Abstract
The objective of the study was to evaluate the frequency and genetic characteristics of ESBL-producing Escherichia coli and Klebsiella spp. and the risk factors associated with a high total bacterial count in bulk tank milk samples of dairy farms in three municipalities of the Antioquia Department, Colombia. Fifteen samples were positive for E. coli and Klebsiella spp. Subsequent analysis of the 16 S rRNA gene sequences confirmed these isolates included E. coli (n = 3), K. oxytoca (n = 11), and K. pneumoniae (n = 1). None of the isolates was positive for ESBL identification by phenotypic methods, but the only the isolate of K. pneumoniae was positive for the blaSHV61 gene by sequence analysis. The antibiotic susceptibility evaluation for all Klebsiella spp. isolates identified resistance to fosfomycin (50%; 6/12) and ampicillin (100%; 12/12). While most of the herds maintain adequate hygienic quality, specific risk factors such as having more than 60 milking cows, frequent changes in milkers, milking in paddocks, and using a chlorinated product for pre-dipping have been identified as associated with a high total bacterial count > 100,000 CFU/mL in bulk tank milk. However, certain variables including the milker being the owner of the animals and the proper washing and disinfection of the milking machine contribute to maintain a high level of hygiene and quality in the raw milk stored in the tanks. In conclusion, the frequency of ESBL producers was relatively low, with only K. pneumoniae testing positive for the blaSHV ESBL type. The presence of these bacteria in milk tanks represents a potential risk to public health for consumers of raw milk and its derivatives.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Raw bovine milk may contain pathogenic microorganisms that originate from multiple sources of contamination. Due to contaminants introduced through cow udder teats, milking system, and farm environment, the final composition of the microorganisms in bulk tank milk is highly diverse [1]. The hygienic quality of bulk tank milk in Colombia is evaluated by estimating the total bacterial count [2]. A high total bacterial value could indicate contamination mainly with environmental bacteria due to poor hygiene measures at the time of milking, contaminated milking equipment, and insufficient or inadequate cooling during transportation and storage [3,4,5]. In addition to the safety concerns associated with raw milk, an increase in the total bacterial count can also adversely affect its organoleptic and nutritional properties, potentially reducing the selling price or rendering the product unfit for human consumption. Moreover, tank milk samples can also be an important and valuable tool for estimating the individual farm situation concerning to overall bacteriological quality of the milk and pathogenic microorganisms, especially antibiotic resistance situations at the herd level in dairy farms [6, 7]. Due to the growing worldwide problem of resistance to broad-spectrum antibiotics such as cephalosporins among Enterobacteriaceae, studies have increased in recent years, particularly focusing on foods of animal origin and dairy farms as possible reservoirs of resistant bacteria that produce extended-spectrum beta-lactamases (ESBLs). Most of these studies were conducted with Escherichia coli and Klebsiella spp., which are generally considered valuable indicators of antibiotic resistance due to their wide entry into the food chain and environment, as well as their potential for gene exchange between humans and animals. Several studies have identified the presence of ESBL-producing Enterobacteriaceae in bovine milk with clinical mastitis [8, 9]. However, only limited data are available on the presence of ESBL-producing Enterobacteriaceae, especially E. coli and Klebsiella spp., in bulk tank milk [10,11,12]. In Colombia, Vásquez-Jaramillo et al. (2017) first reported in a previous local study the presence of ESBL-carrying Enterobacteriaceae in bulk tank milk samples of dairy herds from a municipality in Antioquia. This suggests that ESBLs are circulating among Enterobacteriaceae in the country´s dairy herds, necessitating the development of larger epidemiological studies that can provide more detailed and accurate information on this phenomenon.
Therefore, this study aimed to estimate the frequency and genetic characteristics of ESBL-producing E. coli and Klebsiella spp. in bulk tank milk samples from dairy farms in three geographically different municipalities of the Antioquia Department, Colombia. Additionally, it aimed to identify the risk factors associated with a high total bacterial count.
Materials and methods
Herds and samples
Information on the dairy herds sampled and the procedure for collecting and handling bulk-tank milk samples for microbiological analysis was previously published by Ágredo-Campos et al., 2023. A total of 150 dairy herds were randomly selected from three municipalities (Santa Rosa de Osos, San Pedro de los Milagros, and Entrerríos) in northern Antioquia Department, Colombia, between August 2019 and January 2020. The dairy herds were classified based on the number of lactating cows as follows: small herds (< 30 dairy cows), medium herds (30–60 dairy cows), and large herds (> 60 dairy cows). Most of the dairy cows in the herds were of the Holstein breed, and they were grazing with supplementary feed at the time of milking. The milking systems used in the herds were mainly paddock and parlor mechanical milking systems, with a few herds practicing manual milking. For the microbiological analysis and evaluation of the total bacterial count, two milk samples were taken from each herd’s milk tank. These samples were collected in sterile containers containing the Bronopol preservative, following the standards established by the National Mastitis Council [13]. The samples were immediately transported to the Laboratory of Milk Quality at the Faculty of Agricultural Sciences of the Universidad de Antioquia, maintaining a temperature of 4 °C +/- 2 °C during transportation.
