Abstract
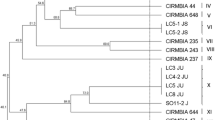
Among the milk contaminating microorganisms, those which are able to form heat-resistant spores are concerning, especially for dairy companies that use ultra-high temperature (UHT) technology. These spores, throughout storage, can germinate and produce hydrolytic enzymes that compromise the quality of the final product. This study evaluated 184 UHT milk samples from different batches collected from seven Brazilian dairy companies with a possible microbial contamination problem. The bacteria were isolated, phenotypically characterized, clustered by REP-PCR, and identified through 16S rDNA sequencing. The presence of Bacillus sporothermodurans was verified using biochemical tests (Gram staining, catalase and oxidase test, glucose fermentation, esculin hydrolysis, nitrate reduction, and urease test). According to these tests, none of the isolates presented typical characteristics of B. sporothermodurans. In sequence, the isolates, that presented rod-shapes, were submitted to molecular analyses in order to determine the microbial biodiversity existing among them. The isolates obtained were grouped into 16 clusters, four of which were composed of only one individual. A phylogenetic tree was constructed using the sequences obtained from the 16S rDNA sequencing and some reference strains of species close to those found using BLAST search in the NCBI nucleotide database. Through this tree, it was possible to verify the division of the isolates into two large groups, the Bacillus subtilis and the Bacillus cereus groups. Furthermore, most isolates are phylogenetically closely related, which makes it even more difficult to identify them at the species level. In conclusion, it was possible to assess, in general, the groups of sporulated contaminants in Brazilian UHT milk produced in the regions evaluated. In addition, it was also possible to determine the biodiversity of spore-forming bacteria found in UHT milk samples, thus opening up a range of possible research topics regarding the effects of the presence of these microorganisms on milk quality.



Similar content being viewed by others
References
ABLV (Associação Brasileira de Leite Longa Vida) (2020) Relatório Anual da Administração – 2020
BRASIL. Ministério da Agricultura Pecuária e Abastecimento (1997) Portaria no 370, de 04 de setembro de 1997. Regulamento Técnico para Fixação de Identidade e Qualidade do Leite U.H.T (U.A.T). Diário Oficial da República Federativa do Brasil 19700
Lorenzo JM, Munekata PE, Dominguez R et al (2018) Main groups of microorganisms of relevance for food safety and stability: general aspects and overall description. In: Innovative technologies for food preservation: Inactivation of spoilage and pathogenic microorganisms. Elsevier, pp 53–107
Pettersson B, Lembke F, Hammer P et al (1996) Bacillus sporothemodurans, a new species producing heat-resistant endospores highly. Int J Syst Bacteriol 46:759–764. https://doi.org/10.1099/00207713-46-3-759
Menezes MF, Simeoni CP, Bortoluzzi D et al (2014) Microbiota e conservação do leite. Revista Eletrônica em Gestão, Educação e Tecnologia Ambiental 18:76–89. https://doi.org/10.5902/2236117013033
Techer C, Baron F, Jan S (2014) Spoilage of animals products | Microbial milk spoilage. In: Encyclopedia of Food Microbiology, 2nd edn. pp 446–452. https://doi.org/10.1016/B978-0-12-384730-0.00443-2
Zacarchenco PB, de Freitas Leitão MF, Destro MT, Andrigheto C (2000) Ocorrência de Bacillus sporothermodurans em leite UAT/UHT brasileiro e a influência do tratamento térmico. Ciênc Tecnol Aliment 20(363):368. https://doi.org/10.1590/S0101-20612000000300014
Zhang D, Li S, Palmer J et al (2020) The relationship between numbers of Pseudomonas bacteria in milk used to manufacture UHT milk and the effect on product quality. Int Dairy J 105:104687. https://doi.org/10.1016/j.idairyj.2020.104687
Elzhraa F, Al-Ashmawy M, El-Sherbini M, Abdelkhalek A (2021) Evaluation of physicochemical properties and microbiological quality of UHT milk regularly introduced to resident patients in Mansoura University hospitals. Veterinarija ir Zootechnika 79:9–16
