Abstract
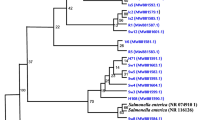
The aim of this study was to identify virulence and antimicrobial resistance profiles and determine the sequence type (ST) by multilocus sequence typing (MLST) of Salmonella enterica isolates from bovine carcasses from slaughterhouse located in Minas Gerais state, Brazil, and its relationship with bovine isolates obtained on the American continent based on sequence type profile. The MLST results were compared with all Salmonella STs associated with cattle on American continent, and a multi-locus sequence tree (MS tree) was built. Among the 17 S. enterica isolates, five ST profiles identified, and ST10 were the most frequent, grouping seven (41.2%) isolates. The isolates presented 11 different profiles of virulence genes, and six different antibiotics resistance profiles. The survey on Enterobase platform showed 333 Salmonella STs from American continent, grouped into four different clusters. Most of the isolates in the present study (13/17), were concentrated in a single cluster (L4) composed by 74 STs. As a conclusion, five different STs were identified, with ST10 being the most common. The isolates showed great diversity of virulence genes and antibiotics resistance profiles. Most of the isolates of this study were grouped into a single cluster composed by 74 STs formed by bovine isolates obtained on the American continent.

Similar content being viewed by others
Data Availability
The dataset analyzed during the current study is available from the corresponding author on reasonable request.
References
Eng SK, Pusparajah P, Ab Mutalib NS, Ser HL, Chan KG, Lee LH (2015) Salmonella: A review on pathogenesis, epidemiology and antibiotic resistance. Front Life Sci 8(3):284–293. https://doi.org/10.1080/21553769.2015.1051243
CDC (2020) Salmonella. Centers for Disease Controle and Prevention. Published 2020. https://www.cdc.gov/salmonella/index.html. Accessed 22 Apr 2024
Monte DF, Lincopan N, Fedorka-Cray PJ, Landgraf M (2019) Current insights on high priority antibiotic-resistant Salmonella enterica in food and foodstuffs: a review. Curr Opin Food Sci 26:35–46. https://doi.org/10.1016/j.cofs.2019.03.004
Ni P, Xu Q, Yin Y et al (2018) Prevalence and characterization of Salmonella serovars isolated from farm products in Shanghai. Food Control 85:269–275. https://doi.org/10.1016/j.foodcont.2017.10.009
Monte DFM, Nethery MA, Barrangou R, Landgraf M, Fedorka-Cray PJ (2020) Whole-genome sequencing analysis and CRISPR genotyping of rare antibiotic-resistant Salmonella enterica serovars isolated from food and related sources. Food Microbiol 2021(93):103601. https://doi.org/10.1016/j.fm.2020.103601
Harhay DM, Weinroth MD, Bono JL, Harhay GP, Bosilevac JM (2021) Rapid estimation of Salmonella enterica contamination level in ground beef – Application of the time-to-positivity method using a combination of molecular detection and direct plating. Food Microbiol 93. https://doi.org/10.1016/j.fm.2020.103615
Jiang Z, Anwar TM, Peng X et al (July2020) Prevalence and antimicrobial resistance of Salmonella recovered from pig-borne food products in Henan. China Food Control 2021(121):107535. https://doi.org/10.1016/j.foodcont.2020.107535
Nair DVT, Venkitanarayanan K, Johny AK (2018) Antibiotic-resistant Salmonella in the food supply and the potential role of antibiotic alternatives for control. Foods 7(10). https://doi.org/10.3390/foods7100167
Abdel Aziz SA, Abdel-Latef GK, Shany SAS, Rouby SR (2018) Molecular detection of integron and antimicrobial resistance genes in multidrug resistant Salmonella isolated from poultry, calves and human in Beni-Suef governorate. Egypt Beni Suef Univ J Basic Appl Sci 7(4):535–542. https://doi.org/10.1016/j.bjbas.2018.06.005
dos Reis RO, Souza MN, Cecconi MCP et al (2018) Increasing prevalence and dissemination of invasive nontyphoidal Salmonella serotype Typhimurium with multidrug resistance in hospitalized patients from southern Brazil. Braz J Infect Dis 22(5):424–432. https://doi.org/10.1016/j.bjid.2018.08.002
