexes required for phagophore/PAS formation (Proikas-Cezanne et al. 2015). The membran source at the initiation of the autophagosome formation delivered by Atg9 vesicles from multiple organelles (Nishimura et al. 2020). The Atg2-Atg18 complex tethers the isolation membrane to the ER (Kotani et al. 2018). These complexes mediate membrane growth and closure, as well as in matured autophagosome-lysosome/vacuole fusion (Nguyen et al. 2016). Other complexes are also involved in the process of autophagosome-lysosome/vacuole fusion. A so-called Mon1-Ccz1 complex activates the RAB GTPase protein Ypt7. The activated Ypt7-GTP binds the hexameric, anchoring HOPS (homotypic vacuole fusion and protein sorting) complex. In higher eukaryotes, more small GTP proteins participate in vesicle transport and fusion events, including Rab2 (Ras-associated binding protein 2), Rab7 (ortholog of Ypt7 in metazoa) and Arl8 (Arf-like protein 8) (Boda et al. 2019). The HOPS complex binds to the vesicles involved in the fusion, thereby forming a bridge between them, and recognizes specific SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins located on the surface of vesicles (Reggiori and Ungermann 2017). During vesicle fusion, autophagosome fuses with a lysosome to form an autolysosome, in which the cargo is degraded by acidic hydrolases such as proteases, lipases, nucleases and glycosidases. Autophagy can even degrade large protein aggregates and entire organelles (e.g., mitochondria-the process is called mitophagy) (Mizushima and Levine 2010).
The role of ATG8/LC3 in autophagy
The autophagic process is mediated by ATG proteins. In yeast, 34 different ATG proteins has been identified so far. One of these proteins is ATG8 that has a pivotal role in autophagosome formation and autophagosome-lysosome fusion. In metazoan systems, generally more than one ATG8 paralogs present whose functions can significantly differ from each other (Shpilka et al. 2011). In this article, we overview the evolutionary and functional diversity of ATG8 paralogs in major genetic models used in autophagy research.
The formation and completion of the phagophore membrane into an autophagosomes are regulated by two distinct protein complexes, the ATG8/LC3 lipidation complexes and the ATG12-ATG5-ATG16 conjugation complex. The major role of ATG8 is to mediate autophagosome membrane growth and elongation, as well as the closure of the isolation membrane into an autophagosome (vesicle). In the absence of ATG8, no matured autophagosome (vesicle) is formed. Another important role of the protein is to mediate selective autophagy, during which a specific component (e.g., macromolecule, organelle or viral/bacterial particle) is sequestered for degradation. Such a component is anchored to the lipid-conjugated ATG8 through an adaptor protein, such as Ref(2)P/p62 in Drosophila melanogaster (Johansen and Lamark 2011; Bjørkøy et al. 2005; Nezis et al. 2008). In the operation of the conjugation complex, ATG4 first activates ATG8. ATG4 is a cysteine protease that cleaves Atg8 to generate a reactive, C-terminal glycine on it (Yu et al. 2012). After the cleavage, the E1-like ATG7 further activates ATG8 under the consumption of ATP. The modified ATG8 is then processed by the E2-like ATG3 (Ichimura et al. 2000). For the ATG-lipid conjugation process, an E3-like enzyme is also required which is provided by the heterotetrameric complex ATG5-ATG12-ATG16 (Tanida et al. 2002). In this complex, ATG12 becomes activated by ATG7 (hence, ATG7 participates in both conjugation systems). Then, the activated ATG12 conjugates to ATG5, mediated by the E2-like enzyme ATG10. This enzymatic chain ensures a covalent bound between ATG8 and the lipid phosphatidyletanolamine (PE). The ATG12-ATG5 dimer becomes tetramerized with ATG16 (in mammals, Apg16L) (Mizushima 2003). In addition to the covalent binding of Atg8 to the lipid, the complex also functions in its own protection. It enables the binding of ATG8 to PE until the closure of the autophagosome becomes completed (Nakatogawa et al. 2012). The ATG5-ATG12-ATG16 heterotetramer finally dissociates from the mature, closed autophagosome, thereby ensuring Atg4 to cleave ATG8 from the outer membrane of the vesicle. So, Atg4 has a dual role in controlling ATG8. First, it activates ATG8, second it inhibits the membrane localization of ATG8 (Tanida et al. 2004). It is intriguing that Drosophila mutants defective for Atg7 function are viable (Juhász et al. 2007). In these mutant animals, however, autophagy still operates because in the fly mid-intestinal cells Atg7 and Atg3 are dispensable for the process. In this case, another E1-like enzyme, Uba1, substitutes the role of Atg7 in the ubiquitin-like conjugation and lipidization complexes (Chang et al. 2013) (Fig. 1). As mentioned above, Atg8 is also involved in selective autophagy. In mammals, p62 can bind endoplasmic reticulum- (ER) derived autophagic membrane even in the absence of LC3 (Itakura and Mizushima 2011). p62 can therefore function in the lysosomal degradation of selectively anchored cytoplasmic constituents independently of LC3. These results suggest that during autophagy p62 is also able to bind other proteins than LC3.
The role of ubiquitin-like lipidization and conjugation complexes in anchoring ATG8 to the phagophore membrane. The linking of ATG8 to the isolation/phagophore membrane is an essential step of the mechanism of autophagy; it is required for autophagosome formation and completion. This process is mediated by several enzymes. The soluble ATG8 protein (Atg8-I) is first cut by an ATG4 protease to remove the C terminus, thereby liberating a glycine at the C terminal end of the protein. ATG8 is then activated by a E1-like (E - labels different types of ubiquitin-like enzymes) ATG7 protein, which processes it to an E2-like ATG3 protein.