BRASIL. Ministério da Agricultura Pecuária e Abastecimento (2003) Instrução Normativa no 62, de 26 de agosto de 2003. Oficializa os Métodos Analíticos Oficiais para Análises Microbiológicas para Controle de Produtos de Origem Animal e Água. Diário Oficial da República Federativa do Brasil. Brasília, DF, 14, sessão 1
Doll EV, Scherer S, Wenning M (2017) Spoilage of microfiltered and pasteurized extended shelf life milk is mainly induced by psychrotolerant spore-forming bacteria that often originate from recontamination. Front Microbiol 8:135. https://doi.org/10.3389/fmicb.2017.00135
Dal Bello B, Rantsiou K, Bellio A et al (2010) Microbial ecology of artisanal products from North West of Italy and antimicrobial activity of the autochthonous populations. LWT 43:1151–1159. https://doi.org/10.1016/j.lwt.2010.03.008
Felske A, Rheims H, Wokerink A et al (1997) Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology (N Y) 143:2983–2989. https://doi.org/10.1099/00221287-143-9-2983
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027. https://doi.org/10.1093/molbev/msab120
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376. https://doi.org/10.1007/BF01734359
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution (N Y) 39:783–791. https://doi.org/10.2307/2408678
Hasegawa M, Kishino H, Yano T (1985) Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22:160–174. https://doi.org/10.1007/BF02101694
Letunic I, Bork P (2021) Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:w293–w296. https://doi.org/10.1093/nar/gkab301
BRASIL. Ministério da Agricultura Pecuária e Abastecimento (2019) Instrução Normativa n° 60, de 23 de dezembro de 2019. Listas de padrões microbiológicos para alimentos prontos para oferta ao consumidor. Diário Oficial da República Federativa do Brasil. Brasília, DF, 133
Guinebretière MH, Auger S, Galleron N et al (2013) Bacillus cytotoxicus sp. nov. is a novel thermotolerant species of the Bacillus cereus group occasionally associated with food poisoning. Int J Syst Evol Microbiol 63:31–40. https://doi.org/10.1099/ijs.0.030627-0
Hall BG (2013) Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol 30:1229–1235. https://doi.org/10.1093/molbev/mst012
Coelho PS, Silva N, Brescia MV, Siqueira AP (2001) Avaliação da qualidade microbiológica do leite UAT integral comercializado em Belo Horizonte. Arq Bras Med Vet Zootec 53:256–262. https://doi.org/10.1590/S0102-09352001000200021
Bersot LDS, Galvão JA, Raymundo NKL et al (2010) Evaluation of microbiological and physical-chemical quality of UHT milk produced in the Parana State – Brazil. Semin Cienc Agrar 31(645):652
Souza LV, da S Meloni VA, de Souza Batista C et al (2014) Avaliação da qualidade microbiológica e físico-química de leite UHT integral processado em indústrias do estado de Minas Gerais, Brasil. Revista Brasileira de Agropecuária Sustentável (RBAS) 4(6):15
Pinto CLO, Souza LV, Meloni VAS et al (2018) Microbiological quality of Brazilian UHT milk: Identification and spoilage potential of spore-forming bacteria. Int J Dairy Technol 71:20–26. https://doi.org/10.1111/1471-0307.12339
BRASIL. Ministério da Agricultura Pecuária e Abastecimento (2018) Instrução Normativa no 30, de 26 de junho de 2018. Oficializa os métodos constantes do Manual de Métodos Oficiais para Análise de Alimentos de Origem Animal. Diário Oficial da República Federativa do Brasil 9
Chavan RS, Chavan SR, Khedkar CD, Jana AH (2011) UHT milk processing and effect of plasmin activity on shelf life: a review. Compr Rev Food Sci Food Saf 10:251–268. https://doi.org/10.1111/j.1541-4337.2011.00157.x
Pereira JR, Tamanini R, Rios EA et al (2013) Microbiota mesófila aeróbia contaminante do leite UHT. Revista do Instituto de Laticínios Cândido Tostes 68:25–31. https://doi.org/10.5935/2238-6416.20130039
Costa EA, Carioca LJ, Freitas VV et al (2017) Avaliação da eficiência de sanitizantes sobre bactérias esporuladas isoladas de leite UHT integral. Revista do Instituto de Laticínios Cândido Tostes 71:01. https://doi.org/10.14295/2238-6416.v71i1.442