Chang YC, Scaria J, Ibraham M et al (2016) Distribution and factors associated with Salmonella enterica genotypes in a diverse population of humans and animals in Qatar using multi-locus sequence typing (MLST). J Infect Public Health 9(3):315–323. https://doi.org/10.1016/j.jiph.2015.10.013
Lytsy B, Engstrand L, Gustafsson Å, Kaden R (2017) Time to review the gold standard for genotyping vancomycin-resistant enterococci in epidemiology: comparing whole-genome sequencing with PFGE and MLST in three suspected outbreaks in Sweden during 2013–2015. Infect Genet Evol 54:74–80. https://doi.org/10.1016/j.meegid.2017.06.010
Jain P, Sudhanthirakodi S, Chowdhury G et al (2018) Antimicrobial resistance, plasmid, virulence, multilocus sequence typing and pulsed-field gel electrophoresis profiles of Salmonella enterica serovar Typhimurium clinical and environmental isolates from India. PLoS ONE 13(12):1–16. https://doi.org/10.1371/journal.pone.0207954
Zhao X, Li W, Hou S et al (2022) Epidemiological investigation on drug resistance of Salmonella isolates from duck breeding farms in Shandong Province and surrounding areas, China. Poult Sci 101(8):101961. https://doi.org/10.1016/j.psj.2022.101961
Zahli R, Scheu AK, Abrini J, et al (2022) Salmonella spp: prevalence, antimicrobial resistance and molecular typing of strains isolated from poultry in Tetouan-Morocco. Lwt 153. https://doi.org/10.1016/j.lwt.2021.112359
Feijao P, Yao HT, Fornika D et al (2018) MentaLiST - A fast MLST caller for large MLST schemes. Microb Genom 4(2):1–8. https://doi.org/10.1099/mgen.0.000146
Paudyal N, Pan H, Elbediwi M et al (2019) Characterization of Salmonella Dublin isolated from bovine and human hosts. BMC Microbiol 19(1):1–8. https://doi.org/10.1186/s12866-019-1598-0
Vilela FP, dos Prazeres RD, Costa RG, Casas MRT, Falcão JP, Campioni F (2020) High similarity and high frequency of virulence genes among Salmonella Dublin strains isolated over a 33-year period in Brazil. Braz J Microbiol 51(2):497–509. https://doi.org/10.1007/s42770-019-00156-5
Yamba K, Kapesa C, Mpabalwani E et al (2022) Antimicrobial susceptibility and genomic profiling of Salmonella enterica from bloodstream infections at a tertiary referral hospital in Lusaka, Zambia, 2018–2019. IJID Regions 3(March):248–255. https://doi.org/10.1016/j.ijregi.2022.04.003
WHO (2020) Antimicrobial resistance. World Health Organization. Published 2020. https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance. Accessed 22 Apr 2024
Cossi MVC, Burin RCK, Camargo AC et al (2014) Low occurrence of Salmonella in the beef processing chain from Minas Gerais state, Brazil: from bovine hides to end cuts. Food Control 40(1):320–323. https://doi.org/10.1016/j.foodcont.2013.12.018
Alikhan NF, Zhou Z, Sergeant MJ, Achtman M (2018) A genomic overview of the population structure of Salmonella. PLoS Genet 14(4):e1007261. https://doi.org/10.1371/JOURNAL.PGEN.1007261
Zhou Z, Alikhan NF, Mohamed K, Fan Y, Achtman M (2020) The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res 30(1):138–152. https://doi.org/10.1101/GR.251678.119/-/DC1
Francisco AP, Bugalho M, Ramirez M, Carriço JA (2009) Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics 10(1):1–15. https://doi.org/10.1186/1471-2105-10-152/FIGURES/5
Francisco AP, Vaz C, Monteiro PT, Melo-Cristino J, Ramirez M, Carriço JA (2012) PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics 13(1):1–10. https://doi.org/10.1186/1471-2105-13-87/FIGURES/5
CLSI (2020) M100 Performance Standards for Antimicrobial Susceptibility Testing, vol 8, 30th edn. Institute C and LS. https://www.nih.org.pk/wp-content/uploads/2021/02/CLSI-2020.pdf. Accessed 22 Apr 2024
Magiorakos A, Srinivasan A, Carey RB et al (2012) Bacteria : an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18(3):268–281. https://doi.org/10.1111/j.1469-0691.2011.03570.x
Achtman M, Wain J, Weill FX, et al (2012) Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog 8(6). https://doi.org/10.1371/journal.ppat.1002776