Microbiological analyses
The microbiological analysis for the detection of E. coli and Klebsiella spp. followed the protocols established by the National Mastitis Council [13]. Briefly, 0.01 mL of milk samples was streaked on MacConkey agar (Merck, Darmstadt, Germany) using a sterile calibrated loop (Biologix® Group Ltd., Shandong, China). After incubation at 37 °C for 24 h, the presumptive E. coli and Klebsiella spp. colonies grown on the agar were subjected to Gram staining and the oxidase strip test (Merck) for initial identification. Gram-negative and oxidase-negative colonies were further subjected to biochemical tube tests, including triple sugar iron agar, indole, citrate, urea, and lysine tests. Subsequently, the isolates were cultured by streaking on Columbia agar® (BioMérieux, Marcy-l’Étoile, France), supplemented with 5% sheep blood, for further analyses.
Bacterial confirmation
For the extraction of bacterial DNA, the commercial kit DNeasy UltraClean bacteria (Qiagen, Hilden, Germany) was used following the manufacturer’s instructions. Species-specific identification of isolates was conducted by amplification and sequencing of the 16 S rRNA gene, following the method reported by Kuhnert et al. (1996). PCR products were purified using the QIAquick PCR kit (Qiagen) according to the manufacturer’s recommendations and were subsequently sequenced by Microsynth Sequence Laboratories, Göttingen, Germany under standard conditions. The DNA sequences obtained were then subjected to homology searches using the Basic Local Alignment Search Tool (BLAST) provided by the National Center for Biotechnology Information (NCBI) (available at http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Phenotypic detection of ESBL
The isolates were screened for ESBL detection using the double-disc synergy test. For this purpose, 1–2 colonies were inoculated in sterile saline solution and adjusted to a density of 0.5 according to the McFarland scale standard (1 × 108 CFU/mL). A sterile swab was dipped into the culture suspension tube, and then a Mueller-Hinton agar plate (BioMérieux) was inoculated by uniformly streaking the swab in three planes at an angle of 60° to each other. Finally, a circle was made to correct any excess on the agar plate. Zones of inhibition were determined for each isolate using antibiotic disks, each containing 30 µg of aztreonam (ATM), cefotaxime (CTX), ceftazidime (CAZ), ceftriaxone (CRO), or cefepime (FEP) (Oxoid, United Kingdom), either alone or in combination with 10 µg of clavulanic acid, with a disc distance of 20 mm. The results were evaluated according to the cut-off points established by the Clinical Laboratory Standard Institute [14]. As negative and positive controls, the susceptible E. coli strain ATCC 25,922 and the ESBL-producing K. pneumoniae strain ATCC 700,603 were used, respectively.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was carried out using the Vitek® 2 system (BioMérieux) with the AST-N271 according to the manufacturer’s instructions. MIC breakpoints for 17 antibiotics, including ampicillin, ampicillin/sulbactam, cephalothin, cefuroxime, cefotaxime, ceftazidime, ceftriaxone, cefepime, ertapenem, meropenem, amikacin, gentamicin, ciprofloxacin, norfloxacin, fosfomycin, nitrofurantoin, and trimethoprim/sulfamethoxazole, were set according to the Clinical and Laboratory Standards Institute (CLSI) breakpoint tables for the interpretation of MICs and zone diameters (M100: Performance Standards for Antimicrobial Susceptibility test. 30th edition) [14].
Molecular determination of ESBL
The isolates were subjected to conventional PCR analysis for the identification of β-lactamase (bla) genes belonging to the ESBL-subgroup, including TEM, SHV, and CTX-M (CTXM groups 1, 2, 8, 9, and 25), following the methods established by Batchelor et al. (2005) and Pitout et al. (1998). For quality control, positive controls were used, including K. pneumoniae strain ATCC 700,603 (harboring blaSHV gene) and a strain of K. pneumoniae (harboring blaCTX-M and blaTEM genes), while E. coli ATCC 25,922 was used as the negative control for all PCR amplification tests. The resulting amplicons were purified using the PCR Purification Kit (Qiagen), and sequencing was performed at SeqLab in Goettingen, Germany. The obtained sequences were evaluated using the BLAST algorithm available at http://blast.ncbi.nlm.nih.gov/Blast.cgi.