In yeast, there is single ATG8 protein
In the Saccharomyces cerevisiae, ATG8 was described in 1993 when Ohsumi and colleagues identified 15 different autophagy defective mutant strains, which accumulated defective autophagic vacuoles during starvation (Tsukada and Ohsumi 1993). The identified genes were originally called apg1-15. Other laboratories also performed genetic screens in yeast to identify genes implicated in autophagy. As a result, ATG8 was named in different ways: Apg8 and Aut7 (Thumm et al. 1994), Cvt5 (Harding et al. 1995), and Paz2 (Mukaiyama et al. 2002). The unionization of the nomenclature was taken in 2003, giving the final name Atg8 (Klionsky et al. 2003). In yeast, there is a single ATG8 protein, no other paralog exists (Kumar et al. 2020; Weiergräber et al. 2013).
The conserved parts of Atg8 involve a so-called (AIM (Atg8-family interacting motif)) binding site, a C-terminal ubiquitin-like domain and an N-terminal helix domain. The latter contains an alpha helix and an exposed beta-strand that is characteristic to the ATG8 family proteins specifically. Atg8 also possesses two hydrophobic motifs, W-site and L-site, which are required for recognizing tryptophan and leucine in the interaction motif (WXXL). Although the hydrophobic motifs are conserved, L-site slightly differs among the Atg8 homologs (Noda et al. 2010). The yeast ATG8 also acts in the vacuolar fusion process. To perform this function, the C-terminal activity of the protein is needed, while its lipidization is indispensable (Tamura et al. 2010).
Divergence of ATG8 proteins in metazoan systems
According to phylogenetic analyses, 3 different ATG8 paralogous subfamilies emerged when multicellular organisms raised (Lee and Lee 2016). These are represented by MAP1LC3 (microtubule-associated protein 1 light chain 3–hereafter referred as to LC3), GABARAP (γ-aminobutyric acid receptor-associated protein) and GATE-16 (Golgi-associated ATPase enhancer of 16 kDa protein). Each of these protein subfamilies were identified in bilateral organisms by sequence analysis, but also present in lower ranked eukaryotic taxa such as Sponges and Cnidaria (Shpilka et al. 2011). Despite identifying them in higher ranked eukaryotic taxa (for example, there are 2 GABARAP orthologs in Drosophila–Srivastava et al. 2010), the functional analysis of each of the 3 subfamilies has remained unresolved. In the next chapters, we introduce the functional diversity of Atg8 paralogs in 3 different genetic models used most frequently in autophagy research, the nematode Caenorhabditis elegans, fruit fly Drosophila melanogaster and human Homo sapiens (Table 1). The evolutionarily divergence of ATG8 superfamily proteins can be seen in Figure 2.
The evolutionarily relationships of Atg8 orthologs. Atg8 proteins in yeast, nematodes, flies, and humans. The scheme is based on a phylogenetic tree calculated from protein multiple alignment of Atg8 proteins from representative species with complete genome data. The human GABARAP family, including the GATE-16 protein, is closely related to the yeast Atg8. The two D. melanogaster orthologs and C. elegans LGG-1 have higher similarities to GABARAP proteins, while LGG-2 is closely related to LC3. The tree was calculated using the MEGAX program version 11.0.11. with the UPGMA (unweighted pair group method with arithmetic mean) algorithm.
LC3 and GABARAP paralogs in Caenorhabditis elegans
In C. elegans, two Atg8 paralogs have been identified so far. The sequence of LGG-1 (LC3, GABARAP and GATE-16 family) shows a strong similarity to the mammalian GABARAP protein family, while LGG-2 is most strongly related to LC3 (Jenzer et al. 2014). These proteins function in aging control (life span determination) and dauer larva (a developmental diapause that is triggered by crowding in the wild type) formation. The role of these paralogs in these processes is additive, so they act in parallel to control aging and development (Alberti et al. 2010). LGG-1 affects the designation of cytoplasmic materials destined for degradation and autophagosome formation via modulating the UNC-51/EPG-1 complex (uncoordinated-51/ectopic P granules-1; UNC-51 is orthologous to Atg1, while EPG-1 is similar to mammalian Atg14) (Wu et al. 2015). Being an interaction partner of Vps34/LET-512 (lethal), LGG-2 plays a role in autophagosome maturation and autophagosome-lysosome fusion process (Manil-Ségalen et al. 2014). It anchors the LGG-3/ATG12-ATG-5-ATG-16 complex to the isolation membrane, and this step ensures the tight closure of the autophagosome (Wu et al. 2015). In addition, LGG-2 regulates the size of autophagosomes during early embryonic development. Prior to the 200-cell-embryonic stage, the protein controls selective autophagy independently of scaffold proteins (Wu et al. 2015).