Cai Y, Tao J, Jiao Y et al (2016) Phenotypic characteristics and genotypic correlation between Salmonella isolates from a slaughterhouse and retail markets in Yangzhou. China Int J Food Microbiol 222:56–64. https://doi.org/10.1016/j.ijfoodmicro.2016.01.020
Li Y, Cai Y, Tao J et al (2016) Salmonella isolated from the slaughterhouses and correlation with pork contamination in free market. Food Control 59:591–600. https://doi.org/10.1016/j.foodcont.2015.06.040
Li YC, Pan ZM, Kang XL et al (2014) Prevalence, characteristics, and antimicrobial resistance patterns of Salmonella in retail pork in Jiangsu province, eastern China. J Food Prot 77(2):236–245. https://doi.org/10.4315/0362-028X.JFP-13-269
Cao Y, Shen Y, Cheng L et al (2017) Combination of multilocus sequence typing and pulsed-field gel electrophoresis reveals an association of molecular clonality with the emergence of extensive-drug resistance (XDR) in Salmonella. Microbiol Res 2018(207):170–176. https://doi.org/10.1016/j.micres.2017.12.001
Lamas A, Miranda JM, Regal P, Vázquez B, Franco CM, Cepeda A (2017) A comprehensive review of non-enterica subspecies of Salmonella enterica. Microbiol Res 2018(206):60–73. https://doi.org/10.1016/j.micres.2017.09.010
Hounmanou YMG, Baniga Z, García V, Dalsgaard A (2022) Salmonella Salamae and S. Waycross isolated from Nile perch in Lake Victoria show limited human pathogenic potential. Sci Rep 2022 12:1 12(1):1–11. https://doi.org/10.1038/s41598-022-08200-5
Ilyas B, Tsai CN, Coombes BK (2017) Evolution of Salmonella-host cell interactions through a dynamic bacterial genome. Front Cell Infect Microbiol. 7(SEP). https://doi.org/10.3389/fcimb.2017.00428
Abatcha MG, Effarizah ME, Rusul G (2018) Antibiotic susceptibility and molecular characterization of Salmonella enterica serovar Paratyphi B isolated from vegetables and processing environment in Malaysia. Int J Food Microbiol 2019(290):180–183. https://doi.org/10.1016/j.ijfoodmicro.2018.09.021
Borges KA, Furian TQ, Souza SN, Salle CTP, Moraes HLS, Nascimento VP (2019) Antimicrobial resistance and molecular characterization of salmonella enterica serotypes isolated from poultry sources in brazil. Rev Bras Cienc Avic 21(1). https://doi.org/10.1590/1806-9061-2018-0827
Fardsanei F, SoltanDallal MM, Douraghi M et al (2017) Genetic diversity and virulence genes of Salmonella enterica subspecies enterica serotype Enteritidis isolated from meats and eggs. Microb Pathog 107:451–456. https://doi.org/10.1016/j.micpath.2017.04.026
Viana C, Sereno MJ, Pegoraro K et al (2019) Distribution, diversity, virulence genotypes and antibiotic resistance for Salmonella isolated from a Brazilian pork production chain. Int J Food Microbiol 310(July):108310. https://doi.org/10.1016/j.ijfoodmicro.2019.108310
Alix E, Miki T, Felix C et al (2008) Interplay between MgtC and PagC in Salmonella enterica serovar Typhimurium. Microb Pathog 45(3):236–240. https://doi.org/10.1016/j.micpath.2008.06.001
Crago AM, Koronakis V (1999) Binding of extracellular matrix laminin to Escherichia coli expressing the Salmonella outer membrane proteins Rck and PagC. FEMS Microbiol Lett 176(2):495–501. https://doi.org/10.1016/S0378-1097(99)00279-7
Wannaprasat W, Padungtod P, Chuanchuen R (2011) Class 1 integrons and virulence genes in Salmonella enterica isolates from pork and humans. Int J Antimicrob Agents 37(5):457–461. https://doi.org/10.1016/j.ijantimicag.2010.12.001
Bakowski MA, Braun V, Lam GY et al (2010) The phosphoinositide phosphatase SopB manipulates membrane surface charge and trafficking of the Salmonella-containing vacuole. Cell Host Microbe 7(6):453–462. https://doi.org/10.1016/j.chom.2010.05.011
Giacomodonato MN, Llana MN, del Castañeda MRA et al (2014) Dam methylation regulates the expression of SPI-5-encoded sopB gene in Salmonella enterica serovar Typhimurium. Microbes and infection / Institut Pasteur 16(8):615–622. https://doi.org/10.1016/j.micinf.2014.03.009
Patel JC, Hueffer K, Lam TT, Galán JE (2009) Diversification of a Salmonella virulence protein function by ubiquitin-dependent differential localization. Cell 137(2):283–294. https://doi.org/10.1016/j.cell.2009.01.056