Detection of shiga toxin-producing E. coli
The genes for the two main Shiga toxins, STX1 and STX2, of Shiga Toxin-producing E. coli (STEC) were amplified using conventional PCR with primers MK1 and MK2, following previously established protocols by Karch and Meyer (1989).
Evaluation of total bacterial count
The total bacterial count in samples was determined using the flow cytometry method (ISO 21187:2021/ IDF 196:2021) on the Bactoscan FC + instrument (Foss Electric, Hillerød, Denmark).
Case definition
A herd was considered positive for the total bacterial count if the value obtained was greater than 100,000 CFU/mL, and it was considered negative if the value was equal to or less than 100,000 CFU/mL, based on the criteria established by the Commission Regulation (CE) No. 853/2004 [15].
Epidemiological questionnaire
A questionnaire was used to collect data on herd general characteristics, milking management practices, and antibiotic use. The method of application and general findings were previously published [16] and are available upon request. For the determination of the association of general variables and herd management practices with the response variable total bacterial count > 100,000 CFU/mL, the variables “adequate washing and disinfection of the milking machine” and “adequate washing and disinfection of the bulk tank” were constructed. These processes were considered adequate when the herd had a disinfection protocol for the machine and bulk tank, and this protocol was elaborated by an external provider.
Statistical analysis
Descriptive statistic was applied for the determination of measures of central tendency and frequency distribution to the variables resistance or susceptibility to the evaluated antibiotics, detection of ESBL-producing bacteria, total bacterial count, and to the variables adequate washing and disinfection of the milking machine and adequate washing and disinfection of the bulk tank.
For the total bacterial count variable, the geometric and arithmetic mean with its 95% CI and statistical significance were estimated. The latter was estimated using the Mann-Whitney U test since the variables did not follow a normal distribution according to the Kolmogorov-Smirnov test.
For the selection of the variables to be included in the multivariate model, those that in the bivariate analysis had a p < 0.25 value or that had been previously reported were used. For the dichotomized total bacterial count variable, binomial regression or Poisson regression was used when the former did not reach convergence. All primary exposures of interest were adjusted for confounding variables and the interaction was assessed by including the term in the model whenever significant. For the selection of the final model, the AIC criterion (Akaike Information Criterion) was used. Adjusted RR with 95%CI was reported and a p < 0.05 value was significant. All analyses were performed with Stata 16.0 (StataCorp LLC, Texas, USA).
Results
Microbiological isolation and species identification of Klebsiella spp. and E. coli
After the initial screening of samples, a total number of 15 morphologically different Gram-negative colonies from equal number of bulk-tank milk samples were obtained. For each morphologically distinct type of colony, one isolate representing each bulk-tank milk sample/dairy herd was selected. The isolates were then biochemically identified as E. coli (n = 3), and Klebsiella spp. (n = 12). Further, 16 S rRNA gene sequence analysis confirmed these isolates on species level as E. coli (n = 3), K. oxytoca (n = 11), and K. pneumoniae (n = 1) (Table 1).
Phenotypic and genotypic detection of ESBL production
None of the E. coli and Klebsiella spp. isolates were ESBL positive by phenotypic methods. ESBL production, however, was genotypically confirmed only in one isolate (K. pneumoniae #11) from the bulk-milk tank from a dairy farm in the municipality Entrerríos (1/150; 0.7%). The isolate K. pneumoniae #11 hosted blaSHV gene, based on PCR, and subsequently the sequence analysis showed strong homology with blaSHV-61 variant. None of the isolates was positive for the blaTEM and blaCTX-M genes in the genotypic tests (Table 2).
Antimicrobial susceptibility
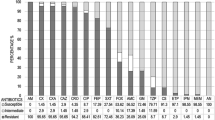
Besides the β-lactam resistances, the isolates were also tested for resistance to other antibiotics. Antibiotic susceptibility evaluated by MIC showed resistance to ampicillin in 12 (80%) isolates, of which all were Klebsiella spp. In addition, six Klebsiella spp. isolates were resistant to fosfomycin. For the remaining antibiotics evaluated, all isolates were sensitive. Resistance to two antimicrobial groups´ ampicillin and fosfomycin was detected only in six Klebsiella spp. isolates, including five K. oxytoca and one K. pneumoniae (Table 2).
Detection of shiga toxin-producing E. coli
All E. coli isolates obtained from three bulk-milk samples were negative for both stx1 and stx2 genes.
Evaluation of total bacterial count
The mean value for total bacterial count was 6.97 × 106 CFU/mL (95% CI 6.55 × 106 − 2.05 × 107 CFU/mL) and a geometric mean of 2.16 × 104 CFU/mL (95% CI 1.62 × 104– 28.83 × 104 CFU/mL). A total bacterial count value ≤ 100,000 CFU/mL was found in 84% (126/150) of the herds. According to these results, most of the herds had adequate hygienic quality following Colombian legislation [2].