Atg8a and Atg8b have different roles in Drosophila melanogaster
In Drosophila, Atg8 is also a member of membrane conjugation complexes, which has an important role in the biogenesis and elongation of autophagic membranes (Mizushima et al. 1998; Ichimura et al. 2000). The Drosophila Atg8 protein family includes two members, Atg8a (CG32672) and Atg8b (CG12334). The function of Drosophila Atg8a can be associated with basal and starvation-induced autophagy mainly which is similar to mammalian LC3 (Scott et al. 2004). Although Atg8a was identified functionally as an LC3 ortholog, based on protein sequence it is more related to GABARAPL1 (Figure 2). In addition to basal and starvation-induced autophagy, Atg8a, which is a ubiquitin-like protein, also participates in developmental autophagy, for example, it is important in the formation of the eye disc (Billes et al. 2018) or in the degradation of larval salivary gland and fat body during metamorphosis (Berry and Baehrecke 2007) (Figure 3). The protein is also involved in the determination of life span (Cho et al. 2021). Similar to classical ubiquitination, E1-E3-like activation enzymes mediate the procession of Atg8a. Contrary to ubiquitination, however, the main target of Atg8a is the membranous PE. When Atg8a is covalently linked to the isolation membrane, the protein enables phagophore elongation, contributes to phagophore closure into an autophagosomal structure, and serves as an anchor for various selective autophagy receptors, motor protein adaptors and factors mediating autophagosome-lysosome fusion (Ichimura et al. 2000). Atg8a is recruited to the phagophore, thus it can be used as key marker for monitoring the autophagic process (Scott et al. 2004). The presence of Atg8a was detected in aging neurons, and its role in these cells is required for resistance against oxidative stress and normal life span (Simonsen et al. 2008). The later function of the protein is established through the pathway mediated by the TOR (target of rapamycin) kinase (Vellai 2021). Ref(2)P, the Drosophila ortholog of mammalian p62, becomes degraded in an Atg8a-dependent way. Thus, in the absence of Atg8a function, Ref(2)P accumulates in the form of aggregates in the brain (Bhukel et al. 2019). The later function of the protein is established through the pathway mediated by the TOR (target of rapamycin) kinase. During this process, Atg8a is regulated developmentally, that is specific HOX proteins (master regulators of early development), such as Lab, influence Atg8a expression in the morphogenetic furrow enabling differentiation signal (Billes et al. 2018; Curtiss et al. 2002). It was shown in the Drosophila larval fat body that the spatiotemporal expression of Hox genes determines autophagic activity during different developmental stages (Banreti et al. 2014). In the feeding larval stage (L3F) Deformed, Ultrabithorax and AbdominalB downregulate autophagy while during in the wandering larval stage (L3W) these HOX proteins do not influence the process because they become downregulated in the latter stage.

source and moves away for molting), which triggers starvation-induced autophagy (a large amount of red foci in the fluorescent figure). When animals at the L3F are exposed to food deprivation, starvation-induced autophagy also becomes induced (numerous red foci in the middle panel). mCherry-Atg8a marker labels early and late autophagic structures (red foci – phagophores, autophagosomes and autolysosomes), Hoechst staining (blue) indicates nuclei. Figures were taken with the same exposure time.
Autophagic activity in Drosophila larval fat body cells at different developmental stages. The L3 larval stage in Drosophila can be divided into two distinct stages: first, the L3 feeding (L3F) stage (left and middle panels), then, the L3 wandering (L3W) stage (right panel). In the latter, the animal does not feed (the animal leaves the food
Autophagy is also implicated in programmed cell death that occurs during Drosophila development. In this process, Atg8a play a pivotal role. During methamorphosis, degradation of larval mid-gut is established by autophagy in a caspase-independent way (Xu et al. 2015). In this process, autophagy operates independently of Atg3 and Atg7 function (Chang et al. 2013). An additional interesting phenomenon is that the degradation of mid-gut caeca is independent of the lipidation of Atg8a through its C-terminal glycine (Jipa et al. 2021).
Atg8b is specific to Drosophila, and its expression is restricted to males only, mainly in the testis (Vedelek et al. 2018). Based on the evolutionary development of Atg8 family proteins, in most species of Diptera order, the second was lost; however, in Drosophilidae their second Atg8a gene was originated by duplication resulting in the appearance of a new paralog, the Drosophilidae-specific Atg8b. This evolutionary phenomenon had a retrotransposone-mediated event, because Atg8b lacks introns. On an aminoacid sequence level the insect Atg8a and the Drosophilidae-specific Atg8b are more similar to human GABARAP subfamily rather than MAP1LC3 proteins. The protein is important in maintaining male fertility by controlling postmeiotic spermatide generation and mobility. Atg8b deficiency leads to male sterility. The role of Atg8b is independent of autophagy (Jipa et al. 2021).
Roles of the mammals Atg8 protein family
While in yeast there is only a single Atg8 protein, in mammals at least eight Atg8 paralogs have been identified (Shpilka et al. 2011). Based on their amino acid sequences, the mammalian ATG8 proteins can be classified into three subfamilies: LC3 (LC3A-C-LC3A has two alternative splice variants), GAGARAP (GABARAP, GABARAP-L1 and GABARAP-L3) as well as GATE-16 (it is also called GABARAPL2) (Xie et al. 2008; Srivastava et al. 2010). Among these proteins, LC3B is the most intensely investigated factor. These protein subfamilies differ from each other in both function and expression/activity level. In the LC3 subfamily, LC3C has the lowest expression activity, which mainly is restricted to the lung. The expression of GABARAP and GATE-16 proteins is more evident in the central nervous system (Tolle et al. 2008; Sagiv et al. 2000). Interestingly, GABARAP-L1 expression is restricted to the somatomotoric and endocrine regulatory (pons, diencephalon) areas. In contrast, GABARAP accumulation is more evident in endocrine glands (Nemos et al. 2003).
Weiberg and colleagues showed that each LC3 paralog exerts an activity in autophagy, but they have a role in different stages of autophagosome formation (Weidberg et al. 2010). Both downregulation and hyperactivation of LC3 and GABARAP significantly alter the size of the phagophore in an opposite way; downregulation of GABARAP increases while LC3 inactivation decreases phagophore size as compared with control. The Atg8 subfamilies investigated so far influence only autophagosome formation and maturation, but do not alter lysosomal characteristics (acidification, size, enzyme activity) (Weidberg et al. 2010). Based on these features, members of the LC3 subfamily has a primary role in phagophore elongation, while GABARAP and GATE-16 paralogs function mainly in the later stages of autophagosome maturation (Albanesi et al. 2015). GABARAP influences the function of the HOPS complex through binding PLEKHM1 (Pleckstrin homology domain containing protein member 1) (McEwan et al. 2015). Thus, GABARAP proteins also function in the autophagosome-lysosome fusion event.