Gopal N, Hill C, Ross PR et al (2015) The prevalence and control of Bacillus and related spore-forming bacteria in the dairy industry. Front Microbiol 6:1418. https://doi.org/10.3389/fmicb.2015.01418
Velázquez-Ordoñez V, Valladares-Carranza B, Tenorio-Borroto E et al (2019) Microbial contamination in milk quality and health risk of the consumers of raw milk and dairy products. In: Nutrition in Health and Disease-Our Challenges Now and Forthcoming Time, vol 11. pp 181–205
Ledina T, Djordjevic J, Bulajic S (2021) Spore-forming bacteria in the dairy chain. In: IOP Conference Series: Earth and Environmental Science. IOP Publishing Ltd, p 012051
Montanhini MTM, Colombo M, Nero LA, Bersot LS (2013) Short communication: presence of neutral metallopeptidase (npr) gene and proteolytic activity of Bacillus cereus isolated from dairy products. J Dairy Sci 96:5641–5643. https://doi.org/10.3168/jds.2013-6886
Rezer APDS (2010) Avaliação da qualidade microbiológica e físico-química do leite UHT integral comercializado no Rio Grande do Sul. Universidade Federal de Santa Maria, p 73
Busatta B, Valdruga C, Cansian E, Luis R (2005) Ocorrência de Bacillus sporothermodurans em leite UAT integral e desnatado. Food Sci Technol 25:408–411. https://doi.org/10.1590/S0101-20612005000300003
Kmiha S, Aouadhi C, Klibi A et al (2017) Seasonal and regional occurrence of heat-resistant spore-forming bacteria in the course of ultra-high temperature milk production in Tunisia. J Dairy Sci 100:6090–6099. https://doi.org/10.3168/jds.2016-11616
Baldauf SL (2003) Phylogeny for the faint of heart: a tutorial. Trends Genet 19:345–351. https://doi.org/10.1016/S0168-9525(03)00112-4
Senesi S, Ghelardi E (2010) Production, secretion and biological activity of Bacillus cereus enterotoxins. Toxins (Basel) 2:1690–1703. https://doi.org/10.3390/toxins2071690
Vidal-Martins AMC, Rossi OD Jr, Rezende-Lago NC (2005) Microrganismos heterotróficos mesófilos e bactérias do grupo do Bacillus cereus em leite integral submetido a ultra alta temperatura. Arq Bras Med Vet Zootec 57:396–400. https://doi.org/10.1590/S0102-09352005000300019
Rezende-Lago NCM, Rossi OD Jr, Vidal-Martins AMC, Amaral LA (2007) Occurrence of Bacillus cereus in Whole milk and enterotoxigenic potential of the isolated strains. Arq Bras Med Vet Zootec 59:1563–1569. https://doi.org/10.1590/S0102-09352007000600032
Montanhini MTM, Pinto JPDAN, Bersot LDS (2012) Ocorrência de Bacillus cereus em leite comercializado nos estados do Paraná, Santa Catarina e São Paulo. UNOPAR Cientifíca - Ciências biológicas e da Saúde 14:155–158. https://doi.org/10.17921/2447-8938.2012v14n3p%25p
de Rezende NCM, Rossi OD, doAmaral LA (2000) Ocorrência de bactérias do grupo do Bacillus cereus em leite UHT integral (Uitra-High-Temperature). Revista Brasileira de Ciência Veterinária 7:162–166. https://doi.org/10.4322/rbcv.2015.205
Wong H-C, Chang M-H, Fan J-Y (1988) Incidence and characterization of Bacillus cereus isolates contaminating dairy products. Appl Environ Microbiol 54:699–702. https://doi.org/10.1128/aem.54.3.699-702.1988
Bartoszewicz M, Hansen BM, Swiecicka I (2008) The members of the Bacillus cereus group are commonly present contaminants of fresh and heat-treated milk. Food Microbiol 25:588–596. https://doi.org/10.1016/j.fm.2008.02.001
Zhou G, Liu H, He J et al (2008) The occurrence of Bacillus cereus, B. thuringiensis and B. mycoides in Chinese pasteurized full fat milk. Int J Food Microbiol 121:195–200. https://doi.org/10.1016/j.ijfoodmicro.2007.11.028
Zhou G, Zheng D, Dou L et al (2010) Occurrence of psychrotolerant Bacillus cereus group strains in ice creams. Int J Food Microbiol 137:143–146. https://doi.org/10.1016/j.ijfoodmicro.2009.12.005
Liu Y, Du J, Lai Q et al (2017) Proposal of nine novel species of the Bacillus cereus group. Int J Syst Evol Microbiol 67:2499–2508. https://doi.org/10.1099/ijsem.0.001821
Molva C, Sudagidan M, Okuklu B (2009) Extracellular enzyme production and enterotoxigenic gene profiles of Bacillus cereus and Bacillus thuringiensis strains isolated from cheese in Turkey. Food Control 20:829–834. https://doi.org/10.1016/j.foodcont.2008.10.016