Risk factors associated with total bacterial count > 100,000 CFU/mL
The bivariate analysis identified several factors that were significantly associated with the increase in total bacterial count (p < 0.25) and were included in the final model: the milker was the owner of the cows (p = 0.018), milking site (p = 0.022), use of automatic separators (p = 0.039), disinfection of the milking system (p = 0.212), type of disinfection protocol of the milking machine (p = 0.091), adequate washing and disinfection of the milking machine (p = 0.142) (Table 3). Concerning the final model, four variables were found to be associated with the risk of presenting CFU > 100,000 CFU/mL: Herds with more than 60 milking cows (RR = 1.35; 95% CI 1.35–1.35), changing milker in the last month (RR = 2.31; IC 95% 1.54–3.47), milking in the paddock (RR = 3.95; IC 95% 1.29–12.09) and use a chlorinated product for pre-dipping (RR = 1.35; IC 95% 1.06–1.73). The variables considered a protective factor to reduce the total bacterial count were the following: when the milker was the owner of the cows (RR = 0.40; IC 95% 0.23–0.71) and adequate washing and disinfection of the milking machine (RR = 0.79; IC 95% 0.71–0.87). There was an interaction between changing milker in the last month and adequate washing and disinfection of the milking machine (p < 0.05) (Table 4).
Discussion
A total of 15 bulk tank milk samples, collected from an equal number of dairy farms in three municipalities including Santa Rosa de Osos, San Pedro de los Milagros, and Entrerríos located in Northern Antioquia were confirmed as positive by bacteriological examination. From these samples, one isolate representing each sample/dairy herd, 11 K. oxytoca, one K. pneumonia, and three E. coli were isolated. Notably, among these isolates, only the isolate K. pneumoniae #11 was blaSHV-61 positive, which was isolated from a dairy farm in the municipality Entrerríos. In the previous study carried out in 2017 exclusively in the single municipality Entrerríos, about 3% of randomly selected dairy herds were found to have ESBL-producing Enterobacteriaceae present [17]. The low frequency of BLEE producers determined in this study (< 1%) is not surprising, since it is in agreement with another study carried out in the area [17]; however, there are still few studies on the frequency of BLEE-producing bacteria at the dairy farm level in Colombia that provide absolute clarity on the subject. We hypothesize that in addition to the low use of antibiotics at farm level together with adequate hygiene standards required by local dairy processing plants could be the reason for this favourable situation. Supporting this view, bonuses offered by some milk processing plants for raw milk with total bacterial count values of less than 175,000 CFU/mL stimulate dairy farms in the implementation of adequate hygiene practices at the time of milking, washing equipment, and promote the cold chain, and therefore reduce the occurrence of environmental pathogens and total bacteria values.
On the other hand, the mean total bacterial count was lower than that found previously in Colombia [18] and most herds (84%) had a total bacterial count value ≤ 100,000 CFU/mL, indicating adequate hygienic quality. However, the bacterial count value is still high in 16% of the herds, which indicates the need of improving practices aiming the hygienic quality in the herds. The variables associated with increased bacterial count in raw milk could be explained by the short time the milker has to implement the necessary measures to control quality at milking, the choice of cleaning materials and utensils, the cleaning of the cows, as well as the lack of training of the milkers [19].
The milk samples of the present study were examined using a non-selective method, in which the samples were cultured on non-selective media for the detection of EBSL-producing Enterobacteriaceae and a randomly selected subset of E. coli and Klebsiella spp. isolates were then subjected to susceptibility testing. It should be noted that ESBL-producing organisms, which might be present as minor constituents of the total bacterial microbiota in tank milk, could potentially go undetected due to the lower sensitivity (10 CFU/mL) of this approach. This stands in contrast to the selective enrichment method which has been previously employed for tank milk samples in various reports [11, 12, 20]; large differences in sensitivity are to be expected between these methods. The results of this study suggest that the sensitivity of this non-selective method for detecting ESBLs might be comparatively lower.
With regard to the ESBL types detected, it was expected to identify clones that harbor the CTX-M gene based on findings of a previous study in the same municipality Entrerríos [17]. Instead, the blaSHV gene in a K. pneumoniae isolate was identified, suggesting that mobile genetic elements or different types of clones circulate in the study area. Since in the present study, EBSL was detected only in K. pneumoniae, it has been reported more frequently in recent years that K. pneumoniae can develop multiple mechanisms to become resistant to antibiotics including the production of antibiotic inactivating enzymes such as β-lactamases and ESBLs [21]. This finding is comparable to what has been found in different studies from other countries in bulk-tank milk samples [12, 20, 22], and in milk samples from cows with bovine mastitis [23,24,25]. In Colombia, ESBL of the SHV type has already been detected in K. pneumoniae in clinical patients [26,27,28], in community environments [29], also in Salmonella from retail broiler meat samples [30], and in E. coli from different sources in the poultry production [31]. However, the distribution and frequency of ESBL types can vary considerably depending on the source of isolates and geographic variations [32,33,34,35,36,37,38].