Previously, LC3/ATG8 has been known to have important function during formation and maturation of autophagosome and it also has role in selective autophagy. A recent study shows a new function of LC3B. It binds directly to a specific AAUAAA sequence on mRNAs. The activation of autophagy leads to the degradation of the mRNAs targeted by LC3B. The CCR4-NOT complex influences the elimination and it happens before the formation of autolysosomes. A specific target of LC3B is PRMT1 mRNA which is translated into an autophagy inhibitor protein. If PRMT1 mRNA is degraded, the autophagy will be induced (Hwang et al. 2022).
In mammals, the ATG8 protein subfamilies also have roles that are independent of autophagy. GABARAP and GATE-16 members participate in the transport of plasma membrane proteins (e.g., receptor trafficking) in an autophagy-independent manner (Wang et al. 1999; Leil et al. 2004). In addition, these ATG8 subfamilies are implicated in ER-Golgi transport through controlling NSF (N-ethylmaleimide sensitive factor) (Sagiy et al. 2000; Legesse-Miller et al. 1998; Muller et al. 2002). NSF dictates the dissociation of the SNARE complex. So, GABARAP and GATE-16 family proteins modulate membrane fusion processes as well. Moreover, GABARAP subfamily proteins have a role in the ER degradation pathway (ERAD–ER- associated degradation), which is a checkpoint system in the breakdown of misfolded proteins. EDEM1 is the effector protein of the process which, in case of steady stage, rapidly dissociates from the ER. Fractions containing EDEM1 are surrounded by soluble LC3 (LC3-I), thereby easily distinguishing these vesicles from LC3-II-associated autophagosomes. Membrane of vesicles generated during phagocytosis can contain LC3 (Schaaf et al. 2016).
The ATG8 family of proteins display a tumour suppression function, which mainly results from the selective degradation of oncogenic proteins and damaged organelles. It is also possible that the cytoplasmic form of ATG8, which is not conjugated to autophagic vesicles, has a potent tumour suppression function (Poillet-Perez et al. 2017). These proteins may influence signaling pathways; for example, GABARAP is capable of blocking cell cycle by activating p53, thereby lowering cell survival, movement and progression in ovarian and breast tumours. Furthermore, they act negatively on the phosphatidylinositol-3 kinase/Akt signaling pathway to inhibit tumour angiogenesis and cell division in ovarian cancer (Jacquet et al. 2021). GABARAP-L1 is associated with various neurodegenerative pathologies, such as Alzheimer’s, Parkinson’s and Huntington’s diseases (Le Grand et al. 2013).
Scaffold proteins involved in selective autophagy are evolutionarily conserved
Scaffold and adaptor proteins are required for the effective and selective transformation of information during signal transduction. Ref(2)P/p62 scaffold protein plays a role in different signaling systems and binds to Atg8 homologs. Thus, the protein is under a continuous degradation by selective autophagy (Moscat et al. 2007).
Autophagic receptors have a characteristic structure, which consists of the following three specific domains, LIR (LC3 interacting region)/AIM (Atg8 interacting motif), UBA (ubiquitin-associated) and PB1 (Phox and Bem1p). UBA domain makes possible the autophagic degradation of ubiquitinated proteins. This process is ensured by specific autophagy receptors. Receptors bind with the LIR domain to the LDS (LIR docking site) domain of LC3. p62/SQSTM1 (sequestosome-1) and NBR1 (neighbor of BRCA1 gene 1) were first identified as interaction partners (Johansen and Lamark 2011). To date, several LIR domain-containing proteins have been described as important adaptors in selective autophagy, e.g., OPTIN (optineurin), NDP52 (Nuclear dot protein 52 kDa) and BNIP3L/NIX (BCL2/adenovirus E1B 19 kDa-interacting protein 3-like). It is worth mentioning that different adaptor proteins bind to ATG8 subfamily proteins with different affinities. For instance, NDKP52 prefers LC3 rather than GABARAP and GABARAP-L1, while BNIP3L binds stronger to GABARAP-L1 that LC3 (Lee and Lee 2016).
Drosophila Atg8a interacts with Sir2 deacetylase to become deacetylated during starvation, thereby promoting the activation of autophagy. Expression of Atg8 gene is controlled at the acetylation level. Atg8a interacts with Seqouia (CG32904), which is a transcription factor, that negatively regulates autophagy. YL-1, a component of a nuclear acetyltransferase complex, and Sir2, which deacetylates Atg8a during starvation, induce autophagy (Jacomin et al. 2020). The Sir2 protein is able to bind the ubiquitinated structure to membrane surface covered by Atg8. Such a domain is found in p62, which mediates the degradation of misfolded proteins, or in NBR1 (Lamark et al. 2009) and optaneurine (Wild et al. 2011). Mammalian p62 is a multifunctional scaffold protein that binds to polyubiquitinated aggregates. Its Drosophila ortholog, Ref(2)P, has a key role in the autophagic degradation of ubiquitinated, supramolecular complexes (He and Klionsky 2009). In addition to its interaction with atypical PKC (protein kinase C) kinases, it has effects on signaling systems, such as Toll and Nrf2/Keap1 (Avila et al. 2002; Jiang et al. 2015), Ref(2)P/p62 can directly bind to Atg8 (Nezis et al. 2008). In case of deleting the LIR motif of Drosophila Ref(2)P, it is observed accumulated in the system, thus these aggregates induce the cnc/NFE2L2/Nrf2 antioxidant pathway (Bhattacharjee et al. 2022). p62 operates as an adaptor protein; it binds ubiquitinated aggregates and facilitates their transport to autophagosomes through binding to Atg8 (Pankiv et al. 2007). p62 levels are established by autophagy because the protein serves as a substrate for the process. Its levels are actually indicative for the flux of autophagy (Komatsu et al. 2007).