Zhou D, Galán J (2001) Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect 3(14–15):1293–1298. https://doi.org/10.1016/S1286-4579(01)01489-7
Graziani C, Busani L, Dionisi AM et al (2011) Virulotyping of Salmonella enterica serovar Napoli strains isolated in Italy from human and nonhuman sources. Foodborne Pathog Dis 8(9):997–1003. https://doi.org/10.1089/fpd.2010.0833
Huehn S, La Ragione RM, Anjum M et al (2010) Virulotyping and antimicrobial resistance typing of Salmonella enterica serovars relevant to human health in Europe. Foodborne Pathog Dis 7(5):523–535. https://doi.org/10.1089/fpd.2009.0447
Kuang D, Xu X, Meng J et al (2015) Antimicrobial susceptibility, virulence gene profiles and molecular subtypes of Salmonella Newport isolated from humans and other sources. Infect Genet Evol 36:294–299. https://doi.org/10.1016/j.meegid.2015.10.003
Asakura H, Ekawa T, Sugimoto N et al (2012) Membrane topology of Salmonella invasion protein SipB confers osmotolerance. Biochem Biophys Res Commun 426(4):654–658. https://doi.org/10.1016/j.bbrc.2012.09.012
Hauser E, Hebner F, Tietze E et al (2011) Diversity of Salmonella enterica serovar Derby isolated from pig, pork and humans in Germany. Int J Food Microbiol 151(2):141–149. https://doi.org/10.1016/j.ijfoodmicro.2011.08.020
Boyd EF, Jia LI, Ochman H, Selander RK (1997) Comparative genetics of the inv-spa invasion gene complex of Salmonella enterica. J Bacteriol 179(6):1985–1991. https://doi.org/10.1128/jb.179.6.1985-1991.1997
Penheiter KL, Mathur N, Giles D, Fahlen T, Jones BD (1997) Non-invasive Salmonella typhimurium mutants are avirulent because of an inability to enter and destroy M cells of ileal peyer’s patches. Mol Microbiol 24(4):697–709. https://doi.org/10.1046/j.1365-2958.1997.3741745.x
Cossi MVC, Burin RCK, Lopes DA et al (2013) Antimicrobial resistance and virulence profiles of Salmonella isolated from butcher shops in minas gerais. Brazil J Food Prot 76(9):1633–1637. https://doi.org/10.4315/0362-028X.JFP-13-119
Singh S, Agarwal RK, Tiwari SC, Himanshu S (2012) Antibiotic resistance pattern among the Salmonella isolated from human, animal and meat in India. Trop Anim Health Prod 44:665–674. https://doi.org/10.1007/s11250-011-9953-7
Zubair MS, Anam S, Lallo S (2016) Asian Paci fi c Journal of Tropical Biomedicine extract. Asian Pac J Trop Biomed 6(11):962–966. https://doi.org/10.1016/j.apjtb.2016.04.014
Chen MH, Hwang WZ, Wang SW, Shih YC, Tsen HY (2011) Pulsed field gel electrophoresis (PFGE) analysis for multidrug resistant Salmonella enterica serovar Schwarzengrund isolates collected in six years (2000–2005) from retail chicken meat in Taiwan. Food Microbiol 28(3):399–405. https://doi.org/10.1016/j.fm.2010.10.002
Possebon FS, Tiba Casas MR, Nero LA, Yamatogi RS, Araújo JP, de Pinto JPAN (2020) Prevalence, antibiotic resistance, PFGE and MLST characterization of Salmonella in swine mesenteric lymph nodes. Prev Vet Med 179(April):105024. https://doi.org/10.1016/j.prevetmed.2020.105024
Yang X, Huang J, Zhang Y et al (2020) Prevalence, abundance, serovars and antimicrobial resistance of Salmonella isolated from retail raw poultry meat in China. Sci Total Environ 713:136385. https://doi.org/10.1016/j.scitotenv.2019.136385
Portes AB, Rodrigues G, Leitão MP, Ferrari R, Conte Junior CA, Panzenhagen P (2022) Global distribution of plasmid-mediated colistin resistance mcr gene in salmonella: A systematic review. J Appl Microbiol 132(2):872–889. https://doi.org/10.1111/jam.15282
de Saraiva MMS, Lim K, do Monte DFM et al (2022) Antimicrobial resistance in the globalized food chain: a One Health perspective applied to the poultry industry. Braz J Microbiol 53(1):465–486. https://doi.org/10.1007/s42770-021-00635-8
Yang X, Huang J, Wu Q et al (2016) Prevalence, antimicrobial resistance and genetic diversity of Salmonella isolated from retail ready-to-eat foods in China. Food Control 60:50–56. https://doi.org/10.1016/j.foodcont.2015.07.019
Rahaman MT, Rahman M, Rahman MB et al (2014) Poultry salmonella specific bacteriophage isolation and characterization. Bangladesh J Vet Med 12:107–114. https://doi.org/10.3329/bjvm.v12i2.21264