In the present study, ESBL-producing K. pneumoniae was not identified by phenotypic methods, but by molecular methods, which is consistent with previous studies that have reported discrepancies between detection methods [39,40,41]. The lack of phenotypic resistance in this study could be due to the bacteria was not being subjected to high selection pressure to produce SHV enzyme in the phenotypic confirmation assay. This would indicate that the isolates do not reveal changes in the phenotypic patterns of resistance, unlike what has been evidenced in other reports in which K. pneumoniae carrying blaSHV presented phenotypic resistance to ampicillin, aztreonam, chloramphenicol, and trimethoprim and inhibitor test sensitivity [37]. This confirms the importance of implementing specific molecular tests for epidemiological monitoring studies due to their high level of specificity [42] since it has been identified that up to 33% of positive strains are not detected with phenotypic methods [43].
Regarding to antibiotic susceptibility of isolates evaluated, E. coli isolates obtained were sensitive to all the antibiotics, however, other local studies have shown resistance patterns of E. coli isolated from bulk milk samples [17] and from bovine mammary quarter samples [44] in the same region and in other regions of Colombia [45]. In contrast, studies from the same study area on Klebsiella spp. isolated from dairy cattle with clinical and subclinical mastitis showed a 40% susceptibility to ampicillin [44], whereas in other countries the rate of resistance to the same antibiotic was variable [23, 46,47,48,49]. In addition, the fosfomycin resistance detected in this study is consistent with previous studies reporting K. pneumoniae from raw milk [47]. All isolates were susceptible to the ertapenem and meropenem. These findings are not surprising, since commercial preparations containing carbapenems are not approved for use in livestock in Colombia.
Finally, no STEC isolates were found, which relates to the good hygienic quality of raw milk found in most of the herds and the low frequency of E. coli isolated considering that not all E. coli strains present in bovine feces are STEC [50]. However, epidemiological surveillance programs are still required for food of animal origin since STEC strains can contaminate raw milk through a fecal source [6], or environmental [51].
In conclusion, the frequency of ESBL-producing E. coli and Klebsiella spp. in bulk-tank milk samples from three municipalities in northern Antioquia was very low (< 1%). Only K. pneumoniae was positive for the ESBL type blaSHV by sequence analysis. Most dairy herds had adequate hygienic quality and herds with more than 60 milking cows, changing milker, milking in paddocks, and using a chlorinated product for pre-dipping are risk factors that increase the total bacterial count in raw milk. Finally, it was identified that some variables such as the milker being the owner of the animals, and the adequate washing and disinfection of the milking machine promote adequate hygienic milk quality of the raw milk stored in the tanks.
Data availability
All data generated or analyzed during this study are included in this published article.
References
Robles I, Kelton DF, Barkema HW et al (2020) Bacterial concentrations in bedding and their association with dairy cow hygiene and milk quality. Animal 14:1052–1066. https://doi.org/10.1017/S1751731119002787
Ministerio de Agricultura y Desarrollo Rural (2012) Resolucion 000017 de 2012 Sistema de pago Leche Cruda al proveedor
Elmoslemany AM, Keefe GP, Dohoo IR, Jayarao BM (2009) Risk factors for bacteriological quality of bulk tank milk in Prince Edward Island dairy herds. Part 2: Bacteria count-specific risk factors. J Dairy Sci 92:2644–2652. https://doi.org/10.3168/jds.2008-1813
Metz M, Sheehan J, Feng PCH (2020) Use of indicator bacteria for monitoring sanitary quality of raw milk cheeses– A literature review. Food Microbiol 85:103283. https://doi.org/10.1016/j.fm.2019.103283
Ndahetuye JB, Artursson K, Båge R et al (2020) MILK Symposium review: Microbiological quality and safety of milk from farm to milk collection centers in Rwanda. J Dairy Sci 103:9730–9739. https://doi.org/10.3168/jds.2020-18302