Alfy (autophagy-linked FYVE protein) is a 400 kDa-size protein that contains three domains, BEACH (Beige and Chediak-Higashi), WD-40 (short ~40 amino acid motifs, often terminating in a Trp-Asp (W-D) dipeptide) and FYVE (named for the first four proteins in which it was recognized, Fab1p, YOTB, Vac1p, and EEA1). The protein is evolutionarily highly conserved: Drosophila [Blue Cheese (BCHS), AE003611], C. elegans (contig of Z99169+AL032675+Z93390), Dictyostelium discoideum (LvsA, AF088979), Arabidopsis thaliana (AC002330) and Schizosaccharomyces pombe (CAA20312). The nematode and fruit fly homologs show a 30% amino acid identity with that of the human Alfy (Simonsen et al. 2004). The selective recruitment and binding of Alfy to LC3B is promoted by GABARAP (Lystad et al. 2014). It is also required for the autophagic degradation of aggregated proteins. Alfy co-localizes and interacts with p62- and NBR1-bounded proteins, and directly binds to Atg5. This interaction makes possible the formation of the Atg12-Atg16L-LC3 complex. Therefore, Alfy exhibits a scaffold function to recruit components of autophagy to a space where aggregated proteins are located. Further experiments demonstrated that Alfy is indispensable for selective autophagy in Drosophila (Filimonenko et al. 2010).
Discussion
The ATG8 family of proteins have a pivotal role in the mechanism of autophagy. While in yeast there is only a single ATG8-like protein, in metazoan systems the ATG8 paralogs can be divided into three subclasses. Components of these subclasses often differ in function and cytoplasmic/tissue localization. This raises the possibility that ATG8 paralogs may have specific functions. Drosophila Atg8b, for example, can exert autophagy-independent cellular activities. Although LC3B has been well characterized functionally, the role of other paralogs has been less understood. Increasing our knowledge about ATG8 proteins is medically highly relevant because, for instance, GABARAP deficiency is linked to the development of several types of cancer and neurodegenerative diseases. In the future, the following questions should be addressed. Where are ATG8 paralogs expressed during development and in the adult body? What is the exact function of specific ATG8 proteins in the mechanism of autophagy? At what extent are the functions of ATG8 paralogs redundant? What is the source of specificity of adaptor proteins to bind specific ATG8 paralogs?
References
Albanesi J, Wang H, Sun HQ, Levine B, Yin H (2015) GABARAP-mediated targeting of PI4K2A/PI4KIIα to autophagosomes regulates PtdIns4P-dependent autophagosome-lysosome fusion. Autophagy 11(11):2127–2129. https://doi.org/10.1080/15548627.2015.1093718
Alberti A, Michelet X, Djeddi A, Legouis R (2010) The autophagosomal protein LGG-2 acts synergistically with LGG-1 in dauer formation and longevity in C. elegans. Autophagy 6(5):622–633. https://doi.org/10.4161/auto.6.5.12252
Avila A, Silverman N, Diaz-Meco MT, Moscat J (2002) The Drosophila atypical protein kinase C-ref(2)p complex constitutes a conserved module for signaling in the toll pathway. Mol Cell Biol 22(24):8787–8795. https://doi.org/10.1128/MCB.22.24.8787-8795.2002
Banreti A, Hudry B, Sass M, Saurin AJ, Graba Y (2014) Hox proteins mediate developmental and environmental control of autophagy. Dev Cell 28(1):56–69. https://doi.org/10.1016/j.devcel.2013.11.024
Berry DL, Baehrecke EH (2007) Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 131(6):1137–1148. https://doi.org/10.1016/j.cell.2007.10.048
Bhattacharjee A, Ürmösi A, Jipa A et al (2022) Loss of ubiquitinated protein autophagy is compensated by persistent cnc/NFE2L2/Nrf2 antioxidant responses. Autophagy. https://doi.org/10.1080/15548627.2022.2037852
Bhukel A, Beuschel CB, Maglione M et al (2019) Autophagy within the mushroom body protects from synapse aging in a non-cell autonomous manner. Nat Commun 10(1):1318. https://doi.org/10.1038/s41467-019-09262-2
Billes V, Kovács T, Manzéger A et al (2018) Developmentally regulated autophagy is required for eye formation in Drosophila. Autophagy 14(9):1499–1519. https://doi.org/10.1080/15548627.2018.1454569
Bjørkøy G, Lamark T, Brech A et al (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171(4):603–614. https://doi.org/10.1083/jcb.200507002
Boda A, Lőrincz P, Takáts S et al (2019) Drosophila Arl8 is a general positive regulator of lysosomal fusion events. Biochim Biophys Acta Mol Cell Res 4:533–544. https://doi.org/10.1016/j.bbamcr.2018.12.011
Chang TK, Shravage BV, Hayes SD et al (2013) Uba1 functions in Atg7- and Atg3-independent autophagy. Nat Cell Biol 15(9):1067–1078. https://doi.org/10.1038/ncb2804
Cho LC, Yu CC, Kao CF (2021) Social perception of young adults prolongs the lifespan of aged Drosophila. NPJ Aging Mech Dis 7(1):21. https://doi.org/10.1038/s41514-021-00073-8
Curtiss J, Halder G, Mlodzik M (2002) Selector and signalling molecules cooperate in organ patterning. Nat Cell Biol 4(3):E48–E51. https://doi.org/10.1038/ncb0302-e48