Yin M, Yang B, Wu Y et al (2016) Prevalence and characterization of Salmonella enterica serovar in retail meats in market place in Uighur, Xinjiang. China Food Control 64:165–172. https://doi.org/10.1016/j.foodcont.2015.12.029
Mohammed M, Delappe N, O’Connor J, McKeown P, Garvey P, Cormican M (2016) Whole genome sequencing provides an unambiguous link between Salmonella Dublin outbreak strain and a historical isolate. Epidemiol Infect 144(3):576–581. https://doi.org/10.1017/S0950268815001636
Zhou Z, Li J, Zheng H et al (2017) Diversity of Salmonella isolates and their distribution in a pig slaughterhouse in Huaian. China Food Control 78:238–246. https://doi.org/10.1016/j.foodcont.2017.02.064
Chao G, Wang C, Wu T et al (2017) Molecular epidemiology and antibiotic resistance phenotypes and genotypes of salmonellae from food supply chains in China. Food Control 77:32–40. https://doi.org/10.1016/j.foodcont.2017.01.022
Zhu A, Zhi W, Qiu Y et al (2019) Surveillance study of the prevalence and antimicrobial resistance of Salmonella in pork from open markets in Xuzhou. China Food Control 98:474–480. https://doi.org/10.1016/j.foodcont.2018.07.035
Antoci S, Iannetti L, Centorotola G, et al (2021) Monitoring Italian establishments exporting food of animal origin to third countries: SSOP compliance and Listeria monocytogenes and Salmonella spp. contamination. Food Control 121(August 2020). https://doi.org/10.1016/j.foodcont.2020.107584
Leekitcharoenphon P, Lukjancenko O, Friis C, Aarestrup FM, Ussery DW (2012) Genomic variation in Salmonella enterica core genes for epidemiological typing. BMC Genomics 13(1). https://doi.org/10.1186/1471-2164-13-88
Keim P (2005) Bacterial pathogens. In: Roger G, Breeze RG, Budowle B, Schutzer SE (eds) Microbial Forensics, 1st edn, pp 99–107. https://doi.org/10.1016/B978-0-12-088483-4.X5000-8
Polveiro RC, Granja MMC, Roldão TCB, et al (2021) Multilocus sequence analysis reveals genetic diversity in Staphylococcus aureus isolate of goat with mastitis persistent after treatment with enrofloxacin. Sci Rep 11(1). https://doi.org/10.1038/s41598-021-96764-z
Pan H, Zhou X, Chai W et al (2019) Diversified sources for human infections by Salmonella enterica serovar newport. Transbound Emerg Dis 66(2):1044. https://doi.org/10.1111/TBED.13099
Acknowledgements
The authors are thankful to National Council of Technological and Scientific Development – CNPq, Brazil, the Foundation for Research Support of the State of Minas Gerais, FAPEMIG, Brazil and the Coordination Office for the Improvement of Higher Educations Personnel (CAPES—(finance code 001)), Brazil.
Author information
Authors and Affiliations
Contributions
Marcus Vinícius Coutinho Cossi: Data curation; Formal analysis; Investigation; Methodology; Project administration; Resources; Supervision; Validation; Visualization; Roles/Writing—original draft; and Writing—review & editing. Richard Costa Polveiro: Formal analysis; Methodology; Visualization; Roles/Writing—original draft. Ricardo Seiti Yamatogi: Formal analysis; Methodology; Visualization; Roles/Writing—original draft. Anderson Carlos Camargo: Formal analysis; Investigation; Methodology; Visualization. Luis Augusto Nero: Conceptualization; Data curation; Funding acquisition; Methodology; Project administration; Resources; Supervision; Validation; Visualization; and Writing—review & editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors of the manuscript declare that we do not have conflict of interest of order: financial, commercial, political academic, and personal.
Additional information
Responsible Editor: Luiz Henrique Rosa.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cossi, M.V.C., Polveiro, R.C., Yamatogi, R.S. et al. Multi-locus sequence typing, antimicrobials resistance and virulence profiles of Salmonella enterica isolated from bovine carcasses in Minas Gerais state, Brazil. Braz J Microbiol (2024). https://doi.org/10.1007/s42770-024-01341-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42770-024-01341-x