Straley BA, Donaldson SC, Hedge NV et al (2006) Public health significance of antimicrobial-resistant gram-negative bacteria in raw bulk tank milk. Foodborne Pathog Dis 3:222–233. https://doi.org/10.1089/fpd.2006.3.222
Berge ACB, Champagne SC, Finger RM, Sischo WM (2007) The use of bulk tank milk samples to monitor trends in antimicrobial resistance on dairy farms. Foodborne Pathog Dis 4:397–407. https://doi.org/10.1089/fpd.2007.0009
Dahmen S, Metayer V, Gay E et al (2013) Characterization of extended-spectrum beta-lactamase (ESBL) -carrying plasmids and clones of Enterobacteriaceae causing cattle mastitis in. Vet Microbiol 162:793–799. https://doi.org/10.1016/j.vetmic.2012.10.015
Ohnishi M, Okatani AT, Harada K et al (2013) Genetic characteristics of CTX-M-type extended-spectrum-β-lactamase (ESBL)-producing Enterobacteriaceae involved in mastitis cases on Japanese dairy farms, 2007 to 2011. J Clin Microbiol 51:3117–3122. https://doi.org/10.1128/JCM.00920-13
Kürekci C, Osek J, Ayd M et al (2019) Evaluation of bulk tank raw milk and raw chicken meat samples as source of ESBL producing Escherichia coli in Turkey: recent insights. J Food Saf 39:1–7. https://doi.org/10.1111/jfs.12605
Odenthal S, Akineden Ö, Usleber E (2016) Extended-spectrum β -lactamase producing Enterobacteriaceae in bulk tank milk from German dairy farms. Int J Food Microbiol 238:72–78. https://doi.org/10.1016/j.ijfoodmicro.2016.08.036
Sudarwanto M, Akineden O, Odenthal S et al (2015) Extended-spectrum b-Lactamase (ESBL)–Producing Klebsiella pneumoniae in Bulk Tank milk from dairy farms in Indonesia. Foodborne Pathog Dis 0. https://doi.org/10.1089/fpd.2014.1895
National Mastitis Council (NMC) (2017) Laboratory Handbook on Bovine Mastitis, Third edit
CLSI (2020) CLSI M100-ED29: 2021 Performance Standards for Antimicrobial Susceptibility Testing, 30th Edition
European Commission (2004) Regulation (EC) N° 853/2004 of the European parlamient and of the Council of 29 April 2004 laying down specific hygiene rules for on the hygiene of foodstuffs. Off J Eur Union L 139:55
Ágredo-campos ÁS, Fernández-Silva JA, Ramírez-Vásquez NF (2023) Staphylococcus aureus, Escherichia coli, and Klebsiella spp. prevalence in bulk tank milk of Colombian herds and associated milking practices. Vet World 16:869–881
Vásquez-Jaramillo L, Ramírez N, Akineden Ö, Fernández-Silva JA (2017) Presence of extended-spectrum beta-lactamase (ESBL)- producing Enterobacteriaceae in bulk-tank milk of bovine dairy farms in Antioquia, Colombia. Rev Colomb Ciencias Pecu 30:85–100. https://doi.org/10.17533/udea.rccp.v30n2a01
Múnera-Bedoya O, Cassoli LD, Machado PF, Cerón-Muñoz MF (2017) Influence of attitudes and behavior of milkers on the hygienic and sanitary quality of milk. PLoS ONE 12:1–13. https://doi.org/10.1371/journal.pone.0184640
Naing YW, Wai SS, Lin TN et al (2019) Bacterial content and associated risk factors influencing the quality of bulk tank milk collected from dairy cattle farms in Mandalay Region. Food Sci Nutr 7:1063–1071. https://doi.org/10.1002/fsn3.945
Bonardi S, Cabassi CS, Fiaccadori E et al (2023) Detection of carbapenemase- and ESBL-producing Klebsiella pneumoniae from bovine bulk milk and comparison with clinical human isolates in Italy. Int J Food Microbiol 387:110049. https://doi.org/10.1016/j.ijfoodmicro.2022.110049
Paterson DL, Bonomo RA (2005) Extended spectrum beta lactamases: a critical update. Multidrug Resist Glob Concern 18:115–129. https://doi.org/10.2174/978160805292911201010115
Nobrega DB, Calarga A, Costa Nascimento L, Gasparotto C et al (2021) Molecular characterization of antimicrobial resistance in Klebsiella pneumoniae isolated from Brazilian dairy herds. J Dairy Sci 104:7210–7224. https://doi.org/10.3168/jds.2020-19569
Yang Y, Peng Y, Jiang J et al (2021) Isolation and characterization of multidrug-resistant Klebsiella pneumoniae from raw cow milk in Jiangsu and Shandong provinces, China. Transbound Emerg Dis 68:1033–1039. https://doi.org/10.1111/tbed.13787
Locatelli C, Scaccabarozzi L, Pisoni G, Moroni P (2010) CTX-M1 ESBL-Producing Klebsiella pneumoniae subsp. pneumoniae isolated from cases of bovine mastitis. J Clin Microbiol 48:3822–3823. https://doi.org/10.1128/JCM.00941-10