Deretic V (2009) Links between autophagy, innate immunity, inflammation and Crohn’s disease. Dig Dis 27(3):246–251. https://doi.org/10.1159/000228557
Filimonenko M, Isakson P, Finley KD et al (2010) The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell 38(2):265–279. https://doi.org/10.1016/j.molcel.2010.04.007
Gozuacik D, Kimchi A (2004) Autophagy as a cell death and tumor suppressor mechanism. Oncogene 23(16):2891–2906. https://doi.org/10.1038/sj.onc.1207521
Harding TM, Morano KA, Scott SV, Klionsky DJ (1995) Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol 131(3):591–602. https://doi.org/10.1083/jcb.131.3.591
He C, Klionsky DJ (2009) Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43:67–93. https://doi.org/10.1146/annurev-genet-102808-114910
Hwang HJ, Ha H, Lee BS, Kim BH, Song HK, Kim YK (2022) LC3B is an RNA-binding protein to trigger rapid mRNA degradation during autophagy. Nat commun 13(1):1436. https://doi.org/10.1038/s41467-022-29139-1
Ichimura Y, Kirisako T, Takao T et al (2000) A ubiquitin-like system mediates protein lipidation. Nature 408(6811):488–492. https://doi.org/10.1038/35044114
Itakura E, Mizushima N (2011) p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol 192(1):17–27. https://doi.org/10.1083/jcb.201009067
Jacomin AC, Petridi S, Di Monaco M et al (2020) Regulation of expression of autophagy genes by Atg8a-interacting partners sequoia, YL-1, and Sir2 in drosophila. Cell Rep 31(8):107695. https://doi.org/10.1016/j.celrep.2020.107695
Jacquet M, Guittaut M, Fraichard A, Despouy G (2021) The functions of Atg8-family proteins in autophagy and cancer: linked or unrelated? Autophagy 17(3):599–611. https://doi.org/10.1080/15548627.2020.1749367
Jenzer C, Manil-Ségalen M, Lefebvre C, Largeau C, Glatigny A, Legouis R (2014) Human GABARAP can restore autophagosome biogenesis in a C. elegans lgg-1 mutant. Autophagy 10(10):1868–1872. https://doi.org/10.4161/auto.29745
Jiang T, Harder B, Rojo de la Vega M, Wong PK, Chapman E, Zhang DD (2015) p62 links autophagy and Nrf2 signaling. Free Radic Biol Med 88(Pt B):199–204. https://doi.org/10.1016/j.freeradbiomed.2015.06.014
Jipa A, Vedelek V, Merényi Z et al (2021) Analysis of Drosophila Atg8 proteins reveals multiple lipidation-independent roles. Autophagy 17(9):2565–2575. https://doi.org/10.1080/15548627.2020.1856494
Johansen T, Lamark T (2011) Selective autophagy mediated by autophagic adapter proteins. Autophagy 7(3):279–296. https://doi.org/10.4161/auto.7.3.14487
Juhász G, Erdi B, Sass M, Neufeld TP (2007) Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev 21(23):3061–3066. https://doi.org/10.1101/gad.1600707
Klionsky DJ, Cregg JM, Dunn WA Jr et al (2003) A unified nomenclature for yeast autophagy-related genes. Dev Cell 5(4):539–545. https://doi.org/10.1016/s1534-5807(03)00296-x
Komatsu M, Waguri S, Koike M et al (2007) Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131(6):1149–1163. https://doi.org/10.1016/j.cell.2007.10.035
Kotani T, Kirisako H, Koizumi M, Ohsumi Y, Nakatogawa H (2018) The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc Natl Acad Sci USA 115(41):10363–10368. https://doi.org/10.1073/pnas.1806727115
Kovács T, Billes V, Komlós M et al (2017) The small molecule AUTEN-99 (autophagy enhancer-99) prevents the progression of neurodegenerative symptoms. Sci Rep 7:42014. https://doi.org/10.1038/srep42014
Kumar S, Jain A, Choi SW et al (2020) Mammalian Atg8-family proteins are upstream regulators of the lysosomalsystem by controlling MTOR and TFEB. Autophagy 16(12):2305–2306. https://doi.org/10.1080/15548627.2020.1837423
Lamark T, Kirkin V, Dikic I, Johansen T (2009) NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle 8(13):1986–1990. https://doi.org/10.4161/cc.8.13.8892
Le Grand JN, Bon K, Fraichard A et al (2013) Specific distribution of the autophagic protein GABARAPL1/GEC1 in the developing and adult mouse brain and identification of neuronal populations expressing GABARAPL1/GEC1. PLoS One 8(5):e63133. https://doi.org/10.1371/journal.pone.0063133
Lee YK, Lee JA (2016) Role of the mammalian ATG8/LC3 family in autophagy: differential and compensatory roles in the spatiotemporal regulation of autophagy. BMB Rep 49(8):424–430. https://doi.org/10.5483/bmbrep.2016.49.8.081
Legesse-Miller A, Sagiv Y, Porat A, Elazar Z (1998) Isolation and characterization of a novel low molecular weight protein involved in intra-Golgi traffic. J Biol Chem 273(5):3105–3109. https://doi.org/10.1074/jbc.273.5.3105
Leil TA, Chen ZW, Chang CS, Olsen RW (2004) GABAA receptor-associated protein traffics GABAA receptors to the plasma membrane in neurons. J Neurosci 24(50):11429–11438. https://doi.org/10.1523/JNEUROSCI.3355-04.2004
Levine B, Kroemer G (2008) Autophagy in the pathogenesis of disease. Cell 132(1):27–42. https://doi.org/10.1016/j.cell.2007.12.018