Heyndrickx M, Scheldeman P (2002) Bacilli associated with spoilage in dairy products and other food. In: Applications and Systematics of Bacillus and Relatives. pp 64–82. https://doi.org/10.1002/9780470696743.ch6
Schallmey M, Singh A, Ward OP (2004) Developments in the use of Bacillus species for industrial production. Can J Microbiol 50:1–17. https://doi.org/10.1139/w03-076
Kai M, Effmert U, Berg G, Piechulla B (2007) Volatiles of bacterial antagonists inhibit mycelial growth of the plant pathogen Rhizoctonia solani. Arch Microbiol 187:351–360. https://doi.org/10.1007/s00203-006-0199-0
Chen H, Xiao X, Wang J et al (2008) Antagonistic effects of volatiles generated by Bacillus subtilis on spore germination and hyphal growth of the plant pathogen, Botrytis cinerea. Biotechnol Lett 30:919–923. https://doi.org/10.1007/s10529-007-9626-9
De Jonghe V, Coorevits A, De Block J et al (2010) Toxinogenic and spoilage potential of aerobic spore-formers isolated from raw milk. Int J Food Microbiol 136:318–325. https://doi.org/10.1016/j.ijfoodmicro.2009.11.007
Reginensi SM, González MJ, Olivera JA et al (2011) RAPD-based screening for spore-forming bacterial populations in Uruguayan commercial powdered milk. Int J Food Microbiol 148:36–41. https://doi.org/10.1016/j.ijfoodmicro.2011.04.020
Dhakal R, Chauhan K, Seale RB et al (2013) Genotyping of dairy Bacillus licheniformis isolates by high resolution melt analysis of multiple variable number tandem repeat loci. Food Microbiol 34:344–351. https://doi.org/10.1016/j.fm.2013.01.006
Westhoff DC, Dougherty SL (1981) Characterization of Bacillus species iIsolated from spoiled ultrahigh temperature processed milk. J Dairy Sci 64:572–580. https://doi.org/10.3168/jds.S0022-0302(81)82614-8
Ruiz-García C, Béjar V, Martínez-Checa F et al (2005) Bacillus velezensis sp. nov., a surfactant-producing bacterium isolated from the river Vélez in Málaga, southern Spain. Int J Syst Evol Microbiol 55:191–195. https://doi.org/10.1099/ijs.0.63310-0
Wang LT, Lee FL, Tai CJ, Kuo HP (2008) Bacillus velezensis is a later heterotypic synonym of Bacillus amyloliquefaciens. Int J Syst Evol Microbiol 58:671–675. https://doi.org/10.1099/ijs.0.65191-0
Jeyaram K, Romi W, Singh TA et al (2011) Distinct differentiation of closely related species of Bacillus subtilis group with industrial importance. J Microbiol Methods 87:161–164. https://doi.org/10.1016/j.mimet.2011.08.011
Elegbeleye JA, Buys EM (2020) Molecular characterization and biofilm formation potential of Bacillus subtilis and Bacillus velezensis in extended shelf-life milk processing line. J Dairy Sci 103:4991–5002. https://doi.org/10.3168/jds.2019-17919
Elegbeleye JA, Buys EM (2022) Potential spoilage of extended shelf-life (ESL) milk by Bacillus subtilis and Bacillus velezensis. LWT 153:112487. https://doi.org/10.1016/j.lwt.2021.112487
McKillip JL, Grutsch A, Wagner ER, Klug C (2016) Bacillus amyloliquefaciens from UHT organic milk produces biofilm and demonstrates virulence potential. J Anim Sci 94:283–283. https://doi.org/10.2527/jam2016-0597
Bulgari O, Caroli AM (2017) Protein profile of ultra high temperature (UHT) milk: occurence of para-kapa-casein in the sediment. Scienza e Tecnica Lattiero-Casearia 68:53–58
Deeth HC (2019) The effect of UHT processing and storage on milk proteins. In: Milk Proteins: From Expression To Food. Elsevier, pp 385–421. https://doi.org/10.1016/B978-0-12-815251-5.00010-4
Acknowledgements
The authors are thankful to Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brasília, DF, Brazil, Financial code 001), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brasília, DF, Brazil), and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG, Belo Horizonte, MG, Brazil).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Responsible Editor: Luis Augusto Nero
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Moreira, I.M.F.B., da Silva Rodrigues, R., Machado, S.G. et al. Phylogenetic characterization and biodiversity of spore-forming bacteria isolated from Brazilian UHT milk. Braz J Microbiol 54, 2153–2162 (2023). https://doi.org/10.1007/s42770-023-01063-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-023-01063-6