Nóbrega DB, Guiduce MVS, Guimarães FF et al (2013) Molecular epidemiology and extended-spectrum β -lactamases production of Klebsiella pneumoniae isolated from three dairy herds. Pesqui Vet Bras 33:855–859
Abril D, Vergara E, Palacios D et al (2021) Within patient genetic diversity of Bla KPC harboring Klebsiella pneumoniae in a Colombian hospital and identification of a new NTE KPC platform . Sci Rep 11:1–16. https://doi.org/10.1038/s41598-021-00887-2
Garcia-fulgueiras V, Zapata Y, Papa-ezdra R et al (2020) First characterization of K. pneumoniae ST11 clinical isolates harboring bla KPC-3 in Latin America. Rev Argent Microbiol 52:211–216. https://doi.org/10.1016/j.ram.2019.10.003
Villegas MV, Lolans K, Correa A et al (2006) First detection of the plasmid-mediated class A carbapenemase KPC-2 in clinical isolates of Klebsiella pneumoniae from South America. Antimicrob Agents Chemoterapy 50:2880–2882. https://doi.org/10.1128/AAC.00186-06
Recalde-reyes DP, Alfonso-ortiz N, Fuentes-quimbayo MF, Ángel-hernández V (2021) Perfil De Resistencia genotípica Y fenotípica presente en bacterias aisladas a partir de fómites en Armenia, Quindío-Colombia período junio-julio 2019. Infect Asoc Colomb Infectología 25:22–27
Castellanos LR, Vander Graaf-van L, Donado-Godoy P et al (2018) Genomic characterization of cephalosporin-resistant Salmonella enterica in the Colombian poultry chain. Front Microbiol 9:1–11. https://doi.org/10.3389/fmicb.2018.02431
Clavijo V, Castellanos LR, Donado-godoy P et al (2017) High heterogeneity of Escherichia coli sequence types harbouring ESBL / AmpC genes on IncI1 plasmids in the Colombian Poultry Chain. PLoS ONE 12:1–15. https://doi.org/10.1371/journal.pone.0170777
Afema JA, Ahmed S, Besser TE et al (2018) Molecular epidemiology of dairy cattle-Associated Escherichia coli carrying blaCTX-M genes in Washington State. Appl Enviromental Microbiol 84:1–12
Mendonc N, Louro D, Canic M (2009) Molecular epidemiology and antimicrobial susceptibility of extended- and broad-spectrum B -lactamase-producing Klebsiella pneumoniae isolated in Portugal. Int J Antimicrob Agents 34:29–37. https://doi.org/10.1016/j.ijantimicag.2008.11.014
Dahmen S, Mansour W, Charfi K et al (2012) Imipenem Resistance in Klebsiella pneumoniae is Associated to the combination of plasmid-mediated CMY-4 AmpC b-Lactamase and loss of an outer membrane protein. Microb Drug Resist 18:479–483. https://doi.org/10.1089/mdr.2011.0214
Dziri O, Dziri R, Maraoub A, Chouchani C (2018) First Report of SHV-148-Type ESBL and CMY-42-Type AmpC b-Lactamase in Klebsiella pneumoniae clinical isolates in Tunisia. Microb Drug Resist 00:1–6. https://doi.org/10.1089/mdr.2018.0073
Cao X, Zhang Z, Shen HAN et al (2014) Genotypic characteristics of multidrug-resistant Escherichia coli isolates associated with urinary tract infections. Acta Pathol Microbiol Immunol Scand 1088–1095. https://doi.org/10.1111/apm.12260
Mondal AH, Siddiqui MT, Sultan I (2019) Prevalence and diversity of bla TEM, Bla SHV and Bla CTX-M variants among multidrug resistant Klebsiella spp. from an urban riverine environment in India. Int J Environ Health Res 29:117–129. https://doi.org/10.1080/09603123.2018.1515425
Mobasseri G, Shuan C, Teh J et al (2019) Molecular characterization of Multidrug-resistant Klebsiella pneumoniae isolated from Swine farms in Malaysia. Microb Drug Resist 00:1–12. https://doi.org/10.1089/mdr.2018.0184
Waworuntu O, Sjahril R, Rasita YD, Munawir M (2020) Characteristic of extended-spectrum-B-lactamase (ESBL) producing Klebsiella pneumoniae at tertiary referral hospital in South Sulawesi, Indonesia. International Journal of Infectious diseases. International Society for Infectious Diseases, p 80
Spanu T, Sanguinetti M, Tumbarello M et al (2006) Evaluation of the new VITEK 2 extended-spectrum beta-lactamase (ESBL) test for rapid detection of ESBL production in Enterobacteriaceae isolates. J Clin Microbiol 44:3257–3262. https://doi.org/10.1128/JCM.00433-06
Espinar M, Rocha R, Ribeiro M et al (2011) Extended-spectrum b -lactamases of Escherichia coli and Klebsiella pneumoniae screened by the VITEK 2 system. J Med Microbiol 60:756–760. https://doi.org/10.1099/jmm.0.024075-0