Lystad AH, Ichimura Y, Takagi K et al (2014) Structural determinants in GABARAP required for the selective binding and recruitment of ALFY to LC3B-positive structures. EMBO Rep 15(5):557–565. https://doi.org/10.1002/embr.201338003
Manil-Ségalen M, Lefebvre C, Jenzer C et al (2014) The C. elegans LC3 acts downstream of GABARAP to degrade autophagosomes by interacting with the HOPS subunit VPS39. Dev Cell 28(1):43–55. https://doi.org/10.1016/j.devcel.2013.11.022
McEwan DG, Popovic D, Gubas A et al (2015) PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell 57(1):39–54. https://doi.org/10.1016/j.molcel.2014.11.006
Meléndez A, Tallóczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B (2003) Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301(5638):1387–1391. https://doi.org/10.1126/science.1087782
Mizushima N, Levine B (2010) Autophagy in mammalian development and differentiation. Nat Cell Biol 12(9):823–830. https://doi.org/10.1038/ncb0910-823
Mizushima N, Noda T, Yoshimori T et al (1998) A protein conjugation system essential for autophagy. Nature 395(6700):395–398. https://doi.org/10.1038/26506
Mizushima N, Kuma A, Kobayashi Y et al (2003) Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci 116(Pt 9):1679–1688. https://doi.org/10.1242/jcs.00381
Mizushima N, Levine B, Cuervo AM, Klionsky DJ (2008) Autophagy fights disease through cellular self-digestion. Nature 451(7182):1069–1075. https://doi.org/10.1038/nature06639
Moscat J, Diaz-Meco MT, Wooten MW (2007) Signal integration and diversification through the p62 scaffold protein. Trends Biochem Sci 32(2):95–100. https://doi.org/10.1016/j.tibs.2006.12.002
Mukaiyama H, Oku M, Baba M et al (2002) Paz2 and 13 other PAZ gene products regulate vacuolar engulfment of peroxisomes during micropexophagy. Genes Cells 7(1):75–90. https://doi.org/10.1046/j.1356-9597.2001.00499.x
Muller JM, Shorter J, Newman R et al (2002) Sequential SNARE disassembly and GATE-16-GOS-28 complex assembly mediated by distinct NSF activities drives Golgi membrane fusion. J Cell Biol 157(7):1161–1173. https://doi.org/10.1083/jcb.200202082
Nakatogawa H, Ishii J, Asai E, Ohsumi Y (2012) Atg4 recycles inappropriately lipidated Atg8 to promote autophagosome biogenesis. Autophagy. 8(2):177–186. https://doi.org/10.4161/auto.8.2.18373
Nemos C, Mansuy V, Vernier-Magnin S, Fraichard A, Jouvenot M, Delage-Mourroux R (2003) Expression of gec1/GABARAPL1 versus GABARAP mRNAs in human: predominance of gec1/GABARAPL1 in the central nervous system. Brain Res Mol Brain Res 119(2):216–219. https://doi.org/10.1016/j.molbrainres.2003.09.011
Nezis IP, Simonsen A, Sagona AP et al (2008) Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol 180(6):1065–1071. https://doi.org/10.1083/jcb.200711108
Nguyen TN, Padman BS, Usher J, Oorschot V, Ramm G, Lazarou M (2016) Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J Cell Biol 215(6):857–874. https://doi.org/10.1083/jcb.201607039
Nishimura T, Tooze SA (2020) Emerging roles of ATG proteins and membrane lipids in autophagosome formation. Cell discov 6(1):32. https://doi.org/10.1038/s41421-020-0161-3
Noda NN, Ohsumi Y, Inagaki F (2010) Atg8-family interacting motif crucial for selective autophagy. FEBS Lett 584(7):1379–1385. https://doi.org/10.1016/j.febslet.2010.01.018
Oliver H, Mohrluder J, Willbol D (2013) Atg8 family proteins—autophagy and beyond. In: Bailly Y (ed) Autophagy-A Double-Edged Sword-Cell Survival or Death? InTech. https://doi.org/10.5772/55647
Pankiv S, Clausen TH, Lamark T et al (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282(33):24131–24145. https://doi.org/10.1074/jbc.M702824200
Poillet-Perez L, Jacquet M, Hervouet E et al (2017) GABARAPL1 tumor suppressive function is independent of its conjugation to autophagosomes in MCF-7 breast cancer cells. Oncotarget 8(34):55998–56020. https://doi.org/10.18632/oncotarget.19639
Proikas-Cezanne T, Takacs Z, Dönnes P, Kohlbacher O (2015) WIPI proteins: essential PtdIns3P effectors at the nascent autophagosome. J Cell Sci 128(2):207–217. https://doi.org/10.1242/jcs.146258
Reggiori F, Ungermann C (2017) Autophagosome maturation and fusion. J Mol Biol 429(4):486–496. https://doi.org/10.1016/j.jmb.2017.01.002
Roberts R, Ktistakis NT (2013) Omegasomes: PI3P platforms that manufacture autophagosomes. Essays Biochem 55:17–27. https://doi.org/10.1042/bse0550017
Sagiv Y, Legesse-Miller A, Porat A, Elazar Z (2000) GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. EMBO J 19(7):1494–1504. https://doi.org/10.1093/emboj/19.7.1494
Schaaf MB, Keulers TG, Vooijs MA, Rouschop KM (2016) LC3/GABARAP family proteins: autophagy-(un)related functions. FASEB J 30(12):3961–3978. https://doi.org/10.1096/fj.201600698R
Scott RC, Schuldiner O, Neufeld TP (2004) Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell 7(2):167–178. https://doi.org/10.1016/j.devcel.2004.07.009
Shpilka T, Weidberg H, Pietrokovski S, Elazar Z (2011) Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol 12(7):226. https://doi.org/10.1186/gb-2011-12-7-226