Thomson KS (2010) Extended-Spectrum-BetaLactamase, AmpC, and Carbapenemase issues. J Clin Microbiol 48:1019–1025. https://doi.org/10.1128/JCM.00219-10
Tepeli SÖ, Zorba NND (2018) Frequency of extended-spectrum β-lactamase (ESBL)– and AmpC β-lactamase– producing Enterobacteriaceae in a cheese production process. J Dairy Sci 101:2906–2914. https://doi.org/10.3168/jds.2017-13878
Ramírez N, Fernández-silva JA, Palacio LG (2018) Tasa de incidencia de mastitis clínica y susceptibilidad antibiótica de patógenos productores de mastitis en ganado lechero del norte de Antioquia, Colombia *. Rev Med Vet (Bogota) 1:75–87
Arenas NE, Abril DA, Valencia P et al (2017) Screening food-borne and zoonotic pathogens associated with livestock practices in the Sumapaz region, Cudinamarca, Colombia. Trop Anim Health Prod 49:739–745. https://doi.org/10.1007/s11250-017-1251-6
Abdi RD, Gillespie BE, Ivey S et al (2021) Antimicrobial Resistance of Major bacterial pathogens from dairy cows with high somatic cell Count and Clinical Mastitis. Animals 11:1–14
Tartor YH, El-aziz NKA, Gharieb RMA (2021) Whole-genome sequencing of Gram-negative Bacteria isolated from bovine mastitis and raw milk: the First Emergence of Colistin mcr– 10 and Fosfomycin fosA5 Resistance genes in Klebsiella pneumoniae in Middle East. Front Microbiol 12. https://doi.org/10.3389/fmicb.2021.770813
Liu K, Zhang L, Gu X, Qu W (2022) The prevalence of Klebsiella spp. Associated with bovine mastitis in China and its Antimicrobial Resistance Rate: a Meta-analysis. Front Vet Sci 9:1–11. https://doi.org/10.3389/fvets.2022.757504
Jong A, De, El F, Simjee S et al (2018) Monitoring of antimicrobial susceptibility of udder pathogens recovered from cases of clinical mastitis in dairy cows across Europe: VetPath results. Vet Microbiol 213:73–81. https://doi.org/10.1016/j.vetmic.2017.11.021
Arimizu Y, Kirino Y, Sato MP et al (2019) Large-scale genome analysis of bovine commensal Escherichia coli reveals that bovine-adapted E. Coli lineages are serving as evolutionary sources of the emergence of human intestinal pathogenic strains. Genome Res 9:1495–1505. https://doi.org/10.1101/gr.249268.119.Freely
Dell’Orco F, Gusmara C, Loiacono M et al (2019) Evaluation of virulence factors profiles and antimicrobials resistance of Escherichia coli isolated from bulk tank milk and raw milk filters. Res Vet Sci 123:77–83. https://doi.org/10.1016/j.rvsc.2018.12.011
Acknowledgements
the authors are grateful to the funders, the Ministry of Science, Technology and Innovation of Colombia– Minciencias– Project: 111580763373, Contract 755–2018– “Molecular epidemiology of mastitis pathogens and determination of antibiotic residues and resistance in herds of the northern dairy region of Antioquia, Colombia: A pilot study”, to the University of Antioquia, and to the Federation of Livestock Associations (FAGA). Also thank the farmers and institutions from Municipalities who helped with the work and left to the researcher to take the samples and information at the herds.
Funding
This work was supported by Ministerio de Ciencia, Tecnología e Innovación of Colombia, Minciencias, Award Number: Project 111580763373, contract 755–2018.
Open Access funding provided by Colombia Consortium
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by NFRV, JAFS, ASAC and ÖA. The first draft of the manuscript was written by ASAC and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Statement of animal rights
This study was approved by the Ethics Committee for Experimentation with Animals of the Universidad de Antioquia, Colombia (Act No. 110 of May 17, 2017). Farmers provided informed consent before data collection.
Conflict of interest
The authors declare no competing interests.
Additional information
Responsible Editor: David Germano Gonçalves Schwarz.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Campos, ÁS.Á., Akineden, Ö., Fernández-Silva, J.A. et al. Extended-spectrum beta-lactamase-producing Klebsiella pneumoniae and risk factors associated with high total bacterial count in bulk tank milk from dairy farms in Colombia. Braz J Microbiol (2024). https://doi.org/10.1007/s42770-024-01396-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42770-024-01396-w