Sigmond T, Barna J, Tóth ML et al (2008) Autophagy in Caenorhabditis elegans. Methods Enzymol 451:521–540. https://doi.org/10.1016/S0076-6879(08)03230-8
Simonsen A, Birkeland HC, Gillooly DJ et al (2004) Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J Cell Sci 117(Pt 18):4239–4251. https://doi.org/10.1242/jcs.01287
Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD (2008) Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 4(2):176–184. https://doi.org/10.4161/auto.5269
Srivastava M, Simakov O, Chapman J et al (2010) The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466(7307):720–726. https://doi.org/10.1038/nature09201
Suzuki K (2013) Selective autophagy in budding yeast. Cell Death Differ 20(1):43–48. https://doi.org/10.1038/cdd.2012.73
Takács-Vellai K, Bayci A, Vellai T (2006) Autophagy in neuronal cell loss: a road to death. Bioessays. 28(11):1126–1131. https://doi.org/10.1002/bies.20489
Tamura N, Oku M, Sakai Y (2010) Atg8 regulates vacuolar membrane dynamics in a lipidation-independent manner in Pichia pastoris. J Cell Sci 123(Pt 23):4107–4116. https://doi.org/10.1242/jcs.070045
Tan CC, Yu JT, Tan MS, Jiang T, Zhu XC, Tan L (2014) Autophagy in aging and neurodegenerative diseases: implications for pathogenesis and therapy. Neurobiol Aging 35(5):941–957. https://doi.org/10.1016/j.neurobiolaging.2013.11.019
Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E (2002) Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J Biol Chem 277(16):13739–13744. https://doi.org/10.1074/jbc.M200385200
Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E (2004) HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem 279(35):36268–36276. https://doi.org/10.1074/jbc.M401461200
Thumm M, Egner R, Koch B et al (1994) Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett 349(2):275–280. https://doi.org/10.1016/0014-5793(94)00672-5
Tolle F, Risold PY, Mansuy-Schlick V et al (2008) Specific regional distribution of gec1 mRNAs in adult rat central nervous system. Brain Res 1210:103–115. https://doi.org/10.1016/j.brainres.2008.02.077
Tsukada M, Ohsumi Y (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333(1–2):169–174. https://doi.org/10.1016/0014-5793(93)80398-e
Vedelek V, Bodai L, Grézal G et al (2018) Analysis of Drosophila melanogaster testis transcriptome. BMC Genom 19(1):697. https://doi.org/10.1186/s12864-018-5085-z
Vellai T (2021) How the amino acid leucine activates the key cell-growth regulator mTOR. Nature 596(7871):192–194. https://doi.org/10.1038/d41586-021-01943-7
Vellai T, Takács-Vellai K (2010) Regulation of protein turnover by longevity pathways. Adv Exp Med Biol. 694:69–80. https://doi.org/10.1007/978-1-4419-7002-2_7
Vellai T, Takács-Vellai K, Sass M, Klionsky DJ (2009) The regulation of aging: does autophagy underlie longevity? Trends Cell Biol 19(10):487–494. https://doi.org/10.1016/j.tcb.2009.07.007
Wang H, Bedford FK, Brandon NJ, Moss SJ, Olsen RW (1999) GABA(A)-receptor-associated protein links GABA(A) receptors and the cytoskeleton. Nature 397(6714):69–72. https://doi.org/10.1038/16264
Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z (2010) LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J 29(11):1792–1802. https://doi.org/10.1038/emboj.2010.74
Wild P, Farhan H, McEwan DG et al (2011) Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333(6039):228–233. https://doi.org/10.1126/science.1205405
Wirth M, Joachim J, Tooze SA (2013) Autophagosome formation–the role of ULK1 and Beclin1-PI3KC3 complexes in setting the stage. Semin Cancer Biol 23(5):301–309. https://doi.org/10.1016/j.semcancer.2013.05.007
Wu F, Watanabe Y, Guo XY et al (2015) Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2. Autophagy Mol Cell 60(6):914–929. https://doi.org/10.1016/j.molcel.2015.11.019
Xie Z, Nair U, Klionsky DJ (2008) Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell 19(8):3290–3298. https://doi.org/10.1091/mbc.e07-12-1292
Xu T, Nicolson S, Denton D, Kumar S (2015) Distinct requirements of Autophagy-related genes in programmed cell death. Cell Death Differ 22(11):1792–1802. https://doi.org/10.1038/cdd.2015.28
Yu ZQ, Ni T, Hong B et al (2012) Dual roles of Atg8-PE deconjugation by Atg4 in autophagy. Autophagy 8(6):883–892. https://doi.org/10.4161/auto.19652
Funding
Open access funding provided by Eötvös Loránd University. Funding was provided by OTKA, K109349, Tibor Vellai, K132439, Tibor Vellai, ELKH/MTA-ELTE Genetics Research Group, 01062, Tibor Vellai.
Author information
Authors and Affiliations
Contributions
VB Varga and F Keresztes were contributed equally.
Corresponding author
Additional information
Virginia B. Varga and Fanni Keresztes these author have equally contributed this work.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Varga, V.B., Keresztes, F., Sigmond, T. et al. The evolutionary and functional divergence of the Atg8 autophagy protein superfamily. BIOLOGIA FUTURA 73, 375–384 (2022). https://doi.org/10.1007/s42977-022-00123-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42977-022-00123-6